Drosophila Protein Kinase CK2: Genetics, Regulatory Complexity and Emerging Roles during Development
Abstract
:1. General Overview
2. Biochemical Properties and Regulation of Dm-CK2
3. Substrates of Dm-CK2
4. Drosophila Genes Encoding Catalytic (α) and Regulatory (β) Subunits
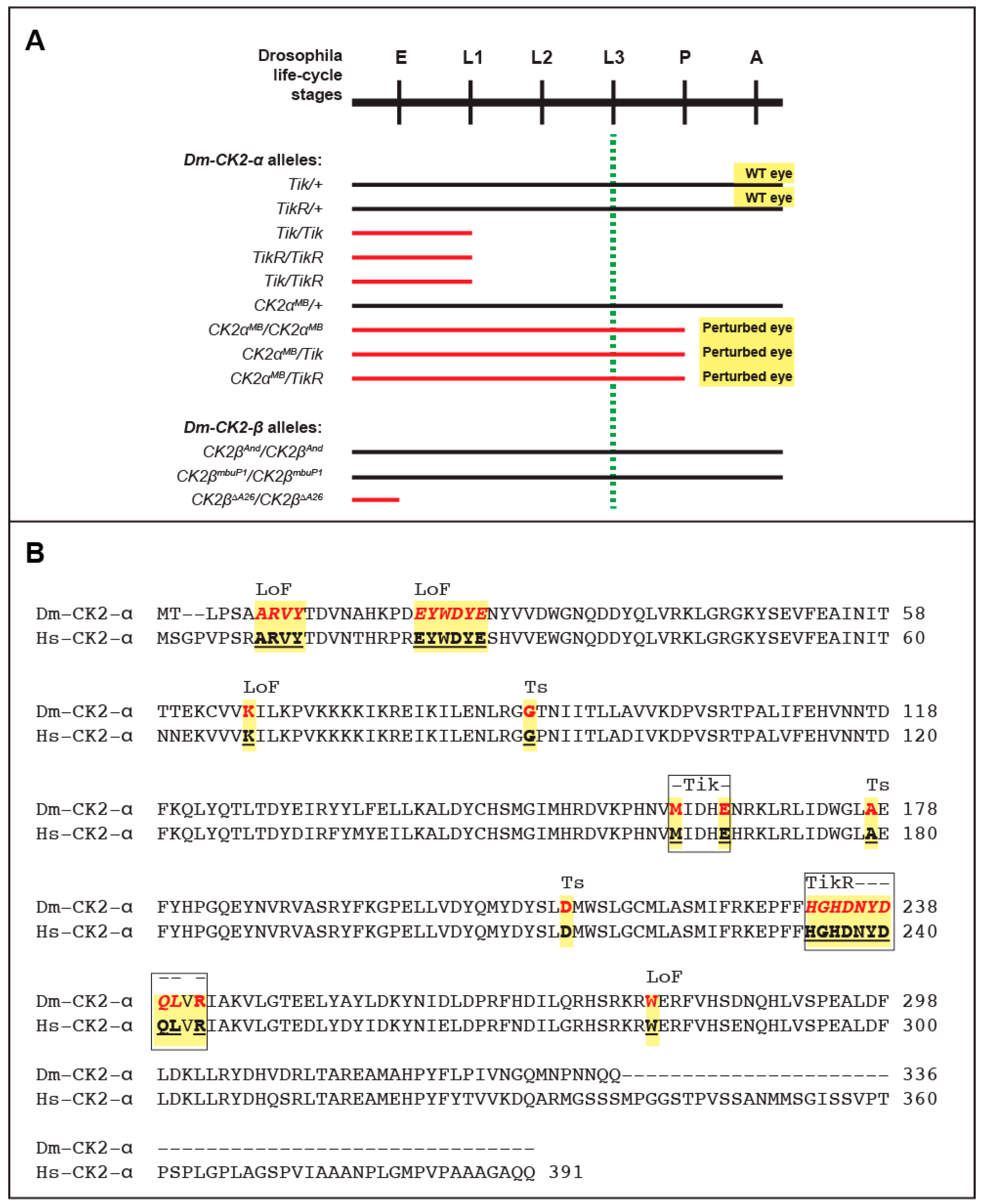
5. Mutations in Catalytic (α) and Regulatory (β) Subunits
6. Multiple Non-Redundant Variants Encoded by the Dm-CK2-β Gene
7. The DPiM Database Provides New Insights into the Dm-CK2 Interactome and Subunit Specific Interactions
7.1. Interactions between Dm-CK2 Subunits
7.2. Dm-CK2-α Interactors
7.3. Interactors of Dm-CK2-β
7.4. Interactors of Dm-CK2-β′
7.5. Interactors Shared between Dm-CK2-β and -β′
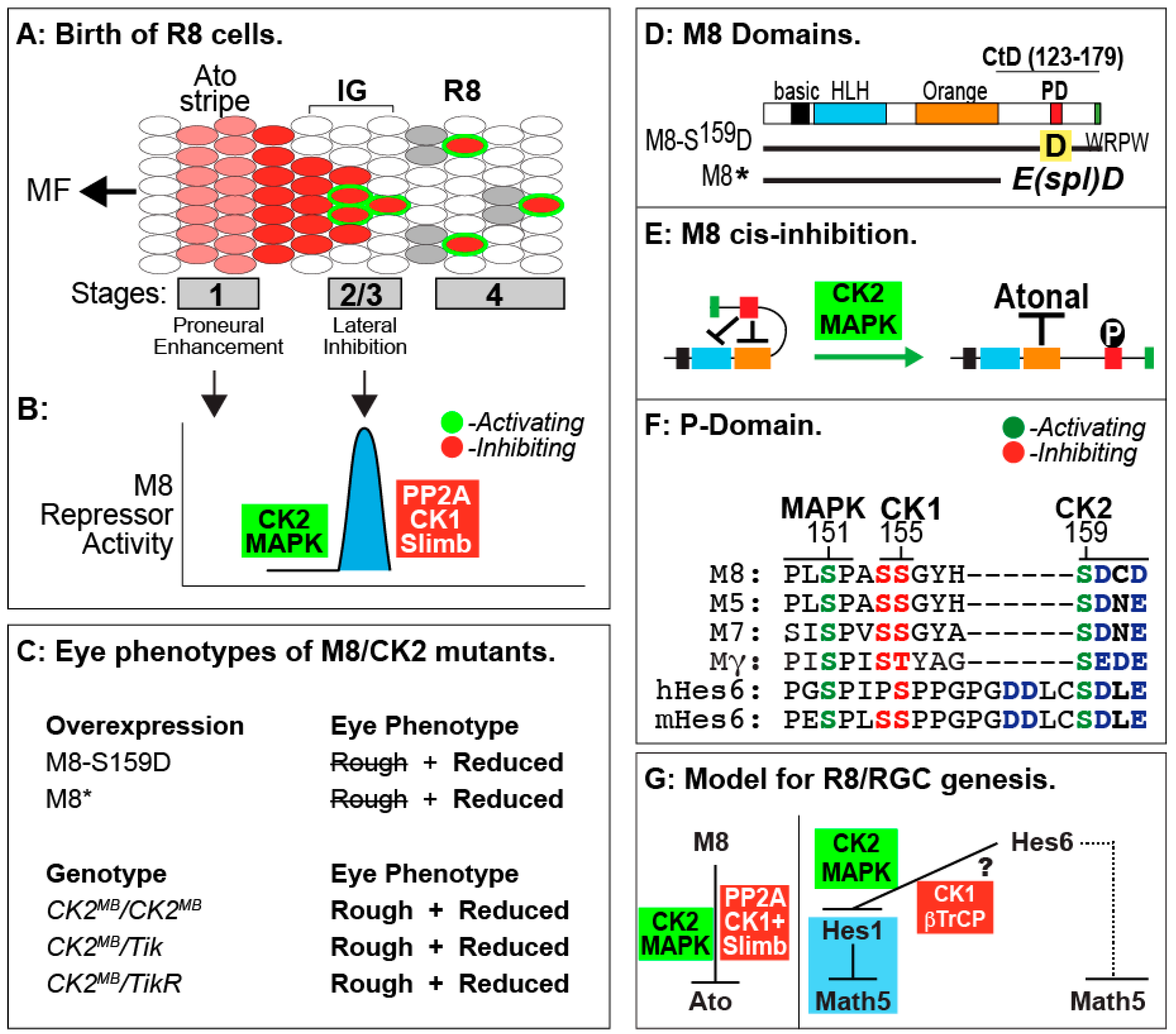
8. Roles of CK2 in Drosophila Eye Development
9. Lessons from Drosophila R8 Cells Applied to Birth of Mammalian RGCs
10. Additional Roles of CK2 during Drosophila Eye Development
11. Potential CK2 Targets Identified Via the Transcriptome of the Developing Eye
11.1. Transcription Factors
11.2. Signaling Pathways
11.3. Protein Turnover
11.4. Regulators of the Cell Cycle, Cell Death, Cell Polarity and Cytoskeleton
12. Summary and Future Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Burnett, G.; Kennedy, E.P. The enzymatic phosphorylation of proteins. J. Biol. Chem. 1954, 211, 969–980. [Google Scholar] [PubMed]
- Krebs, E.G.; Graves, D.J.; Fischer, E.H. Factors affecting the activity of muscle phosphorylase b kinase. J. Biol. Chem. 1959, 234, 2867–2873. [Google Scholar] [PubMed]
- Krebs, E.G. Protein phosphorylation and cellular regulation I (Nobel Lecture). Angew. Chem. 1993, 32, 1122–1129. [Google Scholar] [CrossRef]
- Kennedy, E.P. Sailing to Byzantium. Annu. Rev. Biochem. 1992, 61, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Pinna, L.A. Protein kinase CK2: A challenge to canons. J. Cell Sci. 2002, 115, 3873–3878. [Google Scholar] [CrossRef] [PubMed]
- Venerando, A.; Ruzzene, M.; Pinna, L.A. Casein kinase: The triple meaning of a misnomer. Biochem. J. 2014, 460, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Guerra, B.; Boldyreff, B.; Sarno, S.; Cesaro, L.; Issinger, O.G.; Pinna, L.A. CK2: A protein kinase in need of control. Pharmacol. Ther. 1999, 82, 303–313. [Google Scholar] [CrossRef]
- Pinna, L.A. A historical view of protein kinase CK2. Cell. Mol. Biol. Res. 1994, 40, 383–390. [Google Scholar] [PubMed]
- Filhol, O.; Martiel, J.L.; Cochet, C. Protein kinase CK2: A new view of an old molecular complex. EMBO Rep. 2004, 5, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Cozza, G.; Salvi, M.; Banerjee, S.; Tibaldi, E.; Tagliabracci, V.S.; Dixon, J.E.; Pinna, L.A. A new role for sphingosine: Up-regulation of Fam20C, the genuine casein kinase that phosphorylates secreted proteins. Biochim. Biophys. Acta 2015, 1854, 1718–1726. [Google Scholar] [CrossRef] [PubMed]
- Tagliabracci, V.S.; Wiley, S.E.; Guo, X.; Kinch, L.N.; Durrant, E.; Wen, J.; Xiao, J.; Cui, J.; Nguyen, K.B.; Engel, J.L.; et al. A single kinase generates the majority of the secreted phosphoproteome. Cell 2015, 161, 1619–1632. [Google Scholar] [CrossRef] [PubMed]
- Hathaway, G.M.; Traugh, J.A. Cyclic nucleotide-independant protein kinase from rabbit reticulocytes. Purification of casein kinases. J. Biol. Chem. 1979, 254, 762–768. [Google Scholar] [PubMed]
- Hathaway, G.M.; Lubben, T.H.; Traugh, J.A. Inhibition of casein kinase II by heparin. J. Biol. Chem. 1980, 255, 8038–8041. [Google Scholar] [PubMed]
- Hathaway, G.M.; Zoller, M.J.; Traugh, J.A. Identification of the catalytic subunit of casein kinase II by affinity labeling with 5′-p-fluorosulfonylbenzoyl adenosine. J. Biol. Chem. 1981, 256, 11442–11446. [Google Scholar] [PubMed]
- Glover, C.V.C.; Shelton, E.R.; Brutlag, D.L. Purification and characterization of a type II casein kinase from Drosophila melanogaster. J. Biol. Chem. 1983, 258, 3258–3265. [Google Scholar] [PubMed]
- Meggio, F.; Donella-Deana, A.; Brunati, A.M.; Pinna, L.A. Inhibition of rat liver cytosol casein kinases by heparin. FEBS Lett. 1982, 141, 257–262. [Google Scholar] [CrossRef]
- Kuenzel, E.A.; Krebs, E.G. A synthetic substrate specific for casein kinase II. Proc. Natl. Acad. Sci. USA 1985, 82, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Kuenzel, E.A.; Mulligan, J.A.; Sommercorn, J.; Krebs, E.G. Substrate specificity determinants for casein kinase II as deduced from studies with synthetic peptides. J. Biol. Chem. 1987, 262, 9136–9140. [Google Scholar] [PubMed]
- Meggio, F.; Marchiori, F.; Borin, G.; Chessa, G.; Pinna, L.A. Synthetic peptides including acidic clusters as substrates and inhibitors of rat liver casein kinase TS (type-2). J. Biol. Chem. 1984, 259, 14576–14579. [Google Scholar] [PubMed]
- Marchiori, F.; Meggio, F.; Marin, O.; Borin, G.; Calderan, A.; Ruzza, P.; Pinna, L.A. Synthetic peptide substrates for casein kinase-2: Assessment of minimal structural requirements for phosphorylation. Biochim. Biophys. Acta 1988, 971, 332–338. [Google Scholar] [CrossRef]
- Dahmus, M.E. Purification and properties of calf thymus casein kinase I and II. J. Biol. Chem. 1981, 256, 3319–3325. [Google Scholar] [PubMed]
- Litchfield, D.W.; Lozeman, F.J.; Piening, C.; Sommercorn, J.; Takio, K.; Walsh, K.A.; Krebs, E.G. Subunit structure of casein kinase II from bovine testis. Demonstration that the α and α′ subunits are distinct polypeptides. J. Biol. Chem. 1990, 265, 7638–7644. [Google Scholar] [PubMed]
- Padmanabha, R.; Glover, C.V.C. Casein kinase II of yeast contains two distinct α polypeptides and an unusually large β subunit. J. Biol. Chem. 1987, 262, 1829–1835. [Google Scholar] [PubMed]
- Bidwai, A.P.; Reed, J.C.; Glover, C.V.C. Casein kinase II of Saccharomyces cerevisiae contains two distinct regulatory subunits, β and β′. Arch. Biochem. Biophys. 1994, 309, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Boldyreff, B.; James, P.; Staudenmann, W.; Issinger, O.-G. Ser2 is the autophosphorylation site in the β subunit from bicistronically expressed human casein kinase-2 and from native rat liver casein kinase-2β. Eur. J. Biochem. 1993, 218, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Rigobello, M.P.; Jori, E.; Carignani, G.; Pinna, L.A. Isolation and characterization of a Type II casein kinase (casein kinase-TS) from Saccharomyces cerevisiae. FEBS Lett. 1982, 144, 354–358. [Google Scholar] [CrossRef]
- Cochet, C.; Chambaz, E.M. Oligomeric structure and catalytic activity of G type casein kinase. J. Biol. Chem. 1983, 258, 1403–1406. [Google Scholar] [PubMed]
- Fiege, J.J.; Cochet, C.; Pirollet, F.; Chambaz, E.M. Identification of the catalytic subunit of an oligomeric casein kinase (G type). Biochemistry 1983, 22, 1452–1459. [Google Scholar] [CrossRef]
- Meggio, F.; Brunati, A.M.; Pinna, L.A. Autophosphorylation of type 2 casein kinase TS at both its α- and β- subunits. FEBS Lett. 1983, 160, 203–208. [Google Scholar] [CrossRef]
- Dahmus, G.K.; Glover, C.V.C.; Brutlag, D.; Dahmus, M.E. Similarities in structure and function of calf thymus and Drosophila casein kinase II. J. Biol. Chem. 1984, 259, 9001–9006. [Google Scholar] [PubMed]
- Saxena, A.; Padmanabha, R.; Glover, C.V.C. Isolation and sequencing of cDNA clones encoding α and β subunits of Drosophila melanogaster casein kinase II. Mol. Cell. Biol. 1987, 7, 3409–3417. [Google Scholar] [CrossRef] [PubMed]
- Padmanabha, R.; Chen-Wu, J.L.P.; Hanna, D.E.; Glover, C.V.C. Isolation, sequencing, and disruption of the yeast CKA2 gene: Casein kinase II is essential for viability in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990, 10, 4089–4099. [Google Scholar] [CrossRef] [PubMed]
- Chen-Wu, L.P.J.; Padmanabha, R.; Glover, C.V.C. Isolation, sequencing, and disruption of the CKA1 gene encoding the alpha subunit of yeast casein kinase II. Mol. Cell. Biol. 1988, 8, 4981–4990. [Google Scholar] [CrossRef] [PubMed]
- Roussou, I.; Dretta, G. The Schizosaccharomyces pombe casein kinase II alpha and beta subunits: Evolutionary conservation and positive role for the beta subunits. Mol. Cell. Biol. 1994, 14, 576–586. [Google Scholar] [CrossRef] [PubMed]
- Hu, E.; Rubin, C. Casein kinase II from Caenorhabditis elegans. Cloning, characterization, and developmental regulation of the gene encoding the beta subunit. J. Biol. Chem. 1991, 266, 19796–19802. [Google Scholar] [PubMed]
- Hu, E.; Rubin, C.S. Expression of wild-type and mutated forms of the catalytic (alpha) subunit of Caenorhabditis elegans casein kinase II in Escherichia coli. J. Biol. Chem. 1990, 265, 20609–20615. [Google Scholar] [PubMed]
- Hu, E.; Rubin, C.S. Casein kinase II from Caenorhabditis elegans: Properties and developmental regulation of the enzyme; cloning and sequence analysis of cDNA and the gene for the catalytic subunit. J. Biol. Chem. 1990, 265, 5072–5080. [Google Scholar] [PubMed]
- Dobrowlska, G.; Boldyreff, B.; Issinger, O.G. Cloning and sequencing of the casein kinase 2 alpha subunit from Zea mays. Biochim. Biophys. Acta 1991, 1129, 139–140. [Google Scholar] [CrossRef]
- Jedlicki, A.; Hinrichs, M.V.; Allende, C.C.; Allende, J.E. The cDNAs coding for the alpha- and beta- subunits of Xenopus laevis casein kinase II. FEBS Lett. 1992, 297, 280–284. [Google Scholar] [CrossRef]
- Meisner, H.; Heller Harrison, R.; Buxton, J.; Czech, M.P. Molecular cloning of the human casein kinase II alpha subunit: Evidence for two genes. Biochemistry 1989, 28, 4072–4076. [Google Scholar] [CrossRef] [PubMed]
- Jakobi, R.; Voss, H.; Pyerin, W. Human phosvitin/casein kinase type II: Molecular cloning and sequencing of full-length cDNA encoding subunit beta. Eur. J. Biochem. 1989, 183, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Lozeman, F.J.; Litchfield, D.W.; Piening, C.; Takio, K.; Walsh, K.A.; Krebs, E.G. Isolation and characterization of human cDNA clones encoding the alpha and alpha′ subunits of casein kinase II. Biochemistry 1990, 29, 8436–8447. [Google Scholar] [CrossRef] [PubMed]
- Takio, K.E.; Kuenzel, K.A.; Walsh, K.A.; Krebs, E.G. Amino acid sequence of the b subunit of bovine lung casein kinase II. Proc. Natl. Acad. Sci. USA 1987, 84, 4851–4855. [Google Scholar] [CrossRef] [PubMed]
- Bidwai, A.P.; Reed, J.C.; Glover, C.V.C. The phosphorylation of Calmodulin by the catalytic subunit of casein kinase II is inhibited by the regulatory subunit. Arch. Biochem. Biophys. 1993, 300, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Bidwai, A.P.; Hanna, D.E.; Glover, C.V.C. The free catalytic subunit of casein kinase II is not toxic in vivo. J. Biol. Chem. 1992, 267, 18790–18796. [Google Scholar] [PubMed]
- Birnbaum, M.J.; Wu, J.; O’Reilley, D.R.; Rivera-Marrero, C.A.; Hanna, D.E.; Miller, L.K.; Glover, C.V.C. Expression, purification and characterization of Drosophila casein kinase II using the baculovirus system. Protein Expr. Purif. 1992, 3, 142–150. [Google Scholar] [PubMed]
- Rasmussen, T.; Skjøth, I.H.E.; Jensen, H.H.; Niefind, K.; Boldyreff, B.; Issinger, O.G. Biochemical characterization of the recombinant human Drosophila homologues Timekeeper and Andante involved in the Drosophila circadian oscillator. Mol. Cell. Biochem. 2005, 274, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Meggio, F.; Boldyreff, B.; Issinger, O.-G.; Pinna, L.A. Casein kinase 2 down regulation and activation by polybasic peptides are mediated by acidic residues in the 55–64 region of the b-subunit: A study with calmodulin as phosphorylatable substrate. Biochemistry 1994, 33, 4336–4342. [Google Scholar] [CrossRef] [PubMed]
- Hathaway, G.M.; Traugh, J.A. Interaction of polyamines and magnesium with casein kinase II. Arch. Biochem. Biophys. 1984, 233, 133–138. [Google Scholar] [CrossRef]
- Hathaway, G.M.; Traugh, J.A. Kinetics of activation of casein kinase II by polyamines and reversal of 2,3-bisphosphoglycerate inhibition. J. Biol. Chem. 1984, 259, 2850–2855. [Google Scholar] [PubMed]
- Nuutinen, M.; Londesborough, J. The stimulation of casein kinase II from yeast by polyamines occurs with endogenous substrates at cytosolic salt concentrations. Second Messengers Phosphoprot. 1988, 12, 197–205. [Google Scholar]
- Sarno, S.; Marin, O.; Meggio, F.; Pinna, L.A. Polyamines as negative regulators of casein kinase II: The phosphorylation of calmodulin triggered by polylysine and by the α-[66–86] peptide is prevented by spermine. Biochem. Biophys. Res. Commun. 1993, 194, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Stark, F.; Pfannstiel, J.; Klaiber, I.; Raabe, T. Protein kinase CK2 links polyamine metabolism to MAPK signalling in Drosophila. Cell. Signal. 2011, 23, 876–882. [Google Scholar] [CrossRef] [PubMed]
- Sommercorn, J.; Krebs, E.G. Induction of casein kinase II during differentiation of 3T3-L1 cells. J. Biol. Chem. 1987, 262, 3839–3843. [Google Scholar] [PubMed]
- Sommercorn, J.; Mulligan, J.A.; Lozeman, F.J.; Krebs, E.G. Activation of casein kinase II in response to insulin and to epidermal growth factor. Proc. Natl. Acad. Sci. USA 1987, 84, 8834–8838. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Kauffmann, R.C.; Zhang, J.; Kladny, S.; Carthew, R.W. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell 2000, 103, 87–97. [Google Scholar] [CrossRef]
- Yarden, Y.; Ullrich, A. Growth factor receptor tyrosine kinases. Annu. Rev. Biochem. 1988, 57, 443–478. [Google Scholar] [CrossRef] [PubMed]
- Litchfield, D.W.; Dobrowolska, G.; Krebs, E.G. Regulation of casein kinase II by growth factors: A reevaluation. Cell. Mol. Biol. Res. 1994, 40, 373–381. [Google Scholar] [PubMed]
- Ole-MoiYoi, O.K.; Brown, W.C.; Iams, K.P.; Nayar, A.; Tsukamoto, T.; Macklin, M.D. Evidence for the induction of casein kinase II in bovine lymphocytes transformed by the intracellular protozoan parasite Theileria parva. EMBO J. 1993, 12, 1621–1631. [Google Scholar] [PubMed]
- Seldin, D.C.; Leder, P. Casein kinase II alpha transgene-induced murine lymphoma: Relation to Theileriosis in cattle. Science 1995, 267, 894–897. [Google Scholar] [CrossRef] [PubMed]
- Glover, C.V.C. A filamentous form of Drosophila casein kinase II. J. Biol. Chem. 1986, 261, 14349–14354. [Google Scholar] [PubMed]
- Lolli, G.; Pinna, L.A.; Battistutta, R. Structural determinants of protein kinase CK2 regulation by autoinhibitory polymerization. ACS Chem. Biol. 2012, 7, 1158–1163. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, P.; Glover, C.V.C.; Osheroff, N. Phosphorylation of DNA topoisomerase II by casein kinase II: Modulation of eukaryotic topoismerase II activity in vitro. Proc. Natl. Acad. Sci. USA 1985, 82, 3164–3168. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, P.; Glover, C.V.C.; Osheroff, N. Phosphorylation of DNA topoisomerase II in vivo and in total homogenates of Drosophila Kc cells: The role of casein kinase II. J. Biol. Chem. 1988, 263, 12653–12660. [Google Scholar] [PubMed]
- Cardenas, M.E.; Gasser, S.M. Regulation of toposiomerase II by phosphorylation: A role for casein kinase II. J. Cell Biol. 1993, 104, 219–225. [Google Scholar]
- Cardenas, M.E.; Walter, R.; Hanna, D.; Gasser, S.M. Casein kinase II copurifies with yeast DNA topoisomerase II and re-activates the dephosphorylated enzyme. J. Cell Biol. 1993, 104, 533–543. [Google Scholar]
- Cardenas, M.; Dang, Q.; Glover, C.V.C.; Gasser, S.M. Casein kinase II phosphorylates the eukaryote-specific C-terminal domain of topoisomerase II in vivo. EMBO J. 1992, 11, 1785–1796. [Google Scholar] [PubMed]
- Franchin, C.; Salvi, M.; Arrigoni, G.; Pinna, L.A. Proteomics perturbations promoted by the protein kinase CK2 inhibitor quinalizarin. Biochim. Biophys. Acta 2015, 1854, 1676–1686. [Google Scholar] [CrossRef] [PubMed]
- Venerando, A.; Franchin, C.; Cant, N.; Cozza, G.; Pagano, M.A.; Tosoni, K.; Al-Zahrani, A.; Arrigoni, G.; Ford, R.C.; Mehta, A.; et al. Detection of phospho-sites generated by protein kinase CK2 in CFTR: Mechanistic aspects of Thr1471 phosphorylation. PLoS ONE 2013, 8, e74232. [Google Scholar] [CrossRef] [PubMed]
- Giot, L.; Bader, J.S.; Brouwer, C.; Chaudhuri, A.; Kuang, B.; Li, Y.; Hao, Y.L.; Ooi, C.E.; Godwin, B.; Vitols, E.; et al. A protein interaction map of Drosophila melanogaster. Science 2003, 302, 1727–1736. [Google Scholar] [CrossRef] [PubMed]
- Guruharsha, K.G.; Obar, R.A.; Mintseris, J.; Aishwarya, K.; Krishnan, R.T.; Vijayraghavan, K.; Artavanis-Tsakonas, S. Drosophila Protein interaction Map (DPiM): A paradigm for metazoan protein complex interactions. Fly (Austin) 2012, 6, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Guruharsha, K.G.; Rual, J.F.; Zhai, B.; Mintseris, J.; Vaidya, P.; Vaidya, N.; Beekman, C.; Wong, C.; Rhee, D.Y.; Cenaj, O.; et al. A Protein Complex Network of Drosophila melanogaster. Cell 2011, 147, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, J.; Jiang, W.; Zhou, H.; Wu, P.; Yin, J.C. Phosphorylation of conserved casein kinase sites regulates cAMP-response element-binding protein DNA binding in Drosophila. J. Biol. Chem. 2004, 279, 12117–12125. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Bidwai, A.P.; Glover, C.V.C. Interaction of casein kinase II with ribosomal protein L22 of Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2002, 298, 60–66. [Google Scholar] [CrossRef]
- Bulat, V.; Rast, M.; Pielage, J. Presynaptic CK2 promotes synapse organization and stability by targeting Ankyrin2. J. Cell Biol. 2014, 204, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, L.; Ryoo, H.-D.; Mann, R.S. A role for phosphorylation by casein kinase II in modulating Antennapedia activity in Drosophila. Genes Dev. 1997, 11, 1327–1340. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, S.-I.; Matsuda, Y.; Lee, J.-S.; Matsubayashi, H.; Sese, S.; Kadowaki, T.; Ishimoto, A. Casein kinase I phosphorylates the Armadillo protein and induces its degradation in Drosophila. EMBO J. 2002, 21, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Packman, L.C.; Kubota, K.; Parker, J.; Gay, N.J. Casein kinase II phosphorylates Ser468 in the PEST domain of the Drosophila IkappaB homologue cactus. FEBS Lett. 1997, 400, 45–50. [Google Scholar] [CrossRef]
- Szabo, A.; Papin, C.; Zorn, D.; Ponien, P.; Weber, F.; Raabe, T.; Rouyer, F. The CK2 kinase stabilizes CLOCK and represses its activity in the Drosophila circadian oscillator. PLoS Biol. 2013, 11, e1001645. [Google Scholar] [CrossRef] [PubMed]
- Willert, K.; Brink, M.; Wodarz, A.; Varmus, H.; Nusse, R. Casein kinase 2 associates with and phosphorylates Dishevelled. EMBO J. 1997, 16, 3089–3096. [Google Scholar] [CrossRef] [PubMed]
- Bouazoune, K.; Brehm, A. dMi-2 chromatin binding and remodeling activities are regulated by dCK2 phosphorylation. J. Biol. Chem. 2005, 280, 41912–41920. [Google Scholar] [CrossRef] [PubMed]
- Karandikar, U.; Trott, R.L.; Yin, J.; Bishop, C.P.; Bidwai, A.P. Drosophila CK2 regulates eye morphogenesis via phosphorylation of E(spl)M8. Mech. Dev. 2004, 121, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Trott, R.L.; Kalive, M.; Paroush, Z.; Bidwai, A.P. Drosophila melanogaster casein kinase II interacts with and phosphorylates the basic-helix-loop-helix (bHLH) proteins M5, M7, and M8 derived from the Enhancer of split Complex. J. Biol. Chem. 2001, 276, 2159–2167. [Google Scholar] [CrossRef] [PubMed]
- Bourbon, H.M.; Martin-Blanco, E.; Rosen, D.; Kornberg, T.B. Phosphorylation of the Drosophila engrailed protein at a site outside its homeodomain enhances DNA binding. J. Biol. Chem. 1995, 270, 11130–11139. [Google Scholar] [CrossRef] [PubMed]
- Gelsthorpe, M.E.; Tan, Z.; Phillips, A.; Eissenberg, J.C.; Miller, A.; Wallace, J.; Tsubota, S.I. Regulation of the Drosophila melanogaster protein, enhancer of rudimentary, by casein kinase II. Genetics 2006, 174, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Siomi, M.C.; Higashijima, K.; Ishizuka, A.; Siomi, H. Casein kinase II phosphorylates the fragile X mental retardation protein and modulates its biological properties. Mol. Cell. Biol. 2002, 22, 8438–8447. [Google Scholar] [CrossRef] [PubMed]
- Bonet, C.; Fernandez, I.; Aran, X.; Bernues, J.; Giralt, E.; Azorin, F. The GAGA protein of Drosophila is phosphorylated by CK2. J. Mol. Biol. 2005, 351, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Kwong, P.N.; Chambers, M.; Vashisht, A.A.; Turki-Judeh, W.; Yau, T.Y.; Wohlschlegel, J.A.; Courey, A.J. The central region of the Drosophila co-repressor Groucho as a Regulatory Hub. J. Biol. Chem. 2015, 290, 30119–30130. [Google Scholar] [CrossRef] [PubMed]
- Kahali, B.; Trott, R.; Paroush, Z.; Allada, R.; Bishop, C.P.; Bidwai, A.P. Drosophila CK2 phosphorylates Hairy and regulates its activity in vivo. Biochem. Biophys. Res. Commun. 2008, 373, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Eissenberg, J.C. Phosphorylation of heterochromatin protein 1 by casein kinase II is required for efficient heterochromatin binding in Drosophila. J. Biol. Chem. 1999, 274, 15095. [Google Scholar] [CrossRef] [PubMed]
- Hovhanyan, A.; Herter, E.K.; Pfannstiel, J.; Gallant, P.; Raabe, T. Drosophila mbm is a nucleolar myc and casein kinase 2 target required for ribosome biogenesis and cell growth of central brain neuroblasts. Mol. Cell. Biol. 2014, 34, 1878–1891. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Strand, D.; Krehan, A.; Pyerin, W.; Heid, H.; Neumann, B.; Mechler, B.M. Casein kinase 2 binds and phosphorylates the nucleosomal assembly protein-1 (NAP1) in Drosophila melanogaster. J. Mol. Biol. 1999, 293, 1067–1084. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, R.E.; Cook, O.; Dinur, T.; Pisante, A.; Karandikar, U.C.; Bidwai, A.; Paroush, Z. An EH1-like motif in odd-skipped mediates recruitment of groucho and repression in vivo. Mol. Cell. Biol. 2005, 25, 10711–10720. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.C.; Costa, A.; McLeod, I.; Sarkeshik, A.; Yates, J., III; Kyin, S.; Perlman, D.; Schedl, P. The functioning of the Drosophila CPEB protein Orb is regulated by phosphorylation and requires casein kinase 2 activity. PLoS ONE 2011, 6, e24355. [Google Scholar] [CrossRef] [PubMed]
- Taliaferro, J.M.; Marwha, D.; Aspden, J.L.; Mavrici, D.; Cheng, N.E.; Kohlstaedt, L.A.; Rio, D.C. The Drosophila splicing factor PSI is phosphorylated by casein kinase II and tousled-like kinase. PLoS ONE 2013, 8, e56401. [Google Scholar] [CrossRef] [PubMed]
- Akten, B.; Tangredi, M.M.; Jauch, E.; Roberts, M.A.; Ng, F.; Raabe, T.; Jackson, F.R. Ribosomal s6 kinase cooperates with casein kinase 2 to modulate the Drosophila circadian molecular oscillator. J. Neurosci. 2009, 29, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Liu, Y.; Xia, R.; Tong, C.; Yue, T.; Jiang, J.; Jia, J. Casein kinase 2 promotes Hedgehog signaling by regulating both smoothened and Cubitus interruptus. J. Biol. Chem. 2010, 285, 37218–37226. [Google Scholar] [CrossRef] [PubMed]
- Cartier, E.; Hamilton, P.J.; Belovich, A.N.; Shekar, A.; Campbell, N.G.; Saunders, C.; Andreassen, T.F.; Gether, U.; Veenstra-Vanderweele, J.; Sutcliffe, J.S.; et al. Rare autism-associated variants implicate syntaxin 1 (STX1 R26Q) phosphorylation and the dopamine transporter (hDAT R51W) in dopamine neurotransmission and behaviors. EBioMedicine 2015, 2, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Huang, H.; Li, J.; Yin, M.X.; Lu, Y.; Wu, W.; Zeng, R.; Jiang, J.; Zhao, Y.; Zhang, L. Drosophila casein kinase 2 (CK2) promotes warts protein to suppress Yorkie protein activity for growth control. J. Biol. Chem. 2014, 289, 33598–33607. [Google Scholar] [CrossRef] [PubMed]
- Trott, R.L.; Kalive, M.; Karandikar, U.; Rummer, R.; Bishop, C.P.; Bidwai, A.P. Identification and characterization of proteins that interact with Drosophila melanogaster protein kinase CK2. Mol. Cell. Biochem. 2001, 227, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Bidwai, A.P.; Saxena, A.; Zhao, W.; McCann, R.O.; Glover, C.V.C. Multiple, closely spaced alternative 5′ exons in the DmCKII-β gene of Drosophila melanogaster. Mol. Cell Biol. Res. Commun. 2000, 3, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Shevelyov, Y.Y. Copies of Stellate gene variants are located in the X heterochromatin of Drosophila melanogaster and are probably expressed. Genetics 1992, 132, 1033–1037. [Google Scholar] [PubMed]
- Palumbo, G.; Bonaccorsi, S.; Robbins, L.G.; Pimpinelli, S. Genetic analysis of Stellate elements of Drosophila melanogaster. Genetics 1994, 138, 1181–1197. [Google Scholar] [PubMed]
- Livak, K.J. Detailed structure of the Drosophila melanogaster Stellate genes and their transcripts. Genetics 1990, 124, 303–316. [Google Scholar] [PubMed]
- Bidwai, A.P.; Zhao, W.F.; Glover, C.V.C. A gene located at 56F1–2 in Drosophila melanogaster encodes a novel metazoan β-like subunit of casein kinase II. Mol. Cell Biol. Res. Commun. 1999, 1, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Karandikar, U.; Anderson, S.; Mason, N.; Trott, R.L.; Bishop, C.P.; Bidwai, A.P. The Drosophila SSL gene is expressed in larvae, pupae, and adults, exhibits sexual dimorphism, and mimics properties of the β subunit of casein kinase II. Biochem. Biophys. Res. Commun. 2003, 301, 941–947. [Google Scholar] [CrossRef]
- Kalmykova, A.I.; Shevelyov, Y.Y.; Polesskaya, O.O.; Dobritsa, A.A.; Evstafieva, A.G.; Boldyreff, B.; Issinger, O.-G.; Gvozdev, V.A. CK2βtes gene encodes a testis-specific isoform of the regulatory subunit of casein kinase 2 in Drosophila melanogaster. Eur. J. Biochem. 2002, 269, 1418–1427. [Google Scholar] [CrossRef] [PubMed]
- Kalmykova, A.I.; Dobritsa, A.A.; Gvozdev, V.A. Su(Ste) diverged tandem repeats in a Y Chromosome of Drosophila melanogaster are transcribed and variously processed. Genetics 1998, 148, 243–249. [Google Scholar] [PubMed]
- Kalmykova, A.I.; Shevelyov, Y.Y.; Dobritsa, A.A.; Gvozdev, V.A. Acquisition and amplification of a testis-expressed autosomal gene, SSL, by the Drosophila Y Chromosome. Proc. Natl. Acad. Sci. USA 1997, 94, 6297–6302. [Google Scholar] [CrossRef] [PubMed]
- Jauch, E.; Wecklein, H.; Stark, F.; Jauch, M.; Raabe, T. The Drosophila melanogaster DmCK2β transcription unit encodes for functionally non-redundant protein isoforms. Gene 2006, 374, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Chantalat, L.; Leroy, D.; Filhol, O.; Nueda, A.; Benitez, M.J.; Chambaz, E.M.; Cochet, C.; Dideberg, O. Crystal structure of the human protein kinase CK2 regulatory subunit reveals its zinc finger-mediated dimerization. EMBO J. 1999, 18, 2930–2940. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.M.; Kilman, V.L.; Keegan, K.; Paddock, B.; Emery-Le, M.; Rosbash, M.; Allada, R. A role for casein kinase 2α in the Drosophila circadian clock. Nature 2002, 420, 816–820. [Google Scholar] [CrossRef] [PubMed]
- Kunttas-Tatli, E.; Bose, A.; Kahali, B.; Bishop, C.P.; Bidwai, A.P. Functional dissection of Timekeeper (Tik.) implicates opposite roles for CK2 and PP2A during Drosophila neurogenesis. Genesis 2009, 47, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Niefind, K.; Guerra, B.; Ermakowa, I.; Issinger, O.G. Crystal structure of human protein kinase CK2: Insights into basic properties of the CK2 holoenzyme. EMBO J. 2001, 20, 5320–5331. [Google Scholar] [CrossRef] [PubMed]
- Niefind, K.; Guerra, B.; Pinna, L.A.; Issinger, O.G.; Schomburg, D. Crystal structure of the catalytic subunit of protein kinase CK2 from Zea mays at 2.1 A resolution. EMBO J. 1998, 17, 2451–2462. [Google Scholar] [CrossRef] [PubMed]
- Hanna, D.E.; Rethinaswamy, A.; Glover, C.V.C. Casein kinase II is required for cell cycle progression during G1 and G2/M in Saccharomyces cerevisiae. J. Biol. Chem. 1995, 270, 25905–25914. [Google Scholar] [CrossRef] [PubMed]
- Kuntamalla, P.; Kunttas, E.; Karandikar, U.; Bishop, C.; Bidwai, A. Drosophila protein kinase CK2 is rendered temperature-sensitive by mutations of highly conserved residues flanking the activation segment. Mol. Cell. Biochem. 2009, 323, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Toth-Petroczy, A.; Palmedo, P.; Ingraham, J.; Hopf, T.A.; Berger, B.; Sander, C.; Marks, D.S. Structured states of disordered proteins from genomic sequences. Cell 2016, 167, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Konopka, R.J.; Smith, R.F.; Orr, D. Characterization of Andante, a new Drosophila clock mutant, and its interactions with other clock mutants. J. Neurogenet. 1991, 7, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Konopka, R.J.; Benzer, S. Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1971, 68, 2112–2116. [Google Scholar] [CrossRef] [PubMed]
- Akten, B.; Jauch, E.; Genova, G.K.; Kim, E.Y.; Edery, I.; Raabe, T.; Jackson, F.R. A role for CK2 in the Drosophila circadian oscillator. Nat. Neurosc. 2003, 6, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Buchou, T.; Vernet, M.; Blond, O.; Jensen, H.; Pointu, H.; Olsen, B.; Cochet, C.; Issinger, O.; Boldyreff, B. Disruption of the regulatory beta subunit of protein kinase CK2 in mice leads to a cell-autonomous defect and early embryonic lethality. Mol. Cell. Biol. 2003, 23, 908–915. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Coulombe-Huntington, J.; Kang, S.; Sheynkman, G.M.; Hao, T.; Richardson, A.; Sun, S.; Yang, F.; Shen, Y.A.; Murray, R.R.; et al. Widespread expansion of protein interaction capabilities by alternative splicing. Cell 2016, 164, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Kalive, M.; Trott, R.L.; Bidwai, A.P. A gene located at 72A in Drosophila melanogaster encodes a novel zinc-finger protein that interacts with protein kinase CK2. Mol. Cell. Biochem. 2001, 227, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Halabi, N.; Rivoire, O.; Leibler, S.; Ranganathan, R. Protein sectors: Evolutionary units of three-dimensional structure. Cell 2009, 138, 774–786. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, R.N., Jr.; Poelwijk, F.J.; Raman, A.; Gosal, W.S.; Ranganathan, R. The spatial architecture of protein function and adaptation. Nature 2012, 491, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Mumby, M. PP2A: Unveiling a reluctant tumor suppressor. Cell 2007, 130, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Cho, U.S.; Morrone, S.; Sablina, A.A.; Arroyo, J.D.; Hahn, W.C.; Xu, W. Structural basis of PP2A inhibition by small t antigen. PLoS Biol. 2007, 5, e202. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Hahn, W.C. Involvement of PP2A in viral and cellular transformation. Oncogene 2005, 24, 7746–7755. [Google Scholar] [CrossRef] [PubMed]
- Bose, A.; Majot, A.T.; Bidwai, A.P. The Ser/Thr Phosphatase PP2A regulatory subunit Widerborst inhibits Notch signaling. PLoS ONE 2014, 9, e101884. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Liu, Y.; Yan, W.; Jia, J. PP4 and PP2A regulate Hedgehog signaling by controlling Smo and Ci phosphorylation. Development 2009, 136, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, A.M.; Barrow, C.A.; Davis, A.J.; Mumby, M.C. Actions of PP2A on the MAP kinase pathway and apoptosis are mediated by distinct regulatory subunits. Proc. Natl. Acad. Sci. USA 2002, 99, 4221–4226. [Google Scholar] [CrossRef] [PubMed]
- Banreti, A.; Lukacsovich, T.; Csikos, G.; Erdelyi, M.; Sass, M. PP2A regulates autophagy in two alternative ways in Drosophila. Autophagy 2012, 8, 623–636. [Google Scholar] [CrossRef] [PubMed]
- Margolis, S.S.; Perry, J.A.; Forester, C.M.; Nutt, L.K.; Guo, Y.; Jardim, M.J.; Thomenius, M.J.; Freel, C.D.; Darbandi, R.; Ahn, J.H.; et al. Role for the PP2A/B56delta phosphatase in regulating 14-3-3 release from Cdc25 to control mitosis. Cell 2006, 127, 759–773. [Google Scholar] [CrossRef] [PubMed]
- Heriche, J.K.; Lebrin, F.; Rabilloud, T.; Leroy, D.; Chambaz, E.M.; Goldberg, Y. Regulation of protein phosphatase 2A by direct interaction with casein kinase 2alpha. Science 1997, 276, 952–955. [Google Scholar] [CrossRef] [PubMed]
- Warner, J.R.; McIntosh, K.B. How common are extraribosomal functions of ribosomal proteins. Mol. Cell 2009, 34, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Boldyreff, B.; Issinger, O.G. A-Raf kinase is a new interacting partner of protein kinase CK2 beta subunit. FEBS Lett. 1997, 403, 197–199. [Google Scholar] [CrossRef]
- Meier, U.T.; Blobel, G. Nopp 140 shuttles on tracks between nucleolus and cytoplasm. Cell 1992, 70, 127–138. [Google Scholar] [CrossRef]
- Jenett, A.; Rubin, G.M.; Ngo, T.T.; Shepherd, D.; Murphy, C.; Dionne, H.; Pfeiffer, B.D.; Cavallaro, A.; Hall, D.; Jeter, J.; et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012, 2, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Elliott, D.A.; Brand, A.H. The GAL4 system: A versatile system for the expression of genes. Methods Mol. Biol. 2008, 420, 79–95. [Google Scholar] [PubMed]
- Duffy, J.B. GAL4 system in Drosophila: A fly geneticist’s Swiss army knife. Genesis 2002, 34, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Klambt, C.; Knust, E.; Tietze, K.; Campos-Ortega, J.A. Closely related transcripts encoded by the neurogenic gene complex Enhancer of split of Drosophila melanogaster. EMBO J. 1989, 8, 203–210. [Google Scholar] [PubMed]
- Knust, E.; Schrons, H.; Grawe, F.; Campos-Ortega, J.A. Seven genes of the enhancer of split complex of Drosophila melanogaster encode helix-loop-helix proteins. Genetics 1992, 132, 505–518. [Google Scholar] [PubMed]
- Delidakis, C.; Artavanis-Tsakonas, S. The enhancer of split [E(spl)] locus of Drosophila encodes seven independant helix-loop-helix proteins. Proc. Natl. Acad. Sci. USA 1991, 89, 8731–8735. [Google Scholar] [CrossRef]
- Schweisguth, F.; Gho, M.; Lecourtois, M.; Morel, V. Signalling by Notch family receptors. C. R. Seances Soc. Biol. Fil. 1997, 191, 55–75. [Google Scholar] [PubMed]
- Mumm, J.S.; Kopan, R. Notch signaling: From the outside in. Dev. Biol. 2000, 228, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Blaumuller, C.M.; Artavanis-Tsakonas, S. Comparative aspects of Notch signaling in lower and higher eukaryotes. Perspect. Dev. Neurobiol. 1997, 4, 325–343. [Google Scholar]
- Tien, A.C.; Rajan, A.; Bellen, H.J. A Notch updated. J. Cell Biol. 2009, 184, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Ilagan, M.X.; Kopan, R. Notch signaling pathway. Cell 2007, 128, 1246. [Google Scholar] [CrossRef] [PubMed]
- Louvi, A.; Artavanis-Tsakonas, S. Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 2006, 7, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Jiang, M.M.; Jiang, L.; Salvo, J.S.; Zeng, H.C.; Dawson, B.; Bertin, T.K.; Rao, P.H.; Chen, R.; Donehower, L.A.; et al. Notch activation as a driver of osteogenic sarcoma. Cancer Cell 2014, 26, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Bajard, L.; Oates, A.C. Breathe in and straighten your back: Hypoxia, notch, and scoliosis. Cell 2012, 149, 255–256. [Google Scholar] [CrossRef] [PubMed]
- De la Pompa, J.L.; Epstein, J.A. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev. Cell 2012, 22, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Gridley, T. Notch signaling in the vasculature. Curr. Top. Dev. Biol. 2010, 92, 277–309. [Google Scholar] [PubMed]
- Wu, M.N.; Raizen, D.M. Notch signaling: A role in sleep and stress. Curr. Biol. 2011, R397–R398. [Google Scholar] [CrossRef] [PubMed]
- Treisman, J.E. How to make an eye. Development 2004, 131, 3823–3827. [Google Scholar] [CrossRef] [PubMed]
- Freeman, M. Cell determination strategies in the Drosophila eye. Development 1997, 124, 261–270. [Google Scholar] [PubMed]
- Pichaud, F.; Treisman, J.; Desplan, C. Reinventing a common strategy for patterning the eye. Cell 2001, 105, 9–12. [Google Scholar] [CrossRef]
- Majot, A.T.; Sizemore, T.S.; Bandyopadhyay, M.; Jozwick, L.M.; Bidwai, A.P. Protein kinase CK2: A window into the posttranslational regulation of the E(spl)/HES repressors from invertebrates and vertebrates. In Protein Kinase CK2 Cellular Function in Normal and Disease States; Ahmed, K., Issinger, O.-G., Szyska, R., Eds.; Springer: New York, NY, USA, 2015; Volume 12, pp. 81–108. [Google Scholar]
- Jarman, A.P.; Grell, E.H.; Ackerman, L.; Jan, L.Y.; Jan, Y.N. Atonal is the proneural gene for Drosophila photoreceptors. Nature 1994, 369, 398–400. [Google Scholar] [CrossRef] [PubMed]
- Ghiasvand, N.M.; Rudolph, D.D.; Mashayekhi, M.; Brzezinski, J.A.T.; Goldman, D.; Glaser, T. Deletion of a remote enhancer near ATOH7 disrupts retinal neurogenesis, causing NCRNA disease. Nat. Neurosci. 2011, 14, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Yan, J.; Ma, Z.; Zhou, Y.; Abbood, N.N.; Liu, J.; Su, L.; Jia, H.; Guo, A.Y. Comparative and evolutionary analysis of the HES/HEY gene family reveal exon/intron loss and teleost specific duplication events. PLoS ONE 2012, 7, e40649. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, R.; Ohtsuka, T.; Kobayashi, T. Roles of Hes genes in neural development. Dev. Growth Differ. 2008, 50 (Suppl. 1), S97–S103. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Jan, L.; Jan, Y. Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development 1998, 125, 3731–3740. [Google Scholar] [PubMed]
- Powell, L.M.; Zur Lage, P.I.; Prentice, D.R.; Senthinathan, B.; Jarman, A.P. The proneural proteins Atonal and Scute regulate neural target genes through different E-box binding sites. Mol. Cell. Biol. 2004, 24, 9517–9526. [Google Scholar] [CrossRef] [PubMed]
- Ligoxygakis, P.; Yu, S.Y.; Delidakis, C.; Baker, N.E. A subset of Notch functions during Drosophila eye development require Su(H) and E(spl) gene complex. Development 1998, 125, 2893–2900. [Google Scholar] [PubMed]
- Nolo, R.; Abbott, L.A.; Bellen, H.J. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell 2000, 102, 349–362. [Google Scholar] [CrossRef]
- Pepple, K.L.; Atkins, M.; Venken, K.; Wellnitz, K.; Harding, M.; Frankfort, B.; Mardon, G. Two-step selection of a single R8 photoreceptor: A bistable loop between senseless and rough locks in R8 fate. Development 2008, 135, 4071–4079. [Google Scholar] [CrossRef] [PubMed]
- Axelrod, J.D. Delivering the lateral inhibition punchline: It’s all about the timing. Sci. Signal. 2010, 3, pe38. [Google Scholar] [CrossRef] [PubMed]
- Simpson, P. Lateral inhibition and the development of the sensory bristles of the adult peripheral nervous system of Drosophila. Development 1990, 109, 509–519. [Google Scholar] [PubMed]
- Ligoxygakis, P.; Bray, S.J.; Apidianakis, Y.; Delidakis, C. Ectopic expression of individual E(spl) genes has differential effects on different cell fate decisions and underscores the biphasic requirement for Notch activity in wing margin establishment in Drosophila. Development 1999, 126, 2205–2214. [Google Scholar] [PubMed]
- Lubensky, D.K.; Pennington, M.W.; Shraiman, B.I.; Baker, N.E. A dynamical model of ommatidial crystal formation. Proc. Natl. Acad. Sci. USA 2011, 108, 11145–11150. [Google Scholar] [CrossRef] [PubMed]
- Struhl, G.; Adachi, A. Nuclear access and action of Notch in vivo. Cell 1998, 93, 649–660. [Google Scholar] [CrossRef]
- Nagel, A.C.; Preiss, A. Notchspl is deficient for inductive processes in the eye, and E(spl)D enhances split by interfering with proneural activity. Dev. Biol. 1999, 208, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Giebel, B.; Campos-Ortega, J.A. Functional dissection of the Drosophila enhancer of split protein, a suppressor of neurogenesis. Proc. Natl. Acad. Sci. USA 1997, 94, 6250–6254. [Google Scholar] [CrossRef] [PubMed]
- Nagel, A.; Yu, Y.; Preiss, A. Enhancer of split [E(spl)D] is a gro-independent, hypermorphic mutation in Drosophila. Dev. Genet. 1999, 25, 168–179. [Google Scholar] [CrossRef]
- Welshons, W.J. Dosage experiments with split mutations in the presense of an enhancer of split. Drosoph. Inf. Serv. 1956, 30, 157–158. [Google Scholar]
- Welshons, W.J. Analysis of a gene in Drosophila. Science 1965, 150, 1122–1129. [Google Scholar] [CrossRef] [PubMed]
- Knust, E.; Bremer, K.A.; Vassin, H.; Ziemer, A.; Tepass, U.; Campos-Ortega, J.A. The enhancer of split locus and neurogenesis in Drosophila melanogaster. Dev. Biol. 1987, 122, 262–273. [Google Scholar] [CrossRef]
- Kahali, B.; Kim, J.; Karandikar, U.; Bishop, C.P.; Bidwai, A.P. Evidence that the C-terminal domain (CtD) autoinhibits neural repression by Drosophila E(spl)M8. Genesis 2010, 48, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Bose, A.; Kahali, B.; Zhang, S.; Lin, J.-M.; Allada, R.; Karandikar, U.; Bidwai, A. Drosophila CK2 regulates lateral-inhibition during eye and bristle development. Mech. Dev. 2006, 123, 649–664. [Google Scholar] [CrossRef] [PubMed]
- White, N.; Jarman, A. Drosophila atonal controls photoreceptor R8-specific properties and modulates both receptor tyrosine kinase and Hedgehog signalling. Development 2000, 127, 1681–1689. [Google Scholar] [PubMed]
- Bandyopadhyay, M.; Bishop, C.P.; Bidwai, A.P. The Conserved MAPK Site in E(spl)-M8, an effector of Drosophila Notch signaling, controls repressor activity during eye development. PLoS ONE 2016, 11, e0159508. [Google Scholar] [CrossRef] [PubMed]
- Jhas, S.; Ciura, S.; Belanger-Jasmin, S.; Dong, Z.; Llamosas, E.; Theriault, F.M.; Joachim, K.; Tang, Y.; Liu, L.; Liu, J.; et al. Hes6 inhibits astrocyte differentiation and promotes neurogenesis through different mechanisms. J. Neurosci. 2006, 26, 11061–11071. [Google Scholar] [CrossRef] [PubMed]
- Belanger-Jasmin, S.; Llamosas, E.; Tang, Y.; Joachim, K.; Osiceanu, A.M.; Jhas, S.; Stifani, S. Inhibition of cortical astrocyte differentiation by Hes6 requires amino- and carboxy-terminal motifs important for dimerization and phosphorylation. J. Neurochem. 2007, 103, 2022–2034. [Google Scholar] [CrossRef] [PubMed]
- Gratton, M.-O.; Torban, E.; Jasmin, S.B.; Theriault, F.M.; German, M.S.; Stifani, S. Hes6 promotes cortical neurogenesis and inhibits Hes1 transcription repression activity by multiple mechanisms. Mol. Cell. Biol. 2003, 23, 6922–6935. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.L.; Dagenais, S.L.; Chen, C.M.; Glaser, T. Molecular characterization and mapping of ATOH7, a human atonal homolog with a predicted role in retinal ganglion cell development. Mamm. Genome 2002, 13, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.L.; Kanekar, S.; Vetter, M.L.; Tucker, P.K.; Gemza, D.L.; Glaser, T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development 1998, 125, 4821–4833. [Google Scholar] [PubMed]
- Brown, N.L.; Patel, S.; Brzezinski, J.; Glaser, T. Math5 is required for retinal ganglion cell and optic nerve formation. Development 2001, 128, 2497–2508. [Google Scholar] [PubMed]
- Hsiung, F.; Moses, K. Retinal development in Drosophila: Specifying the first neuron. Hum. Mol. Genet. 2002, 11, 1207–1214. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.W.; Kim, B.S.; Ding, K.; Wang, H.; Sun, D.; Johnson, R.L.; Klein, W.H.; Gan, L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001, 15, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; Ishibashi, M.; Nakahara, K.; Ang, S.L.; Nakanishi, S.; Guillemot, F.; Kageyama, R. Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron 1996, 16, 723–734. [Google Scholar] [CrossRef]
- Wang, V.Y.; Hassan, B.A.; Bellen, H.J.; Zoghbi, H.Y. Drosophila atonal fully rescues the phenotype of Math1 null mice: New functions evolve in new cellular contexts. Curr. Biol. 2002, 12, 1611–1616. [Google Scholar] [CrossRef]
- Malone, C.M.; Domaschenz, R.; Amagase, Y.; Dunham, I.; Murai, K.; Jones, P.H. Hes6 is required for actin cytoskeletal organization in differentiating C2C12 myoblasts. Exp. Cell Res. 2011, 317, 1590–1602. [Google Scholar] [CrossRef] [PubMed]
- Cossins, J.; Vernon, A.E.; Zhang, Y.; Philpott, A.; Jones, P.H. Hes6 regulates myogenic differentiation. Development 2002, 129, 2195–2207. [Google Scholar] [CrossRef] [PubMed]
- Murai, K.; Vernon, A.E.; Philpott, A.; Jones, P. Hes6 is required for MyoD induction during gastrulation. Dev. Biol. 2007, 312, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Pissarra, L.; Henrique, D.; Duarte, A. Expression of Hes6, a new member of the Hairy/Enhancer-of-split family, in mouse development. Mech. Dev. 2000, 95, 275–278. [Google Scholar] [CrossRef]
- Bae, S.; Bessho, Y.; Hojo, M.; Kageyama, R. The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development 2000, 127, 2933–2943. [Google Scholar] [PubMed]
- Haapa-Paananen, S.; Kiviluoto, S.; Waltari, M.; Puputti, M.; Mpindi, J.P.; Kohonen, P.; Tynninen, O.; Haapasalo, H.; Joensuu, H.; Perala, M.; et al. HES6 gene is selectively overexpressed in glioma and represents an important transcriptional regulator of glioma proliferation. Oncogene 2012, 31, 1299–1310. [Google Scholar] [CrossRef] [PubMed]
- Hartman, J.; Lam, E.W.; Gustafsson, J.A.; Strom, A. Hes-6, an inhibitor of Hes-1, is regulated by 17β-estradiol and promotes breast cancer cell proliferation. Breast Cancer Res. 2009, 11, R79. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, F.L.; Marchionni, L.; Gupta, A.; Kummangal, B.A.; Schaeffer, E.M.; Ross, A.E.; Berman, D.M. HES6 promotes prostate cancer aggressiveness independently of Notch signalling. J. Cell. Mol. Med. 2015, 19, 1624–1636. [Google Scholar] [CrossRef] [PubMed]
- Beltrao, P.; Trinidad, J.C.; Fiedler, D.; Roguev, A.; Lim, W.A.; Shokat, K.M.; Burlingame, A.L.; Krogan, N.J. Evolution of phosphoregulation: Comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009, 7, e1000134. [Google Scholar] [CrossRef]
- Levy, E.D.; Michnick, S.W.; Landry, C.R. Protein abundance is key to distinguish promiscuous from functional phosphorylation based on evolutionary information. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 2594–2606. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Pawson, T. Modular evolution of phosphorylation-based signalling systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 2540–2555. [Google Scholar] [CrossRef] [PubMed]
- Hedges, S.B.; Blair, J.E.; Venturi, M.L.; Shoe, J.L. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol. Biol. 2004, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Beverly, S.M.; Wilson, A.C. Molecular evolution in Drosophila and the higher Diptera II. A time scale for fly evolution. J. Mol. Evol. 1984, 21, 1–13. [Google Scholar] [CrossRef]
- Baker, N.E.; Firth, L.C. Retinal determination genes function along with cell-cell signals to regulate Drosophila eye development: Examples of multi-layered regulation by master regulators. Bioessays 2011, 33, 538–546. [Google Scholar] [CrossRef] [PubMed]
- Kumar, J.P. Retinal determination the beginning of eye development. Curr. Top. Dev. Biol. 2010, 93, 1–28. [Google Scholar] [PubMed]
- Kumar, J.P. The molecular circuitry governing retinal determination. Biochim. Biophys. Acta 2009, 1789, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Halder, G.; Callerts, P.; Gehring, W.J. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 1995, 267, 1788–1792. [Google Scholar] [CrossRef] [PubMed]
- Nfonsam, L.E.; Cano, C.; Mudge, J.; Schilkey, F.D.; Curtiss, J. Analysis of the transcriptomes downstream of eyeless and the hedgehog, decapentaplegic and Notch signaling pathways in Drosophila melanogaster. PLoS ONE 2012, 7, e44583. [Google Scholar] [CrossRef] [PubMed]
- Ostrin, E.J.; Li, Y.; Hoffman, K.; Liu, J.; Wang, K.; Zhang, L.; Mardon, G.; Chen, R. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 2006, 16, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Halder, G.; Callaerts, P.; Flister, S.; Walldorf, U.; Kloter, U.; Gehring, W.J. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development 1998, 125, 2181–2191. [Google Scholar] [PubMed]
- Culi, J.; Martin-Blanco, E.; Modolell, J. The EGF receptor and N signalling pathways act antagonistically in Drosophila mesothorax bristle patterning. Development 2001, 128, 299–308. [Google Scholar] [PubMed]
- Sundaram, M.V. The love-hate relationship between Ras and Notch. Genes Dev. 2005, 19, 1825–1839. [Google Scholar] [CrossRef] [PubMed]
- Benhra, N.; Vignaux, F.; Dussert, A.; Schweisguth, F.; Le Borgne, R. Neuralized promotes basal to apical transcytosis of delta in epithelial cells. Mol. Biol. Cell 2010, 21, 2078–2086. [Google Scholar] [CrossRef] [PubMed]
- Lai, E.C.; Rubin, G.M. Neuralized is essential for a subset of Notch pathway-dependent cell fate decisions during Drosophila eye development. Proc. Natl. Acad. Sci. USA 2001, 98, 5637–5642. [Google Scholar] [CrossRef] [PubMed]
- Isaksson, A.; Peverali, F.A.; Kockel, L.; Mlodzik, M.; Bohmann, D. The deubiquitination enzyme fat facets negatively regulates RTK/Ras/MAPK signalling during Drosophila eye development. Mech. Dev. 1997, 68, 59–67. [Google Scholar] [CrossRef]
- Huang, Y.; Fischer-Vize, J. Undifferentiated cells in the developing Drosophila eye influence facet assembly and require the Fat facets ubiquitin-specific protease. Development 1996, 122, 3207–3216. [Google Scholar] [PubMed]
- Chen, X.; Zhang, B.; Fischer, J.A. A specific protein substrate for a deubiquitinating enzyme: Liquid facets is the substrate of Fat facets. Genes Dev. 2002, 16, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita-Kikuta, E.; Yamada, A.; Inoue, C.; Kinoshita, E.; Koike, T. A novel phosphate-affinity bead with immobilized Phos-tag for separation and enrichment of phosphopeptides and phosphoproteins. J. Integr. OMICS 2011, 1, 157–169. [Google Scholar]
- Sopko, R.; Foos, M.; Vinayagam, A.; Zhai, B.; Binari, R.; Hu, Y.; Randklev, S.; Perkins, L.A.; Gygi, S.P.; Perrimon, N. Combining genetic perturbations and proteomics to examine kinase-phosphatase networks in Drosophila embryos. Dev. Cell 2014, 31, 114–127. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.; Hu, Y.; Udeshi, N.D.; Lau, T.Y.; Wirtz-Peitz, F.; He, L.; Ting, A.Y.; Carr, S.A.; Perrimon, N. Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proc. Natl. Acad. Sci. USA 2015, 112, 12093–12098. [Google Scholar] [CrossRef] [PubMed]
| Protein | Function | Effect of Phosphorylation |
|---|---|---|
| Ankyrin-2 | Cytoskeletal Adaptor | Maintenance of synaptic stability |
| Antennapedia | Transcription Factor | Spatial restriction of activity |
| Armadillo | Transcription Factor | Phosphorylation triggers degradation |
| Cactus | Transcription Factor | Required for activity during axis formation |
| Clock | Transcription Factor | Stabilizes Clock |
| CREB2 | Transcription Factor | Inhibits DNA binding |
| Dishevelled (dsh) | Transcription Factor | Influences Wg/Wnt signaling |
| dMi-2 (DPIM) | Chromatin Structure | Inhibits nucleosome-stimulated ATPase |
| E(spl)-M8/M5/M7 | Transcription Factor | Phosphorylation required for repressor activity |
| Engrailed | Transcription Factor | Phosphorylation enhances DNA-binding |
| Enhancer of Rudimentary | Transcription Factor | Promotes and inhibits activity |
| FMR1 (DPIM) | RNA-Binding | Affects dimerization and RNA-binding |
| GAGA factor (519) | Transcription Factor | Reduced DNA binding affinity |
| Groucho | Transcription Factor | Stimulates short range repression |
| Hairy | Transcription Factor | Promotes repressor activity |
| Heterochromatin protein HP1 | Chromatin Structure | Stimulates DNA binding |
| mushroom body miniature | Ribosome Biogenesis | Promotes nucleolar localization |
| NAP1 | Chromatin Structure | Affects degradation and cellular locale |
| Odd | Transcription Factor | Inhibits Groucho binding and repression |
| Orb (CPEB-family) | RNA-Binding | Promotes Orb activity |
| P element Somatic Inhibitor (PSI) | Splicing factor | Modulates interactions with splicing factors |
| Period | Circadian Clock | Promotes nuclear entry |
| Raf | Protein Kinase | Required for ERK activation |
| Ribosomal S6 kinase | Protein Kinase | Required for activity |
| RPL-22 | Ribosome Structure | Unknown |
| Smoothened | Signaling | Stabilizes and promotes Hedgehog signaling |
| Syntaxin-1 | Membrane Protein | Stimulates interaction with Dopamine Transporter |
| Timeless | Circadian Clock | Promotes nuclear entry |
| Topoisomerase II (DPIM) | DNA-replication | Stimulates activity |
| Warts | Protein Kinase | Indirectly promotes Warts suppression of Yorkie |
| Genes Encoding CK2 Subunits in Drosophila | |||
| Gene (Chromosome) | Isoforms | Alleles | Nature |
| CK2α (III) | Single | CK2α-Tik | M161K + E165D |
| CK2α-TikR | Loss of Function | ||
| CK2α-MB00477 | Hypomorphic | ||
| CK2α-G703 | W279G | ||
| CK2α-H3091 | D212N | ||
| CK2β (X) | Multiple | CK2βAndante | M166I |
| CK2βmbuP1 | Hypomorphic | ||
| CK2βmbuΔA26-2L | Loss of Function | ||
| Stellate (X) | Single | None | N/A |
| CK2β′ (II) | Single | None | N/A |
| SSL/CK2-βTes (II) | Single | None | N/A |
| Phenotypes of Ectopic Expression | |||
| Isoform | Tissue | Overexpression Phenotype | |
| CK2α-WT | Proneural cluster | No Effect | |
| CK2α-Tik | Proneural cluster | Impaired Notch Signaling (eye & bristle) | |
| CK2α-M161K | Proneural cluster | Impaired Notch Signaling (eye & bristle) | |
| CK2α-E165D | Proneural cluster | Impaired Notch Signaling (eye & bristle) | |
| CK2β-VII-a | Ubiquitous | No Effect | Rescues CK2β∆A26 |
| CK2β-VII-b | Ubiquitous | Dominant Lethal | Rescues CK2β∆A26 |
| CK2β-VII-c | Ubiquitous | No Effect | Rescues CK2β∆A26 |
| CK2β-VII-d | Ubiquitous | Dominant Lethal | No rescue of CK2β∆A26 |
| CK2β-VII-d-VI | Ubiquitous | No Effect | No rescue of CK2β∆A26 |
| Stellate | Not Tested | N/A | Not tested |
| CK2β′ | Ubiquitous | Dominant Lethal | No rescue of CK2β∆A26 |
| SSL/CK2-βTes | Ubiquitous | Dominant Lethal | No rescue of CK2β∆A26 |
| Dm-CK2-α Interactors in Drosophila S2-Cells (DPiM) | |||
| Protein | Function | Protein | Function |
| CK2-α | CK2 Catalytic Subunit | Lasp | Cytoskeletal organization |
| CK2-β | CK2 Regulatory Subunit | Rump | RNA-binding |
| AGO2 | RNA-binding | Ran | Small GTPase |
| pAbp | Poly-A binding protein | Rack1 | Receptor for activated PKC |
| glo | mRNA binding | Topo2 (Genetic) | DNA Topoisomerase 2 |
| FMR1 (Genetic) | Fragile-X syndrome | p38b | MAP-kinase |
| Dek (CK2-β) | Homeodomain | Wmd | Wing morphogenesis |
| Rasputin | RNA-binding | Mts | PP2A catalytic subunit |
| Dre4 (CK2-β) | Chromatin-binding | Scf (CK2-β) | Chromatin Organization |
| Vig2 | RNA-binding | Su-var(3)9 | Chromatin regulator |
| Ssrp | HMG Box Domain | 14-3-3 | pSer binding |
| Interactions between Dm-CK2-α, Dm-CK2-β and Dm-CK2-β′ | |||
| FLAG-HA-Fusion (Bait) | Interacting Proteins | Interactions Not Detected | |
| CK2-α | CK2-α, CK2-β | CK2-β′ | |
| CK2-β | CK2-α, CK2-β | CK2-β′ | |
| CK2-β′ | CK2-α, CK2-β′ | CK2-β | |
| Interacting Partners Unique to Dm-CK2-β | |||
| Protein | Function | Protein | Function |
| Dek (CK2-α) | Homeodomain | dMi-2 (Genetic) | Nucleosome binding |
| CG13800 | Actin-Binding | Tango7 | Neuron morphogenesis |
| Dre4 (CK2-α) | Chromatin Binding | CG3817 | rRNA processing |
| Ssrp (CK2-α) | HMG Box Domain | CG1677 | Zinc Finger Protein |
| eIF-3-S8 | Translation Factor | CG5525 | HSP60-family |
| CDK12 | Protein Kinase | Xpc/mus210 | Xeroderma pigmentosum-C |
| CG7033 | HSP60-family | Cpb (CG17158) | Actin Capping Protein |
| CG8258 | Unknown | Prp38 | pre-mRNA processing |
| Arp14D | Actin related protein 2 | D1 Chromosomal Protein | Satellite DNA-binding |
| Sop2 | Actin related protein 2/3 | CycK | Cyclin K |
| Cct5 | T-complex Chaperonin 5 | Hyd (Hyperplastic Disc) | E3-Ub-Ligase |
| Int6 | Proto-oncogene | Scf/DCB-45 (CK2-α) | Chromatin Organization |
| Tcp1-ζ | HSP60-family | CG6724 | WD40 repeats similar to Gβ |
| Arc-p34 | Neuronal development | XNP | Neuronal development |
| Smg5 | Nonsense mediated decay | ||
| Interacting Partners Unique to Dm-CK2-β′ | |||
| Protein | Function | Protein | Function |
| Porin | Mitochondrial OM channel | awd/abnormal wing discs | Nucleotide Kinase |
| Chd64 | Juvenile Hormone Signaling | EB1 | Myosin Binding |
| Fimbrin | Female meiosis | Smt3/SUMO | SUMO family |
| FK506-bp2 | DNA Damage Response | Nlp/CRP1 | Nucleoplasmin |
| Annexin B10 | Annexin Family | PCNA/Mus209 | DNA-Replication |
| Interacting Partners Common to Dm-CK2-β and Dm-CK2-β′ | |||
| Protein | Function | Protein | Function |
| Fax | Axon connectivity | Nopp140 | Cajal body protein |
| EloB/Elongin-B | Wing cell identity | Otefin | Germline stem cell renewal |
| Gene | WebLogo of CK2 Site(s) | Function |
|---|---|---|
| Acinus (Acn) |  | RNA splicing |
| Anterior Open (Aop) |  | Transcription Factor |
| Asteroid (Ast) |  | EGFR signaling |
| AXIN1 upregulated 1 (Axud1) |  | Cell proliferation |
| Cadherin 86C (Cad86C) |  | Cell adhesion/signaling |
| Cadherin N (CadN) |  | Cell adhesion/signaling |
| Capicua (Cic) |  | HMG family Transcription Factor |
| Claspin |  | ATR-Chk1 checkpoint pathway |
| Cubitus Interruptus (Ci) |  | Transcription Factor |
| Cullin 1 (Cul1) |  | Ubiquitin Ligase |
| Cullin 3 (Cul3) |  | Ubiquitin Ligase |
| Decapo (Dap) |  | CDK inhibitor |
| Daughter of Sevenless (Dos) |  | Sevenless RTK signaling |
| Decay |  | Regulator of apoptosis |
| Distal Antenna (Dan) |  | Transcription Factor |
| Distal antenna related (Danr) |  | Transcription Factor |
| Domino (Dom) |  | SNF2/RAD54 helicase family |
| Ebi |  | Chromatin binding |
| EGF-Receptor (EGFR) |  | RTK signaling |
| ELAV |  | Neurogenesis |
| Eyegone (Eyg) |  | Pax family transcription factor |
| Eyeless (Ey) |  | Transcription factor |
| Eyes Absent (Eya) |  | Transcription factor |
| Fat Facets (Faf) |  | Ubiquitin Ligase |
| Garnet (G) |  | Clathrin coatomer adaptor |
| Glass (Gl) |  | Transcription factor |
| Golden Goal (Gogo) |  | Axon guidance |
| GP150 |  | Eye development |
| Head involution defective (Hid) |  | Cell death |
| Homeodomain interacting Kinase (HipK) |  | Eye development |
| IP3-Receptor |  | Inositol 1,4,5-tris-phosphate Receptor |
| Kismet (Kis) |  | Transcription factor |
| Klarsicht (Klar) |  | Kinesin binding |
| Klumpfuss (Klu) |  | Zinc finger protein |
| Liprin-γ |  | Sterile α motif |
| Liquid Facets (Lqf) |  | Ubiquitin binding and eye development |
| Neuralized (Neur) |  | E3 ubiquitin ligase |
| Osa |  | Transcription coactivator |
| PDGF/VEGF related factor 1 |  | Cell signaling |
| Pointed (Pnt) |  | Transcription factor |
| Prickle (Pk) |  | Regulates planar cell polarity |
| RapGAP1 |  | GTPase activating protein |
| Regulator of eph expression (Reph) |  | Ephrin signaling |
| Ret Oncogene (Ret) |  | RTK signaling |
| Scribbler (Sbb) |  | Transcription factor |
| Serrate (Ser) |  | Notch signaling |
| Seven in absentia (Sina) |  | Regulation of R7 differentiation |
| Shaven (dPax2) |  | D-Pax2 family transcription factor |
| Snf5-related 1 (Snr1) |  | Chromatin structure |
| Sine Oculis (SO) |  | Transcription factor |
| SoxNeuro (SoxN) |  | Transcription factor |
| Spineless |  | Regulates Rhodopsin expression |
| Spinster |  | Regulates TGF-β/BMP signaling |
| Star |  | EGF signaling and eye development |
| Sugarless |  | Signaling in eye development |
| Target of wit (Twit) |  | Eye development |
| Terribly reduced optic lobe (Trol) |  | Cell polarity and signaling |
| Tolkin (Tok) |  | Negative regulator of gene expression |
| α-catenin |  | Actin binding |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bandyopadhyay, M.; Arbet, S.; Bishop, C.P.; Bidwai, A.P. Drosophila Protein Kinase CK2: Genetics, Regulatory Complexity and Emerging Roles during Development. Pharmaceuticals 2017, 10, 4. https://doi.org/10.3390/ph10010004
Bandyopadhyay M, Arbet S, Bishop CP, Bidwai AP. Drosophila Protein Kinase CK2: Genetics, Regulatory Complexity and Emerging Roles during Development. Pharmaceuticals. 2017; 10(1):4. https://doi.org/10.3390/ph10010004
Chicago/Turabian StyleBandyopadhyay, Mohna, Scott Arbet, Clifton P. Bishop, and Ashok P. Bidwai. 2017. "Drosophila Protein Kinase CK2: Genetics, Regulatory Complexity and Emerging Roles during Development" Pharmaceuticals 10, no. 1: 4. https://doi.org/10.3390/ph10010004
APA StyleBandyopadhyay, M., Arbet, S., Bishop, C. P., & Bidwai, A. P. (2017). Drosophila Protein Kinase CK2: Genetics, Regulatory Complexity and Emerging Roles during Development. Pharmaceuticals, 10(1), 4. https://doi.org/10.3390/ph10010004





