Role of CYP2C9, CYP2C19 and EPHX Polymorphism in the Pharmacokinetic of Phenytoin: A Study on Uruguayan Caucasian Subjects
Abstract
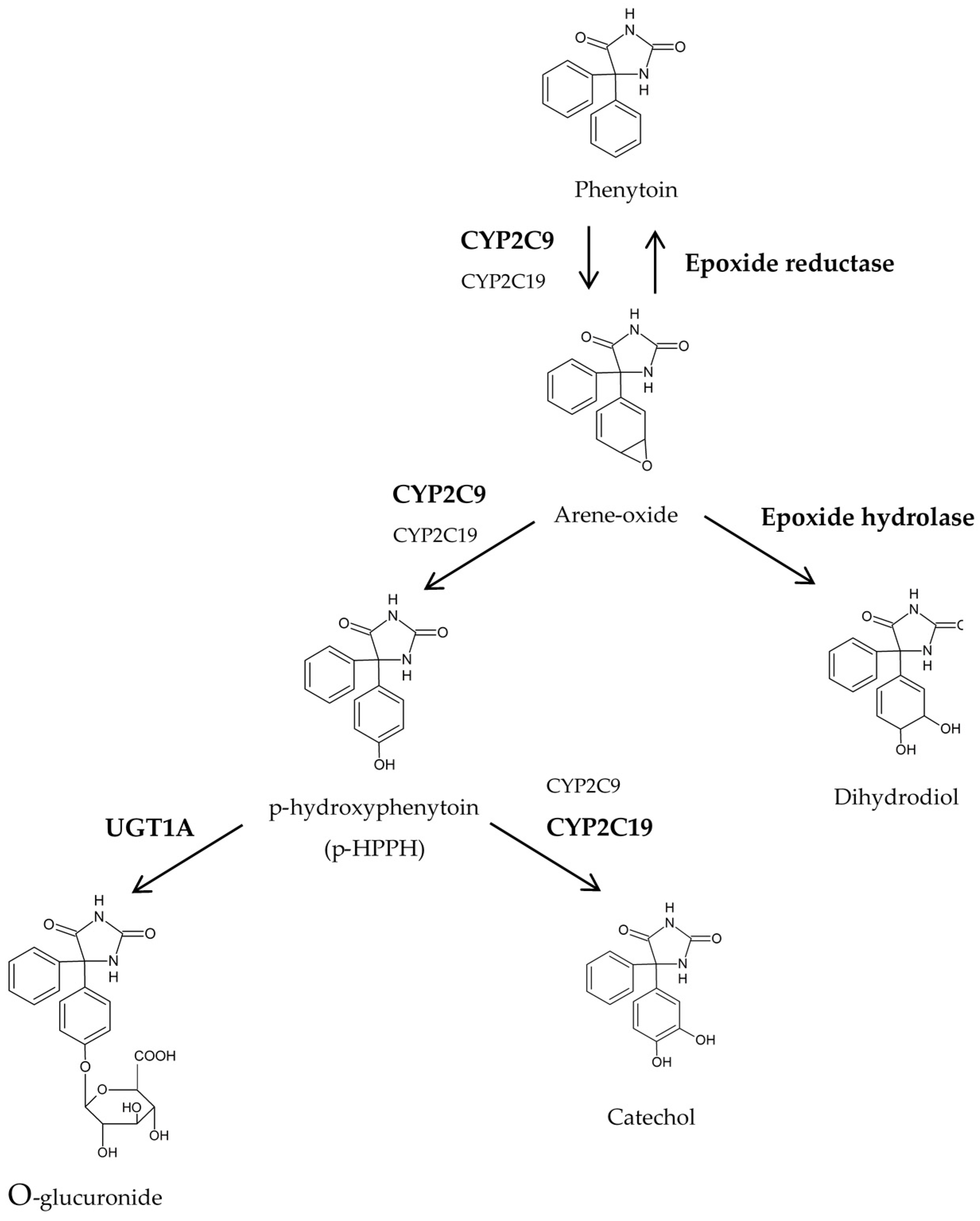
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Subjects and Design
4.2. Genotyping Procedure of EPHX, CYP2C9 and CYP2C19
4.3. Measurement of PHT and p-HPPH Plasma Concentrations
4.4. Statistical Analysis
Author Contributions
Conflicts of Interest
References
- Thorn, C.F.; Whirl-Carrillo, M.; Leeder, J.S.; Klein, T.E.; Altman, R.B. PharmGKB summary: Phenytoin pathway. Pharmacogenet. Genom. 2012, 22, 466–470. [Google Scholar] [CrossRef] [PubMed]
- Argikar, U.A.; Cloyd, J.C.; Birnbaum, A.K.; Leppik, I.E.; Conway, J.; Kshirsagar, S.; Oetting, W.S.; Klein, E.C.; Remmel, R.P. Paradoxical urinary phenytoin metabolite (S)/(R) ratios in CYP2C19*1/*2 patients. Epilepsy Res. 2006, 71, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Caudle, K.E.; Rettie, A.E.; Whirl-Carrillo, M.; Smith, L.H.; Mintzer, S.E.; Lee, M.T.M.; Klein, T.E.; Callaghan, J.T. Clinical pharmacogenetics implementation consortium (CPIC) guidelines for CYP2C9 and HLA-B genotype and phenytoin dosing. Clin. Pharmacol. Ther. 2014, 96, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Eadie, M.J. Phenytoin. In The Treatment of Epilepsy, 3rd ed.; Blackwell Publishing Ltd.: Oxford, UK, 2009; pp. 605–618. ISBN 978-1-405-18383-3. [Google Scholar]
- Eadie, M.J. Metabolism of anticonvulsant drugs. Drug Metab. Drug Interact. 1981, 3, 317–347. [Google Scholar] [CrossRef]
- Ingelman-Sundberg, M.; Gaedigk, A.; Brockmöller, J.; Goldstein, J.A.; Gonzalez, F.J.; Meyer, U.A.; Nelson, D.R.; Wedell, A.; Zanger, U.M. The Human Cytochrome P450 (CYP) Allele Nomenclature Database. Available online: http://www.cypalleles.ki.se (accessed on 30 May 2017).
- Lee, C.R.; Goldstein, J.A.; Pieper, J.A. Cytochrome P450 2C9 polymorphisms: A comprehensive review of the in-vitro and human data. Pharmacogenetics 2002, 12, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Booven, D.V.; Marsh, S.; McLeod, H.; Whirl-Carrillo, M.; Sangkuhl, K.; Klein, T.E.; Altman, R.B. Cytochrome P450 2C9-CP2C9. Pharmacogenet. Genom. 2010, 20, 277–281. [Google Scholar] [CrossRef]
- Wormhoudt, L.W.; Commandeur, J.N.M.; Vermeulen, N.P.E. Genetic polymorphisms of human N-acetyltransferase, cytochrome P450, glutathione-S-transferase, and epoxide hydrolase enzymes: Relevance to xenobiotic metabolism and toxicity. Crit. Rev. Toxicol. 1999, 29, 59–124. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.C.; Lin, C.J.; Chen, C.C.; Chang, C.J.; Liou, H.H. Dosage recommendation of phenytoin for patients with epilepsy with different CYP2C9/CYP2C19 polymorphisms. Ther. Drug Monit. 2004, 26, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, K.; Narayan, S.K.; Shewade, D.G.; Chandrasekaran, A. Influence of CYP2C9 genetic polymorphism and undernourishment on plasma-free phenytoin concentrations in epileptic patients. Ther. Drug Monit. 2010, 32, 762–766. [Google Scholar] [CrossRef] [PubMed]
- Hamdy, S.I.; Hiratsuka, M.; Narahara, K.; Elenany, M.; Moursi, N.; Ahmed, M.S.; Mizugaki, M. Allele and genotype frequencies of polymorphic cytochromes P450 (CYP2C9, CYP2C19, CYP2E1) and dihydropyrimidine dehydrogenase (DPYD) in the Egyptian population. Br. J. Clin. Pharmacol. 2002, 53, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Mamiya, K.; Ieiri, I.; Shimamoto, J.; Yukawa, E.; Imai, J.; Ninomiya, H.; Yamada, H.; Otsubo, K.; Higuchi, S.; Tashiro, N. The effects of genetic polymorphisms of CYP2C9 and CYP2C19 on phenytoin metabolism in Japanese adult patients with epilepsy: Studies in stereoselective hydroxylation and population pharmacokinetics. Epilepsia 1998, 39, 1317–1323. [Google Scholar] [CrossRef] [PubMed]
- Rettie, A.E.; Haining, R.L.; Bajpai, M.; Levy, R.H. A common genetic basis for idiosyncratic toxicity of warfarin and phenytoin. Epilepsy Res. 1999, 35, 253–255. [Google Scholar] [CrossRef]
- Odani, A.; Hashimoto, Y.; Otsuki, Y.; Uwai, Y.; Hattori, H.; Furusho, K.; Inui, K.I. Genetic polymorphism of the CYP2C subfamily and its effect on the pharmacokinetics of phenytoin in Japanese patients with epilepsy. Clin. Pharmacol. Ther. 1997, 62, 287–292. [Google Scholar] [CrossRef]
- Caudle, K.E.; Dunnenberger, H.M.; Freimuth, R.R.; Peterson, J.F.; Burlison, J.D.; Whirl-Carrillo, M.; Scott, S.A.; Rehm, H.L.; Williams, M.S.; Klein, T.E.; et al. Standardizing terms for clinical pharmacogenetic test results: Consensus terms from the clinical pharmacogenetics implementation consortium (CPIC). Genet. Med. 2017, 19, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Pinarbasi, H.; Silig, Y.; Pinarbasi, E. Microsomal epoxide hydrolase polymorphisms. Mol. Med. Rep. 2010, 3, 723–727. [Google Scholar] [CrossRef] [PubMed]
- Hassett, C.; Aicher, L.; Sidhu, J.S.; Omiecinski, C.J. Human microsomal epoxide hydrolase: Genetic polymorphism and functional expression in vitro of amino acid variants. Hum. Mol. Genet. 1994, 3, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Azzato, E.M.; Chen, R.A.; Wacholder, S.; Chanock, S.J.; Klebanoff, M.A.; Caporaso, N.E. Maternal EPHX1 polymorphisms and risk of phenytoin-induced congenital malformations. Pharmacogenet. Genom. 2010, 20, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Zusterzeel, P.L.M.; Peters, W.H.M.; Visser, W.; Hermsen, K.J.M.; Roelofs, H.M.J.; Steegers, E.A. A polymorphism in the gene for microsomal epoxide hydrolase is associated with pre-eclampsia. J. Med. Genet. 2001, 38, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, M.; Fagiolino, P.; Alvariza, S.; Ibarra, M.; Maldonado, C.; Gonzalez, R.; Laborde, A.; Uria, M.; Carozzi, A.; Azambuja, C. Skin reactions associated to phenytoin administration: Multifactorial cause. Clin. Pharmacol. Biopharm. 2014, 3. [Google Scholar] [CrossRef]
- Kerb, R.; Aynacioglu, A.S.; Brockmöller, J.; Schlagenhaufer, R.; Bauer, S.; Szekeres, T.; Hamwi, A.; Fritzer-Szekeres, M.; Baumgartner, C.; Öngen, H.Z.; et al. The predictive value of MDR1, CYP2C9, and CYP2C19 polymorphisms for Phenytoin plasma levels. Pharmacogenom. J. 2001, 1, 204–210. [Google Scholar] [CrossRef]
- Hennessy, S.; Leonard, C.E.; Freeman, C.P.; Metlay, J.P.; Chu, X.; Strom, B.L.; Bilker, W.B. CYP2C9, CYP2C19, and ABCB1 genotype and hospitalization for phenytoin toxicity. J. Clin. Pharmacol. 2009, 49, 1483–1487. [Google Scholar] [CrossRef] [PubMed]
- Dorado, P.; López-Torres, E.; Peñas-Lledó, E.M.; Martínez-Antón, J.; Llerena, A. Neurological toxicity after phenytoin infusion in a pediatric patient with epilepsy: Influence of CYP2C9, CYP2C19 and ABCB1 genetic polymorphisms. Pharmacogenom. J. 2013, 13, 359–361. [Google Scholar] [CrossRef] [PubMed]
- Kesavan, R.; Narayan, S.K.; Adithan, C. Influence of CYP2C9 and CYP2C19 genetic polymorphisms on phenytoin-induced neurological toxicity in Indian epileptic patients. Eur. J. Clin. Pharmacol. 2010, 66, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Navarro, H.; Jiménez-Jiménez, F.J.; García-Agúndez, J.A. Papel del polimorfismo genético CYP2C19 en los efectos adversos a fármacos y en el riesgo para diversas enfermedades. Med. Clín. 2006, 126, 697–706. (In Spanish) [Google Scholar] [CrossRef]
- Ragia, G.; Arvanitidis, K.I.; Tavridou, A.; Manolopoulos, V.G. Need for reassessment of reported CYP2C19 allele frequencies in various populations in view of CYP2C19*17 discovery: The case of Greece. Pharmacogenomics 2009, 10, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.C.; Risinger, C.; Dahl, M.-L.; Aklillu, E.; Christensen, M.; Bertilsson, L.; Ingelman-Sundberg, M. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin. Pharmacol. Ther. 2006, 79, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Li-Wan-Po, A.; Girard, T.; Farndon, P.; Cooley, C.; Lithgow, J. Pharmacogenetics of CYP2C19: Functional and clinical implications of a new variant CYP2C19*17. Br. J. Clin. Pharmacol. 2010, 69, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, R.M.; Ohlsson, S.; Pedersen, R.S.; Mwinyi, J.; Ingelman-Sundberg, M.; Eliasson, E.; Bertilsson, L. Increased omeprazole metabolism in carriers of the CYP2C19*17 allele; a pharmacokinetic study in healthy volunteers. Br. J. Clin. Pharmacol. 2008, 65, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Booth, J.; Hewer, A.; Keysell, C.R.; Sims, P. Enzymic reduction of aromatic hydrocarbon epoxides by the microsomal fraction of rat liver. Xenobiotica 1975, 5, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Savio, E.; Fagiolino, P.; Solana, G.; Parente, E.; León, A. Development of water/oil emulsion. Bioavailability in rats. STP Pharm. Sci. 1991, 1, 379–385. [Google Scholar]
| CYP2C9 Genotype | N° of Subjects | Percentage | CYP2C9 Phenotype | N° of Subjects | Percentage |
|---|---|---|---|---|---|
| *1/*1 | 34 | 68.0 | NM | 34 | 68.0 |
| *1/*2 | 11 | 22.0 | IM | 15 | 30.0 |
| *1/*3 | 4 | 8.0 | |||
| *2/*2 | 1 | 2.0 | PM | 1 | 2.0 |
| CYP2C19 Genotype | CYP2C19 Phenotype | N° of Subjects | Percentage |
|---|---|---|---|
| *1/*1 | NM | 40 | 80.0 |
| *1/*2 | IM | 10 | 20.0 |
| Genotype | N° of Subjects | Percentage |
|---|---|---|
| A/A wild-type (Tyr113Tyr) | 21 | 42.0 |
| A/G heterozygous (Tyr113His) | 22 | 44.0 |
| G/G mutated homozygous (His113His) | 7 | 14.0 |
| Genotype | N° of Subjects | Percentage |
|---|---|---|
| T/T wild-type (His139His) | 37 | 74.0 |
| T/C heterozygous (His139Arg) | 11 | 22.0 |
| C/C mutated homozygous (Arg139Arg) | 2 | 4.0 |
| Phenotype | N° of Subjects | Percentage |
|---|---|---|
| Intermediate | 22 | 44.0 |
| Increased | 5 | 10.0 |
| Decreased | 23 | 46.0 |
| CYP2C9 Genotype | CYP2C19 Genotype | N° of Subjects | CYP2C9/CYP2C19 Phenotype | PHT Normalized Dose (mg/kg) | [PHT] (kg/L) | [p-HPPH] (kg/L) |
|---|---|---|---|---|---|---|
| *1/*1 | *1/*1 | 26 | NM/NM | 4.545 (3.771–4.938) | 1.628 (1.207–2.050) | 0.0250 (0.0190–0.0309) |
| *1/*1 | *1/*2 | 8 | NM/IM | 4.5817 (3.3811–5.7823) | 1.644 (1.083–2.204) | 0.0294 (0.0154–0.0434) |
| *1/*2 | *1/*1 | 9 | IM/NM | 4.0893 (3.5590–4.6196) | 1.917 (1.402–2.431) | 0.0206 (0.0169–0.0243) |
| *1/*3 | 4 | |||||
| *1/*2 | *1/*2 | 1 | IM/IM | 4.0813 (3.0489–5.1136) | 2.943 (2.862–3.023) | 0.0170 (−0.0194–0.0533) |
| *1/*3 | 1 | |||||
| *2/*2 | *1/*1 | 1 | PM/NM | 3.2258 | 3.140 | 0.0153 |
| CYP2C9/CYP2C19/EPHX Phenotype | N° of Subjects | Dose (mg/kg) | [PHT] (kg/L) | [p-HPPH] (kg/L) |
|---|---|---|---|---|
| NM/NM/Intermediate | 10 | 4.279 (3.446–5.112) | 1.886 (1.033–2.740) | 0.0256 (0.0193–0.0319) |
| NM/NM/Increased | 4 | 5.391 (1.535–9.247) | 1.020 (0.1884–1.851) | 0.0168 (0.0073–0.0263) |
| NM/NM/Decreased | 12 | 4.485 (3.569–5.401) | 1.616 (0.9670–2.266) | 0.0271 (0.0146–0.0397) |
| NM/IM/Intermediate | 4 | 4.346 (1.655–7.037) | 1.420 (0.7667–2.072) | 0.0325 (−0.0065–0.0715) |
| NM/IM/Decreased | 4 | 4.817 (2.680–6.955) | 1.868 (0.4940–3.241) | 0.0263 (0.0187–0.0339) |
| IM/NM/Intermediate | 7 | 4.213 (3.161–5.264) | 1.673 (0.8725–2.474) | 0.0223 (0.0158–0.0288) |
| IM/NM/Decreased | 6 | 3.945 (3.421–4.470) | 2.201 (1.349–3.052) | 0.0187 (0.0137–0.0237) |
| IM/IM/Increased | 1 | 4.000 | 2.936 | 0.0141 |
| IM/IM/Decreased | 1 | 4.163 | 2.949 | 0.0198 |
| PM/NM/Intermediate | 1 | 3.226 | 3.140 | 0.0153 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guevara, N.; Maldonado, C.; Uría, M.; González, R.; Ibarra, M.; Alvariza, S.; Carozzi, A.; Azambuja, C.; Fagiolino, P.; Vázquez, M. Role of CYP2C9, CYP2C19 and EPHX Polymorphism in the Pharmacokinetic of Phenytoin: A Study on Uruguayan Caucasian Subjects. Pharmaceuticals 2017, 10, 73. https://doi.org/10.3390/ph10030073
Guevara N, Maldonado C, Uría M, González R, Ibarra M, Alvariza S, Carozzi A, Azambuja C, Fagiolino P, Vázquez M. Role of CYP2C9, CYP2C19 and EPHX Polymorphism in the Pharmacokinetic of Phenytoin: A Study on Uruguayan Caucasian Subjects. Pharmaceuticals. 2017; 10(3):73. https://doi.org/10.3390/ph10030073
Chicago/Turabian StyleGuevara, Natalia, Cecilia Maldonado, Manuel Uría, Raquel González, Manuel Ibarra, Silvana Alvariza, Antonella Carozzi, Carlos Azambuja, Pietro Fagiolino, and Marta Vázquez. 2017. "Role of CYP2C9, CYP2C19 and EPHX Polymorphism in the Pharmacokinetic of Phenytoin: A Study on Uruguayan Caucasian Subjects" Pharmaceuticals 10, no. 3: 73. https://doi.org/10.3390/ph10030073
APA StyleGuevara, N., Maldonado, C., Uría, M., González, R., Ibarra, M., Alvariza, S., Carozzi, A., Azambuja, C., Fagiolino, P., & Vázquez, M. (2017). Role of CYP2C9, CYP2C19 and EPHX Polymorphism in the Pharmacokinetic of Phenytoin: A Study on Uruguayan Caucasian Subjects. Pharmaceuticals, 10(3), 73. https://doi.org/10.3390/ph10030073





