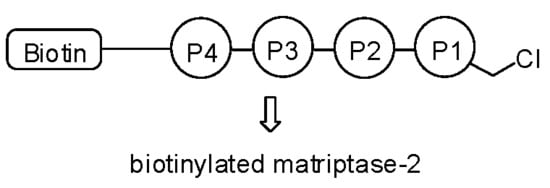
A Short Peptide Inhibitor as an Activity-Based Probe for Matriptase-2
Abstract
:1. Introduction
2. Results
2.1. Kinetic Evaluation of Biotin-RQRR-CMK As an Inhibitor of MT-2
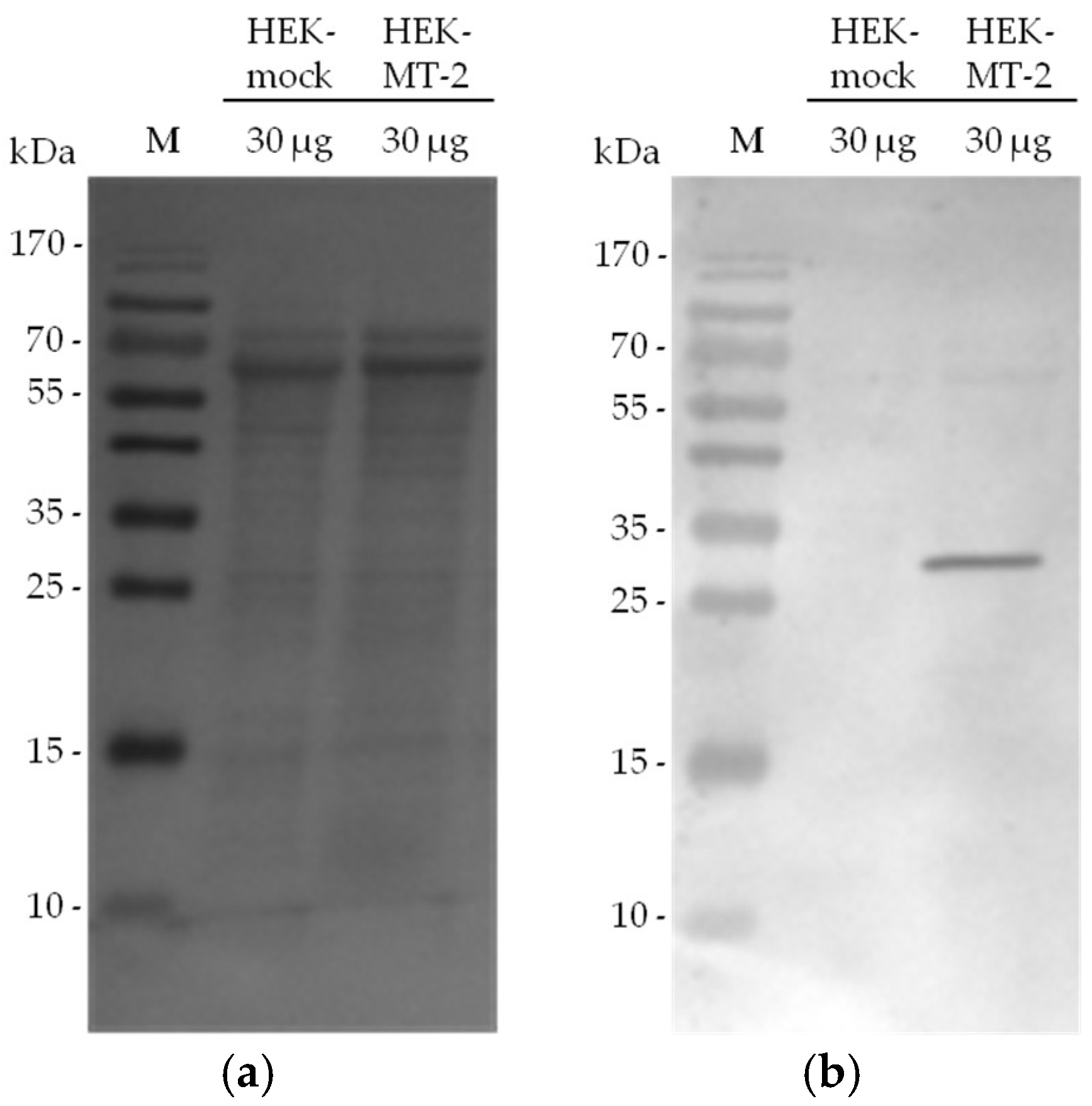
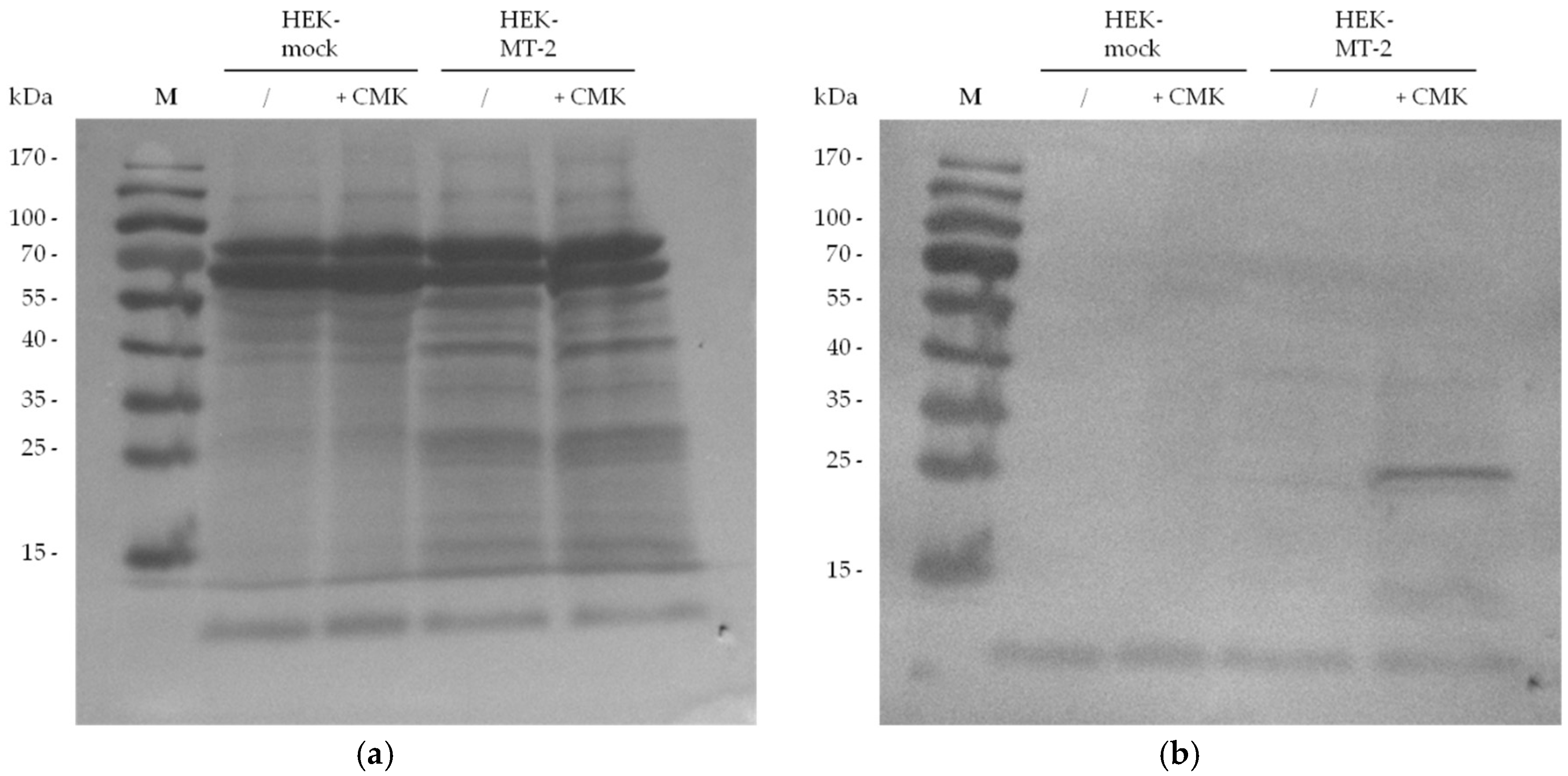
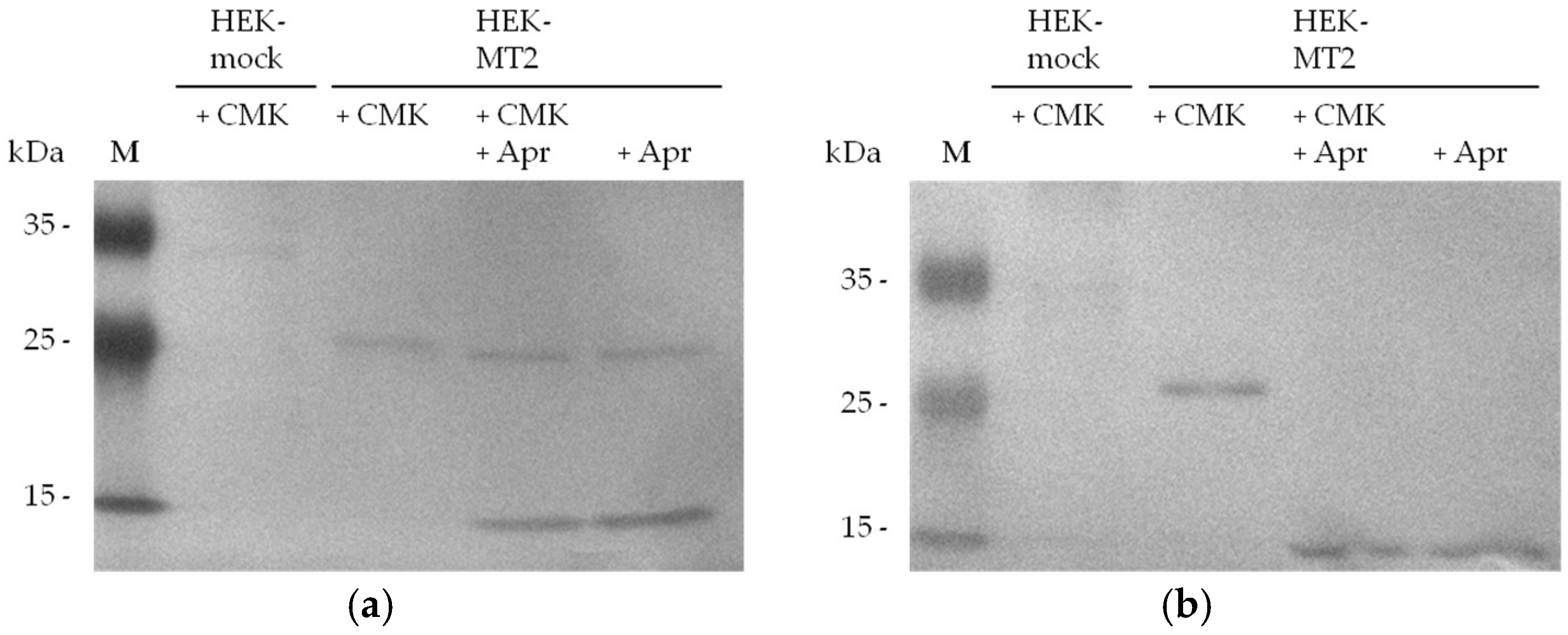
2.2. Detection of Active MT-2 in Western Blot Experiments
3. Discussion and Conclusions
4. Materials and Methods
4.1. Preparation of HEK-Cell Supernatant
4.2. Kinetic Measurement of MT-2 Activity
4.3. Cultivation and Kinetic Measurement of MT-2 Expressing HEK Cells
4.4. Western Blot Detection of Myc-Tagged MT-2
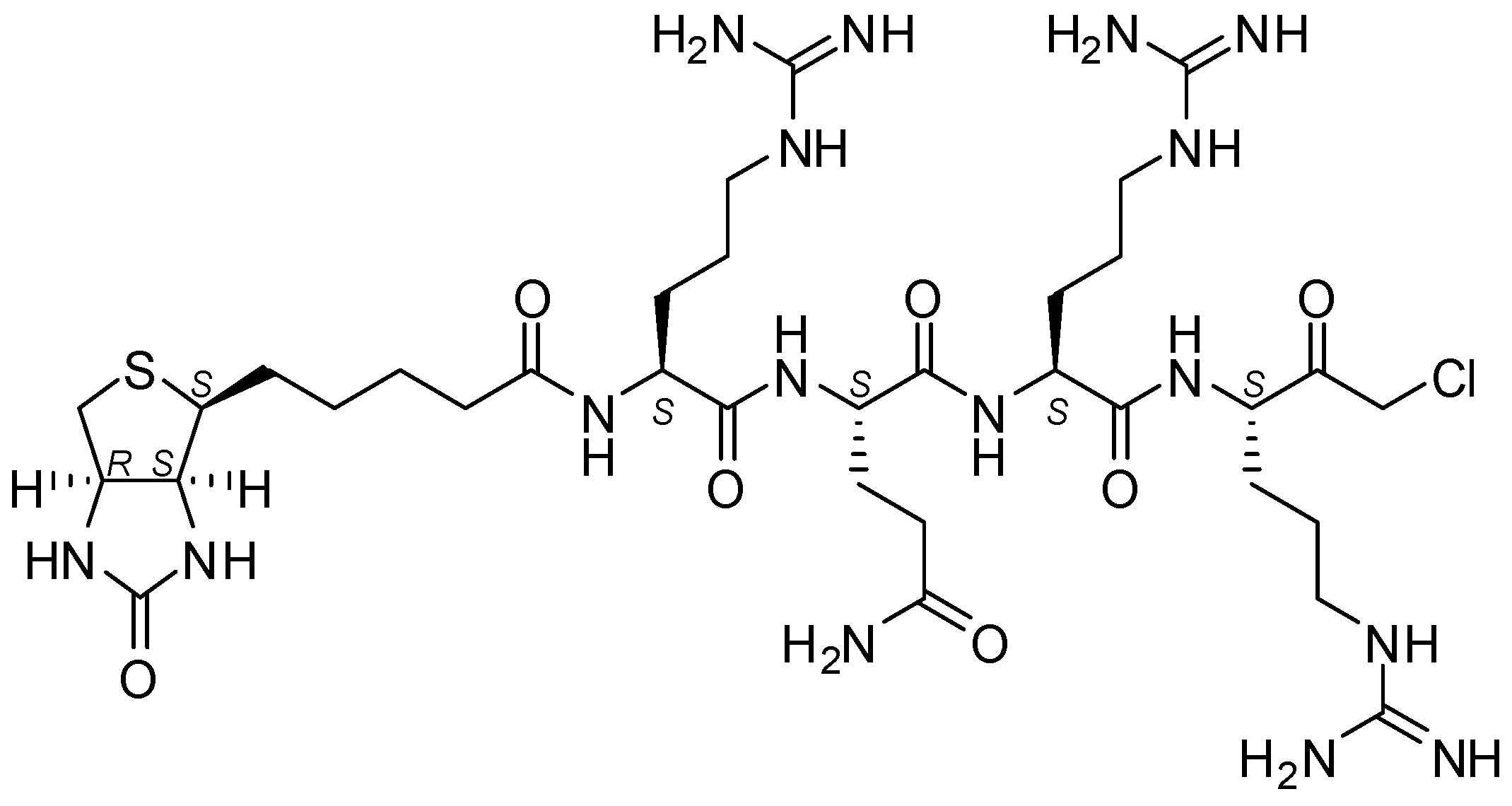
4.5. Biotin-RQRR-CMK
4.6. Western Blot Detection of Biotin-RQRR-CMK-Labeled MT-2
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Velasco, G.; Cal, S.; Quesada, V.; Sánchez, L.M.; López-Otín, C. Matriptase-2, a membrane-bound mosaic serine proteinase predominantly expressed in human liver and showing degrading activity against extracellular matrix proteins. J. Biol. Chem. 2002, 277, 37637–37646. [Google Scholar] [CrossRef] [PubMed]
- Bugge, T.H.; Antalis, T.M.; Wu, Q. Type II transmembrane serine proteases. J. Biol. Chem. 2009, 284, 23177–23181. [Google Scholar] [CrossRef] [PubMed]
- Dion, S.P.; Béliveau, F.; Désilets, A.; Ghinet, M.G.; Leduc, R. Transcriptome analysis reveals TMPRSS6 isoforms with distinct functionalities. J. Cell. Mol. Med. 2018, 22, 2498–2509. [Google Scholar] [CrossRef] [PubMed]
- Folgueras, A.R.; de Lara, F.M.; Pendás, A.M.; Garabaya, C.; Rodriguez, F.; Astudillo, A.; Bernal, T.; Cabanillas, R.; López-Otín, C.; Velasco, G. Membrane-bound serine protease matriptase-2 (Tmprss6) is an essential regulator of iron homeostasis. Blood 2008, 112, 2539–2545. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; She, E.; Gelbart, T.; Truksa, J.; Lee, P.; Xia, Y.; Khovananth, K.; Mudd, S.; Mann, N.; Moresco, E.M.; et al. The serine protease TMPRSS6 is required to sense iron deficiency. Science 2008, 320, 1088–1092. [Google Scholar] [CrossRef] [PubMed]
- Silvestri, L.; Pagani, A.; Nai, A.; De Domenico, I.; Kaplan, J.; Camaschella, C. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab. 2008, 8, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Finberg, K.E.; Whittlesey, R.L.; Fleming, M.D.; Andrews, N.C. Down-Regulation of Bmp/Smad signaling by Tmprss6 is required for maintenance of systemic iron homeostasis. Blood 2010, 115, 3817–3826. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, E.; Tuttle, M.S.; Powelson, J.; Vaughn, M.B.; Donovan, A.; Ward, D.M.; Ganz, T.; Kaplan, J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 2004, 306, 2090–2093. [Google Scholar] [CrossRef] [PubMed]
- Sisay, M.T.; Steinmetzer, T.; Stirnberg, M.; Maurer, E.; Hammami, M.; Bajorath, J.; Gütschow, M. Identification of the first low-molecular-weight inhibitors of matriptase-2. J. Med. Chem. 2010, 53, 5523–5535. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, P.J.; Toudjarska, I.; Sendamarai, A.K.; Racie, T.; Milstein, S.; Bettencourt, B.R.; Hettinger, J.; Bumcrot, D.; Fleming, M.D. An RNAi therapeutic targeting Tmprss6 decreases iron overload in Hfe(−/−) mice and ameliorates anemia and iron overload in murine β-thalassemia intermedia. Blood 2013, 121, 1200–1208. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Casu, C.; Gardenghi, S.; Booten, S.; Aghajan, M.; Peralta, R.; Watt, A.; Freier, S.; Monia, B.P.; Rivella, S. Reducing TMPRSS6 ameliorates hemochromatosis and β-thalassemia in mice. J. Clin. Investig. 2013, 123, 1531–1541. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, P.J.; Fleming, M.D. Modulation of hepcidin as therapy for primary and secondary iron overload disorders: Pre-clinical models and approaches. Hematol. Oncol. Clin. N. Am. 2014, 28, 387–401. [Google Scholar] [CrossRef] [PubMed]
- Finberg, K.E.; Heeney, M.M.; Campagna, D.R.; Aydinok, Y.; Pearson, H.A.; Hartman, K.R.; Mayo, M.M.; Samuel, S.M.; Strouse, J.J.; Markianos, K.; et al. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA). Nat. Genet. 2008, 40, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Stirnberg, M.; Maurer, E.; Horstmeyer, A.; Kolp, S.; Frank, S.; Bald, T.; Arenz, K.; Janzer, A.; Prager, K.; Wunderlich, P.; et al. Proteolytic processing of the serine protease matriptase-2: Identification of the cleavage sites required for its autocatalytic release from the cell surface. Biochem. J. 2010, 430, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Wysocka, M.; Gruba, N.; Miecznikowska, A.; Popow-Stellmaszyk, J.; Gütschow, M.; Stirnberg, M.; Furtmann, N.; Bajorath, J.; Lesner, A.; Rolka, K. Substrate specificity of human matriptase-2. Biochimie 2014, 97, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Béliveau, F.; Désilets, A.; Leduc, R. Probing the substrate specificities of matriptase, matriptase-2, hepsin and DESC1 with internally quenched fluorescent peptides. FEBS J. 2009, 276, 2213–2226. [Google Scholar] [CrossRef] [PubMed]
- Stirnberg, M.; Gütschow, M. Matriptase-2, a regulatory protease of iron homeostasis: Possible substrates, cleavage sites and inhibitors. Curr. Pharm. Des. 2013, 19, 1052–1061. [Google Scholar] [CrossRef] [PubMed]
- Hammami, M.; Rühmann, E.; Maurer, E.; Heine, A.; Gütschow, M.; Klebe, G.; Steinmetzer, T. New 3-amidinophenylalanine-derived inhibitors of matriptase. Med. Chem. Commun. 2012, 3, 807–813. [Google Scholar] [CrossRef]
- Duchêne, D.; Colombo, E.; Désilets, A.; Boudreault, P.L.; Leduc, R.; Marsault, E.; Najmanovich, R. Analysis of subpocket selectivity and identification of potent selective inhibitors for matriptase and matriptase-2. J. Med. Chem. 2014, 57, 10198–10204. [Google Scholar] [CrossRef] [PubMed]
- St-Georges, C.; Désilets, A.; Béliveau, F.; Ghinet, M.; Dion, S.P.; Colombo, É.; Boudreault, P.L.; Najmanovich, R.J.; Leduc, R.; Marsault, É. Modulating the selectivity of matriptase-2 inhibitors with unnatural amino acids. Eur. J. Med. Chem. 2017, 129, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Gitlin, A.; Debowski, D.; Karna, N.; Legowska, A.; Stirnberg, M.; Gütschow, M.; Rolka, K. Inhibitors of matriptase-2 based on the trypsin inhibitor SFTI-1. ChemBioChem 2015, 16, 1601–1607. [Google Scholar] [CrossRef] [PubMed]
- Dosa, S.; Stirnberg, M.; Lülsdorff, V.; Häußler, D.; Maurer, E.; Gütschow, M. Active site mapping of trypsin, thrombin and matriptase-2 by Sulfamoyl Benzamidines. Bioorg. Med. Chem. 2012, 20, 6489–6505. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, A.M.; Gilberg, E.; Gattner, S.; Huang, T.L.; Vanden Eynde, J.J.; Mayence, A.; Bajorath, J.; Stirnberg, M.; Gütschow, M. Evaluation of bisbenzamidines as inhibitors for matriptase-2. Bioorg. Med. Chem. Lett. 2016, 26, 3741–3745. [Google Scholar] [CrossRef] [PubMed]
- Furtmann, N.; Häußler, D.; Scheidt, T.; Stirnberg, M.; Steinmetzer, T.; Bajorath, J.; Gütschow, M. Limiting the number of potential binding modes by introducing symmetry into ligands: Structure-based design of inhibitors for trypsin-like serine proteases. Chem. Eur. J. 2016, 22, 610–625. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.L.; Bacchi, C.J.; Kode, N.R.; Zhang, Q.; Wang, G.; Yartlet, N.; Rattendi, D.; Londono, I.; Mazumder, L.; Vanden Eynde, J.J.; et al. Trypanocidal activity of piperazine-linked bisbenzamidines and bisbenzamidoxime, an orally active prodrug. Int. J. Antimicrob. Agents 2007, 30, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.L.; Vanden Eynde, J.J.; Mayence, A.; Collins, M.S.; Cushion, M.T.; Rattendi, D.; Londono, I.; Mazumder, L.; Bacchi, C.J.; Yarlett, N. Synthesis and SAR of alkanediamide-linked bisbenzamidines with anti-trypanosomal and anti-pneumocystis activity. Bioorg. Med. Chem. Lett. 2009, 19, 5884–5886. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.L.; Mayence, A.; Vanden Eynde, J.J. Some non-conventional biomolecular targets for diamidines. A short survey. Bioorg. Med. Chem. 2014, 22, 1983–1992. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.I.; Vázquez, O.; Vázquez, M.E.; Mascareñas, J.L. Sequence-selective DNA recognition with peptide-bisbenzamidine conjugates. Chem. Eur. J. 2013, 19, 9923–9929. [Google Scholar] [CrossRef] [PubMed]
- Bordello, J.; Sánchez, M.I.; Vázquez, M.E.; Mascareñas, J.L.; Al-Soufi, W.; Novo, M. Fluorescence-labeled bis-benzamidines as fluorogenic DNA minor-groove binders: Photophysics and binding dynamics. Chem. Eur. J. 2015, 21, 1609–1619. [Google Scholar] [CrossRef] [PubMed]
- Godiksen, S.; Soendergaard, C.; Friis, S.; Jensen, J.K.; Bornholdt, J.; Sales, K.U.; Huang, M.; Bugge, T.H.; Vogel, L.K. Detection of active matriptase using a biotinylated chloromethyl ketone peptide. PLoS ONE 2013, 8, e77146. [Google Scholar] [CrossRef] [PubMed]
- Nonboe, A.W.; Krigslund, O.; Soendergaard, C.; Skovbjerg, S.; Friis, S.; Andersen, M.N.; Ellis, V.; Kawaguchi, M.; Kataoka, H.; Bugge, T.H.; et al. HAI-2 stabilizes, inhibits and regulates SEA-cleavage-dependent secretory transport of matriptase. Traffic 2017, 18, 378–391. [Google Scholar] [CrossRef] [PubMed]
- Häußler, D.; Schulz-Fincke, A.C.; Beckmann, A.M.; Keils, A.; Gilberg, E.; Mangold, M.; Bajorath, J.; Stirnberg, M.; Steinmetzer, T.; Gütschow, M. A fluorescent-labeled phosphono bisbenzguanidine as an activity-based probe for matriptase. Chem. Eur. J. 2017, 23, 5205–5209. [Google Scholar] [CrossRef] [PubMed]
- Maurer, E.; Sisay, M.T.; Stirnberg, M.; Steinmetzer, T.; Bajorath, J.; Gütschow, M. Insights into matriptase-2 substrate binding and inhibition mechanisms by analyzing active-site-mutated variants. ChemMedChem 2012, 7, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Kettner, C.; Shaw, E. Synthesis of peptides of arginine chloromethyl ketone. Selective inactivation of human plasma kallikrein. Biochemistry 1978, 17, 4778–4784. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.I.; Dener, J.M.; Molino, B.F.; Gardner, C.J.; D'Alisa, R.; Dunwiddie, C.T.; Kasiewski, C.; Leadley, R.J. O-Benzyl hydroxyproline as a bioisostere for Phe-Pro: Novel dipeptide thrombin inhibitors. Bioorg. Med. Chem. Lett. 1996, 6, 2225–2230. [Google Scholar] [CrossRef]
- Sun, A.; Shoji, M.; Lu, Y.J.; Liotta, D.C.; Snyder, J.P. Synthesis of EF24-tripeptide chloromethyl ketone: A novel curcumin-related anticancer drug delivery system. J. Med. Chem. 2006, 49, 3153–3158. [Google Scholar] [CrossRef] [PubMed]
- Waxler, B.; Rabito, S.F. Aprotinin: A serine protease inhibitor with therapeutic actions: Its interaction with ACE inhibitors. Curr. Pharm. Des. 2003, 9, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, A.M.; Maurer, E.; Lülsdorff, V.; Wilms, A.; Furtmann, N.; Bajorath, J.; Gütschow, M.; Stirnberg, M. En route to new therapeutic options for iron overload diseases: Matriptase-2 as a target for Kunitz-type inhibitors. ChemBioChem 2016, 17, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Oberst, M.D.; Singh, B.; Ozdemirli, M.; Dickson, R.B.; Johnson, M.D.; Lin, C.Y. Characterization of matriptase expression in normal human tissues. J. Histochem. Cytochem. 2003, 51, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
 25 nM;
25 nM;  50 nM;
50 nM;  75 nM;
75 nM;  100 nM); (b) Values kobs plotted vs. probe concentrations.
100 nM); (b) Values kobs plotted vs. probe concentrations.
 25 nM;
25 nM;  50 nM;
50 nM;  75 nM;
75 nM;  100 nM); (b) Values kobs plotted vs. probe concentrations.
100 nM); (b) Values kobs plotted vs. probe concentrations.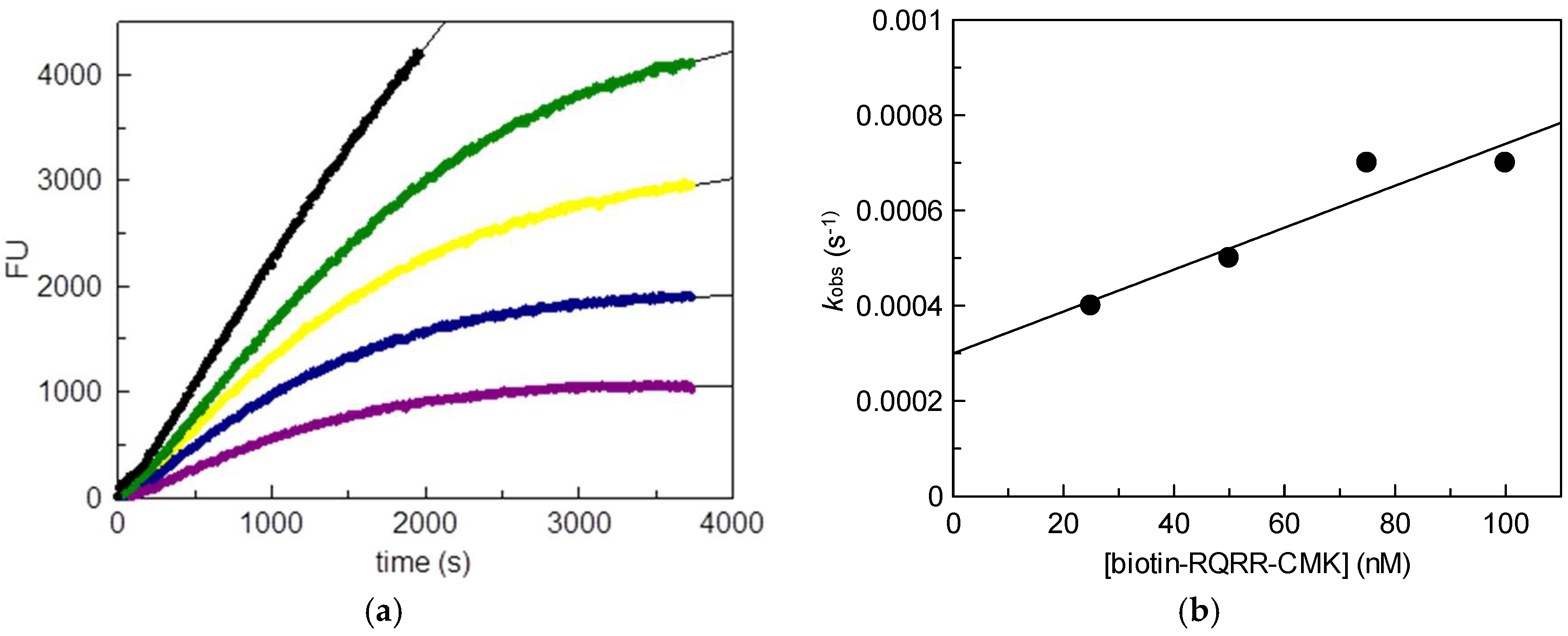
 100 nM;
100 nM;  200 nM;
200 nM;  300 nM;
300 nM;  400 nM;
400 nM;  500 nM;
500 nM;  600 nM); (b) Values vs plotted vs. probe concentrations.
600 nM); (b) Values vs plotted vs. probe concentrations.
 100 nM;
100 nM;  200 nM;
200 nM;  300 nM;
300 nM;  400 nM;
400 nM;  500 nM;
500 nM;  600 nM); (b) Values vs plotted vs. probe concentrations.
600 nM); (b) Values vs plotted vs. probe concentrations.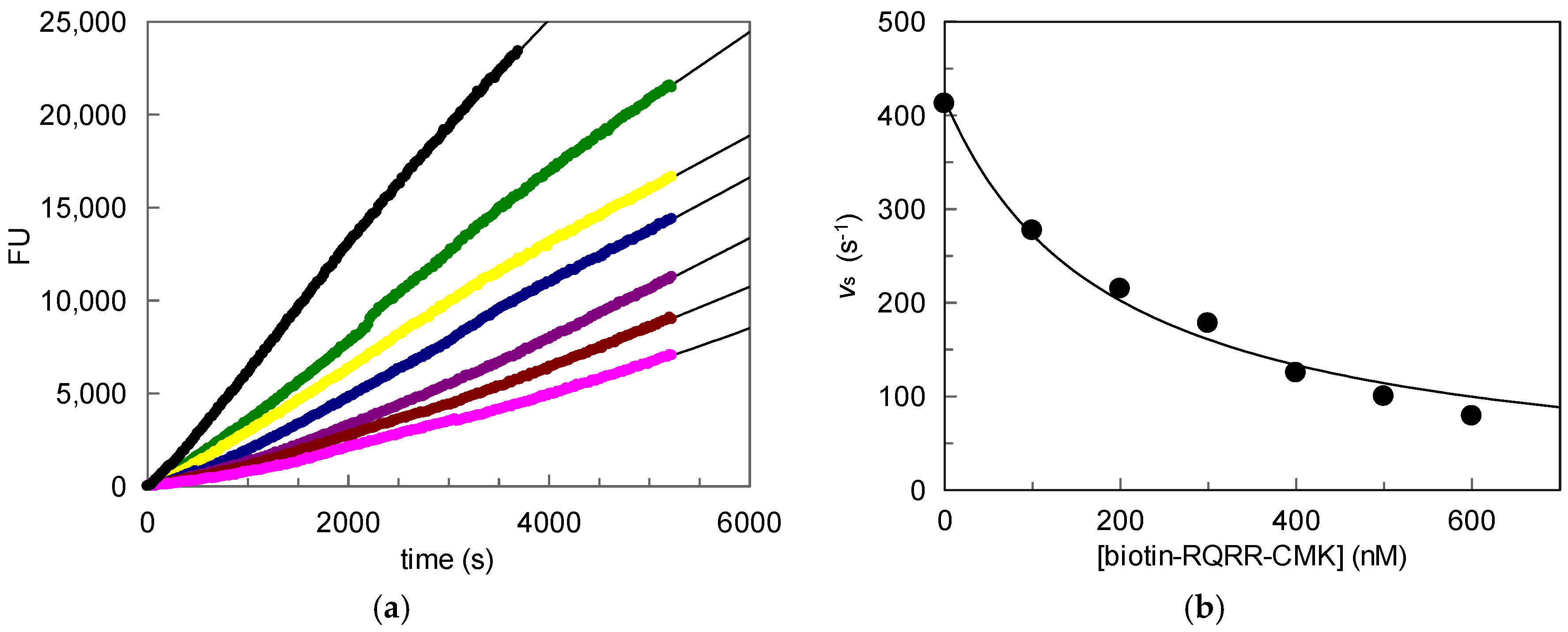
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mangold, M.; Gütschow, M.; Stirnberg, M. A Short Peptide Inhibitor as an Activity-Based Probe for Matriptase-2. Pharmaceuticals 2018, 11, 49. https://doi.org/10.3390/ph11020049
Mangold M, Gütschow M, Stirnberg M. A Short Peptide Inhibitor as an Activity-Based Probe for Matriptase-2. Pharmaceuticals. 2018; 11(2):49. https://doi.org/10.3390/ph11020049
Chicago/Turabian StyleMangold, Martin, Michael Gütschow, and Marit Stirnberg. 2018. "A Short Peptide Inhibitor as an Activity-Based Probe for Matriptase-2" Pharmaceuticals 11, no. 2: 49. https://doi.org/10.3390/ph11020049
APA StyleMangold, M., Gütschow, M., & Stirnberg, M. (2018). A Short Peptide Inhibitor as an Activity-Based Probe for Matriptase-2. Pharmaceuticals, 11(2), 49. https://doi.org/10.3390/ph11020049