Comparative Analysis of Machine Learning Approaches for Fetal Movement Detection with Linear Acceleration and Angular Rate Signals
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Sensor System
2.3. Experimental Protocol
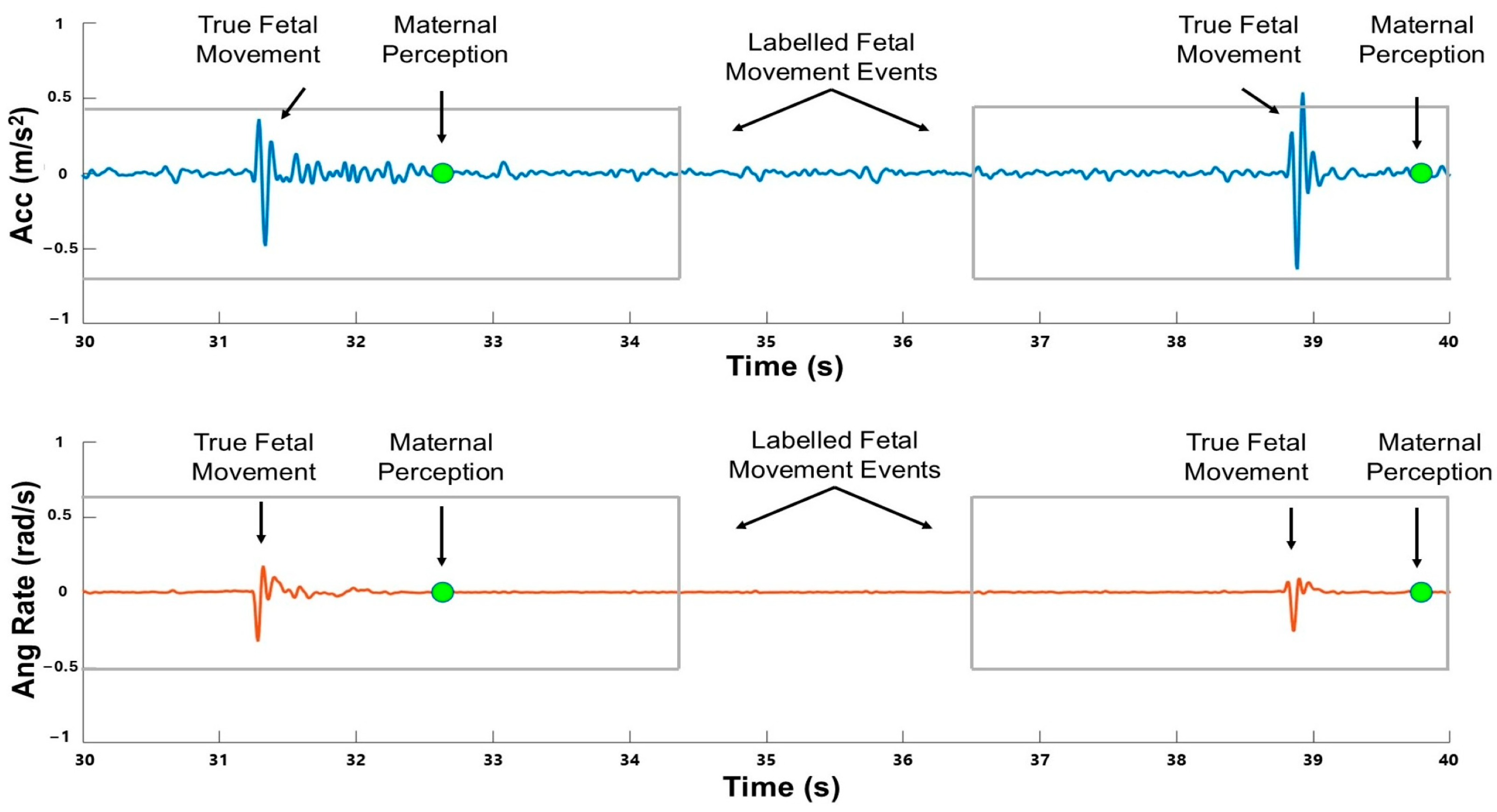
2.4. Signal Processing and Data Labeling
2.5. Model Training and Evaluation
2.6. Feature-Based Approach: Random Forest Model
2.7. Time Series Approach: BiLSTM Model
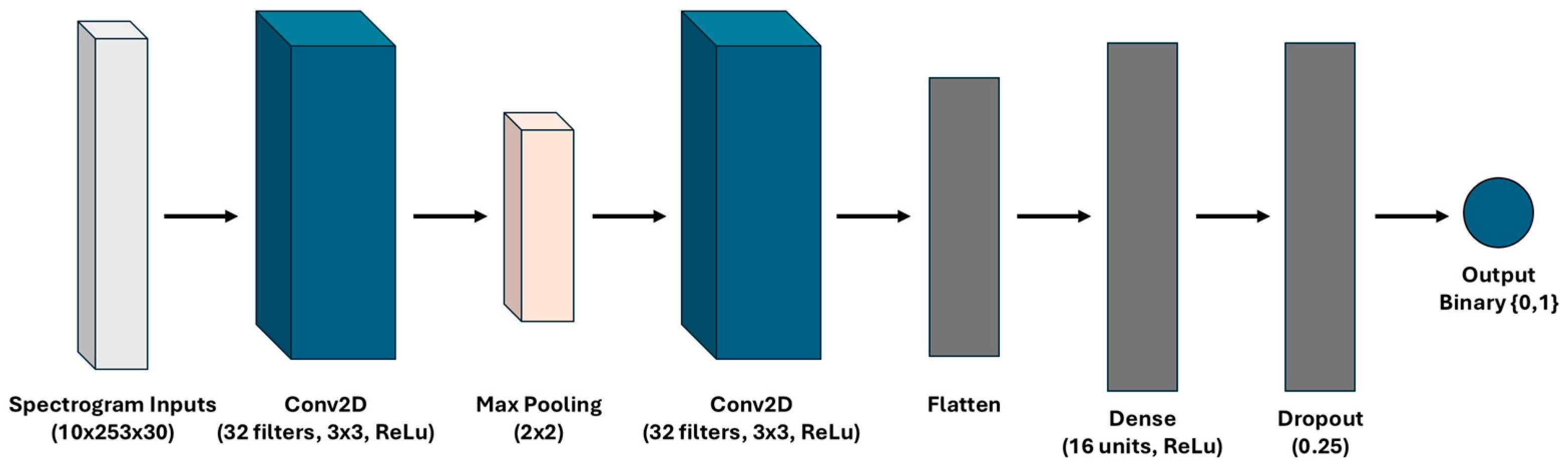
2.8. Time–Frequency Approach: CNN Model
2.9. Thresholding Analysis and Training Set Size Reduction
3. Results
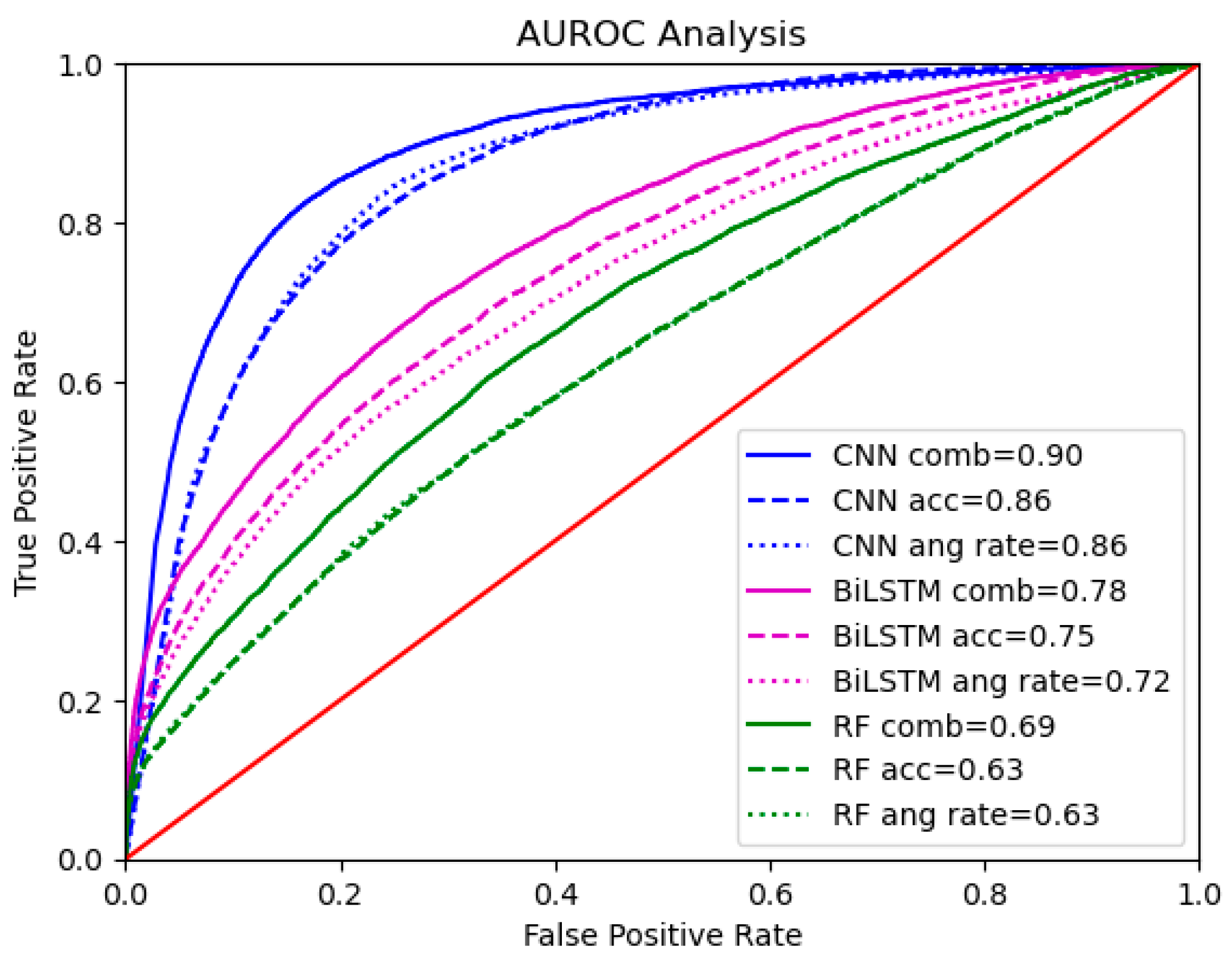
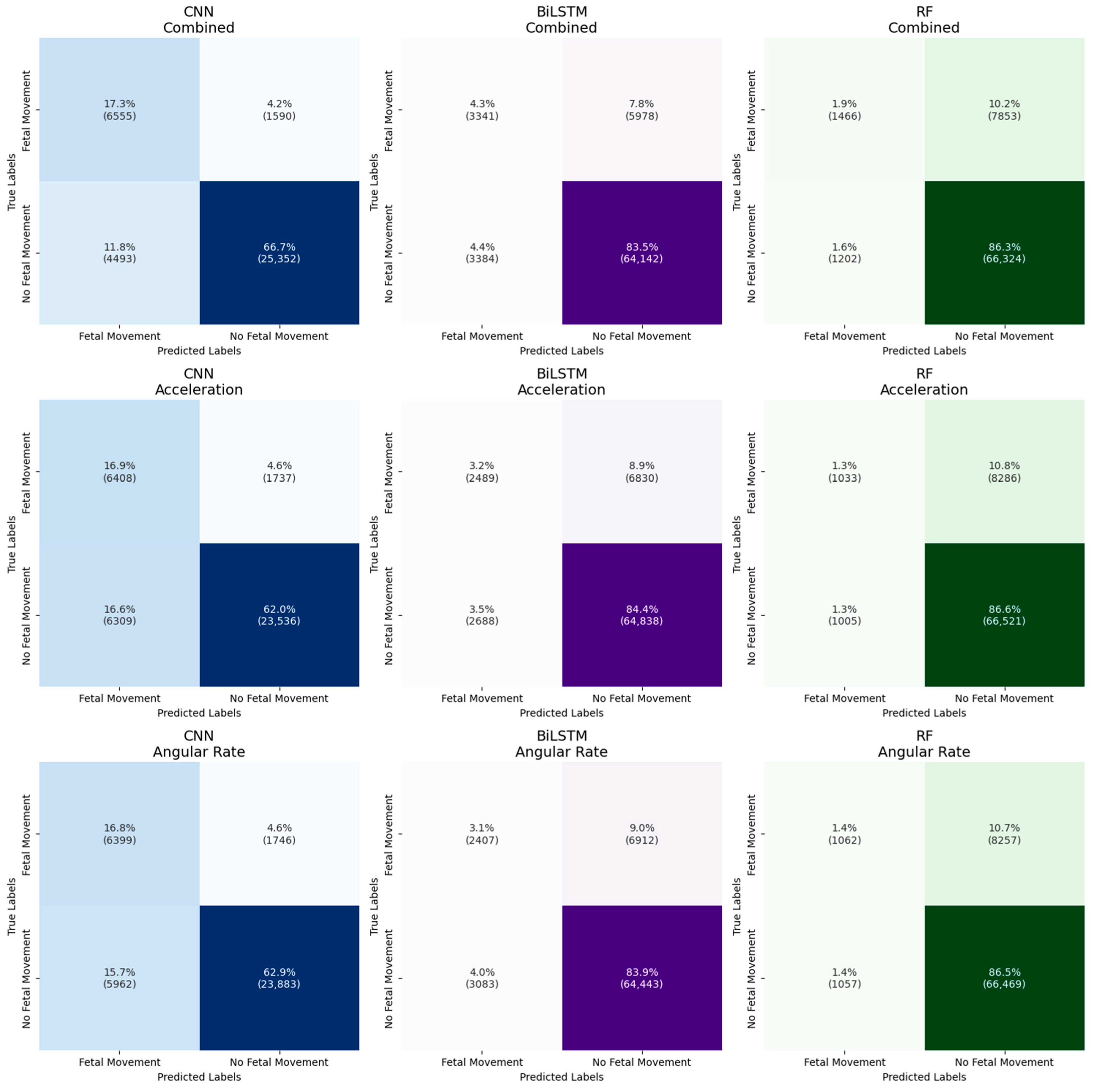
3.1. Summary of the Results
3.2. Model Performance Across Sensor Types
3.3. Model Performance Across Data Representations
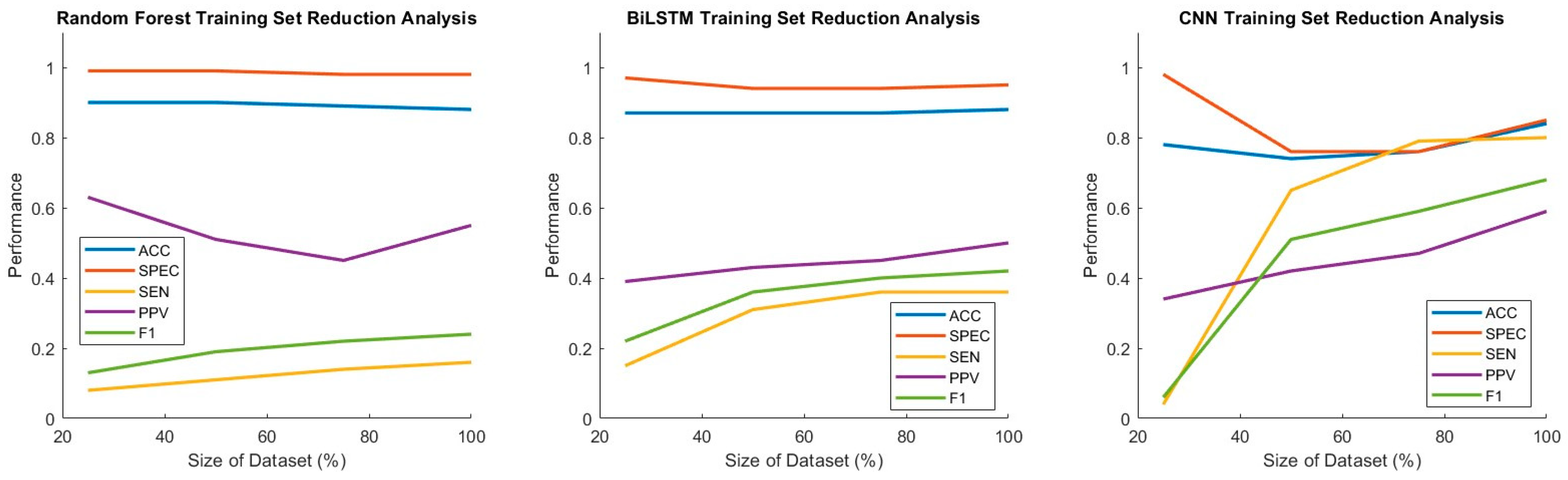
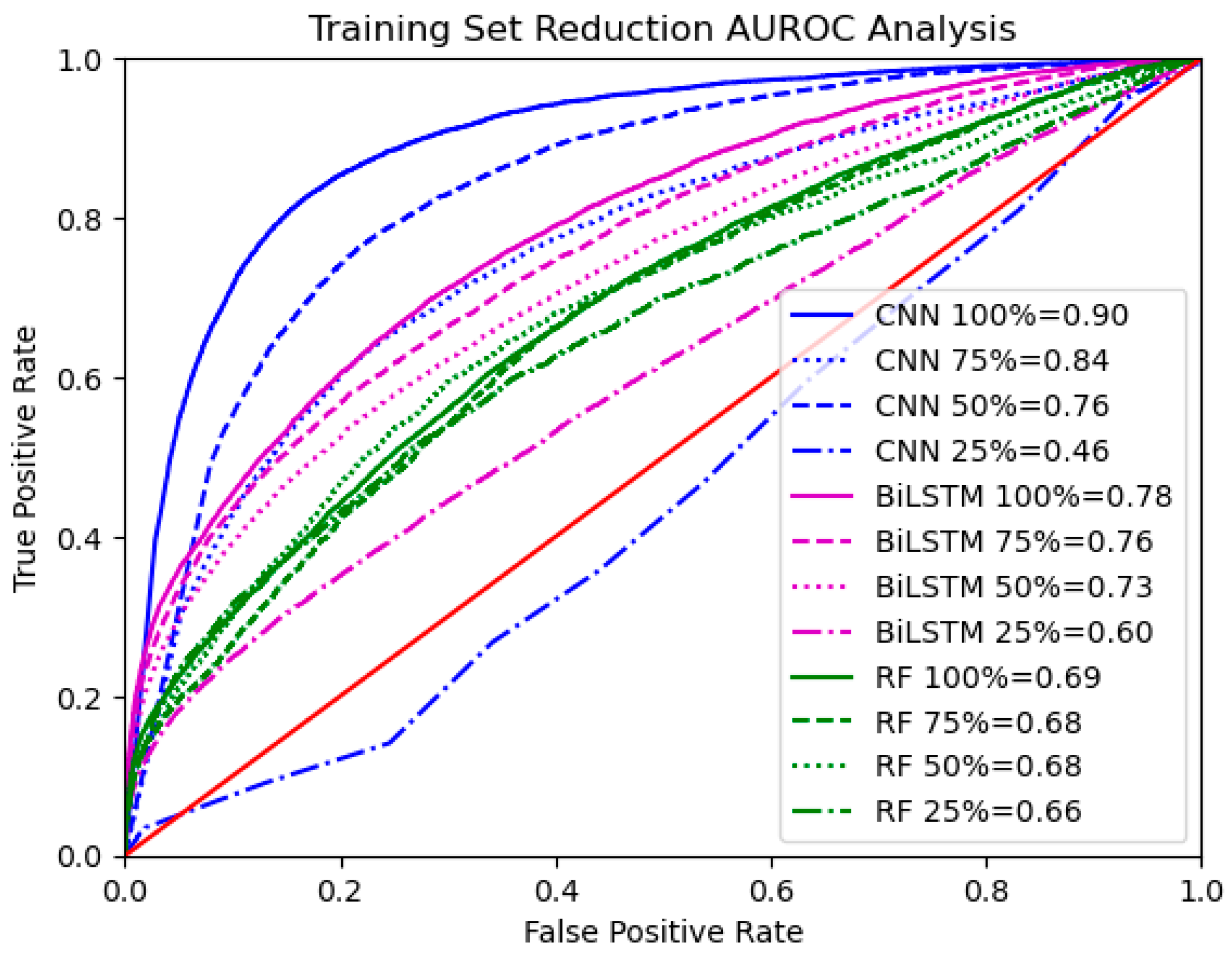
3.4. Thresholding Analysis and Training Set Size Reduction
4. Discussion
4.1. Model Performance Across Sensor Types
4.2. Model Performance Across Data Representations
4.3. Implications for Fetal Movement Detection
4.4. Limitations and Future Work
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Appendix A.1
| Participant Identifier | GA (in Weeks) | Parity | BMI | Time of Day of Data Collection |
|---|---|---|---|---|
| S01 | 32 | Parity ≥ 1 | 37.2 | Evening |
| S02 | 28 | Nulliparous | 38.7 | Morning |
| S03 | 32 | Nulliparous | 31.7 | Afternoon |
| S04 | 32 | Parity ≥ 1 | 36.4 | Evening |
| S05 | 29 | Nulliparous | 28.2 | Morning |
| S06 | 31 | Nulliparous | 28.8 | Afternoon |
| S07 | 26 | Parity ≥ 1 | 36.3 | Afternoon |
| S08 | 26 | Nulliparous | 25.7 | Evening |
| S10 | 25 | Parity ≥ 1 | 27.7 | Morning |
| S11 | 31 | Nulliparous | 25.0 | Evening |
| S12 | 28 | Nulliparous | 23.5 | Evening |
| S13 | 24 | Not reported | 27.1 | Evening |
| S14 | 26 | Parity ≥ 1 | 35.0 | Evening |
| S15 | 28 | Nulliparous | 25.0 | Evening |
| S17 | 32 | Nulliparous | 39.8 | Evening |
| S18 | 27 | Not reported | 30.3 | Evening |
| S19 | 25 | Parity ≥ 1 | 27.1 | Evening |
| S20 | 25 | Parity ≥ 1 | 30.5 | Evening |
| S21 | 30 | Parity ≥ 1 | 35.9 | Evening |
| S22 | 24 | Parity ≥ 1 | 24.8 | Evening |
| S23 | 27 | Nulliparous | 25.8 | Morning |
References
- Walton, J.R.; Peaceman, A.M. Identification, Assessment and Management of Fetal Compromise. Clin. Perinatol. 2012, 39, 753–768. [Google Scholar] [CrossRef] [PubMed]
- Frøen, J.F.; Frøen, J.F. A Kick from Within-Fetal Movement Counting and the Cancelled Progress in Antenatal Care Introduction and History. J. Perinat. Med. 2004, 32, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Dutton, P.J.; Warrander, L.K.; Roberts, S.A.; Bernatavicius, G.; Byrd, L.M.; Gaze, D.; Kroll, J.; Jones, R.L.; Sibley, C.P.; Frøen, J.F.; et al. Predictors of Poor Perinatal Outcome Following Maternal Perception of Reduced Fetal Movements—A Prospective Cohort Study. PLoS ONE 2012, 7, e39784. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.; Johnstone, E.D.; Heazell, A.E.P. Professionals’ Views of Fetal-Monitoring Support the Development of Devices to Provide Objective Longer-Term Assessment of Fetal Wellbeing. J. Matern.-Fetal Neonatal Med. 2016, 29, 1680–1686. [Google Scholar] [CrossRef]
- Minors, D.S.; Waterhouse, J.M. The Effect of Maternal Posture, Meals and Time of Day on Fetal Movements. BJOG Int. J. Obstet. Gynaecol. 1979, 86, 717–723. [Google Scholar] [CrossRef]
- Hijazi, Z.R.; Callan, S.E.; East, C.E. Maternal Perception of Foetal Movement Compared with Movement Detected by Real-Time Ultrasound: An Exploratory Study. Aust. N. Z. J. Obstet. Gynaecol. 2010, 50, 144–147. [Google Scholar] [CrossRef]
- Heazell, A.E.P.; Frøen, J.F. Methods of Fetal Movement Counting and the Detection of Fetal Compromise. J. Obstet. Gynaecol. 2008, 28, 147–154. [Google Scholar] [CrossRef]
- Heazell, A.E.P.; Sumathi, G.M.; Bhatti, N.R. What Investigation Is Appropriate Following Maternal Perception of Reduced Fetal Movements? J. Obstet. Gynaecol. 2005, 25, 648–650. [Google Scholar] [CrossRef]
- Mesbah, M.; Khlif, M.S.; Layeghy, S.; East, C.E.; Dong, S.; Brodtmann, A.; Colditz, P.B.; Boashash, B. Automatic Fetal Movement Recognition from Multi-Channel Accelerometry Data. Comput. Methods Programs Biomed. 2021, 210, 106377. [Google Scholar] [CrossRef]
- Layeghy, S.; Azemi, G.; Colditz, P.; Boashash, B. Non-Invasive Monitoring of Fetal Movements Using Time-Frequency Features of Accelerometry. In Proceedings of the 2014 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Florence, Italy, 4 May 2014; IEEE: New York, NY, USA, 2014; pp. 4379–4383. [Google Scholar]
- Liang, S.; Peng, J.; Xu, Y. Passive Fetal Movement Signal Detection System Based on Intelligent Sensing Technology. J. Healthc. Eng. 2021, 2021, 1745292. [Google Scholar] [CrossRef]
- Zhao, X.; Zeng, X.; Koehl, L.; Tartare, G.; De Jonckheere, J. A Wearable System for In-Home and Long-Term Assessment of Fetal Movement. IRBM 2020, 41, 205–211. [Google Scholar] [CrossRef]
- Khlif, M.S.; Boashash, B.; Layeghy, S.; Ben-Jabeur, T.; Mesbah, M.; East, C.; Colditz, P. Time-Frequency Characterization of Tri-Axial Accelerometer Data for Fetal Movement Detection. In Proceedings of the 2011 IEEE International Symposium on Signal Processing and Information Technology (ISSPIT), Bilbao, Spain, 14 December 2011; IEEE: New York, NY, USA, 2011; pp. 466–471. [Google Scholar] [CrossRef]
- Delay, U.; Nawarathne, T.; Dissanayake, S.; Gunarathne, S.; Withanage, T.; Godaliyadda, R.; Rathnayake, C.; Ekanayake, P.; Wijayakulasooriya, J. Novel Non-Invasive in-House Fabricated Wearable System with a Hybrid Algorithm for Fetal Movement Recognition. PLoS ONE 2021, 16, e0254560. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhao, C.; Ding, B.; Gu, X.; Zeng, W.; Qiu, L.; Yu, H.; Shen, Y.; Liu, H. Fetal Movement Detection by Wearable Accelerometer Duo Based on Machine Learning. IEEE Sens. J. 2022, 22, 11526–11534. [Google Scholar] [CrossRef]
- Altini, M.; Mullan, P.; Rooijakkers, M.; Gradl, S.; Penders, J.; Geusens, N.; Grieten, L.; Eskofier, B. Detection of Fetal Kicks Using Body-Worn Accelerometers During Pregnancy: Trade-Offs Between Sensors Number and Positioning. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16 August 2016; IEEE: New York, NY, USA, 2016; pp. 5319–5322. [Google Scholar]
- Nemati, E.; Zhang, S.; Ahmed, T.; Rahman, M.M.; Kuang, J.; Gao, A. CoughBuddy: Multi-Modal Cough Event Detection Using Earbuds Platform. In Proceedings of the 2021 IEEE 17th International Conference on Wearable and Implantable Body Sensor Networks (BSN), Virtual, 27 July 2021; IEEE: New York, NY, USA, 2021; pp. 1–4. [Google Scholar] [CrossRef]
- Lotfi, R.; Eskicioglu, R.; Tzanetakis, G.; Irani, P. A Comparison between Audio and IMU Data to Detect Chewing Events Based on an Earable Device. In Proceedings of the 11th Augmented Human International Conference, Winnipeg, MB, Canada, 27 May 2020; pp. 1–8. [Google Scholar]
- Gomes, D.; Sousa, I. Real-Time Drink Trigger Detection in Free-Living Conditions Using Intertial Sensors. Sensors 2019, 19, 2145. [Google Scholar] [CrossRef]
- Senyurek, V.Y.; Imtiaz, M.H.; Belsare, P.; Tiffany, S.; Sazonov, E. A CNN-LSTM Neural Network for Recognition of Puffing in Smoking Episodes Using Wearable Sensors. Biomed. Eng. Lett. 2020, 10, 195–203. [Google Scholar] [CrossRef]
- Luinge, H.J.; Veltink, P.H. Measuring Orientation of Human Body Segments Using Miniature Gyroscopes and Accelerometers. Med. Biol. Eng. Comput. 2005, 43, 273–282. [Google Scholar] [CrossRef]
- Varkey, J.P.; Pompili, D.; Walls, T.A. Human Motion Recognition Using a Wireless Sensor-Based Wearable System. Pers. Ubiquitous Comput. 2012, 16, 897–910. [Google Scholar] [CrossRef]
- Kot, A.; Nawrocka, A. Modeling of Human Balance as an Inverted Pendulum. In Proceedings of the 2014 15th International Carpathian Control Conference (ICCC), Velke Karlovice, Czech Republic, 28–30 May 2014; IEEE: New York, NY, USA, 2014; pp. 254–257. [Google Scholar] [CrossRef]
- Noamani, A.; Vette, A.H.; Rouhani, H. Nonlinear Response of Human Trunk Musculature Explains Neuromuscular Stabilization Mechanisms in Sitting Posture. J. Neural Eng. 2022, 19, 026045. [Google Scholar] [CrossRef]
- Alwis, P.; Thilakasiri, I.; Liyanage, S.; Nanayakkara, R.; Godaliyadda, R.; Ekanayake, M.; Rathnayake, C.; Wijayakulasooriya, J.; Herath, V. Application of an LSTM-Based Channel Attention and Classification Mechanism in Fetal Movement Monitoring. TechRxiv 2024. [Google Scholar] [CrossRef]
- Vitali, R.V.; Perkins, N.C. Determining Anatomical Frames via Inertial Motion Capture: A Survey of Methods. J. Biomech. 2020, 106, 109832. [Google Scholar] [CrossRef]
- Bradford, B.F.; Cronin, R.S.; McCowan, L.M.E.; McKinlay, C.J.D.; Mitchell, E.A.; Thompson, J.M.D. Association between Maternally Perceived Quality and Pattern of Fetal Movements and Late Stillbirth. Sci. Rep. 2019, 9, 9815. [Google Scholar] [CrossRef] [PubMed]
- Bradford, B.F.; Cronin, R.S.; McKinlay, C.J.D.; Thompson, J.M.D.; Mitchell, E.A.; Stone, P.R.; McCowan, L.M.E. A Diurnal Fetal Movement Pattern: Findings from a Cross-Sectional Study of Maternally Perceived Fetal Movements in the Third Trimester of Pregnancy. PLoS ONE 2019, 14, e0217583. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.; Woodward, R.; Alexandrov, Y.; ain Munnee, Q.; Lees, C.C.; Vaidyanathan, R.; Nowlan, N.C. Performance of a Wearable Acoustic System for Fetal Movement Discrimination. PLoS ONE 2018, 13, e0195728. [Google Scholar] [CrossRef] [PubMed]
- Spicher, L.; Bell, C.; Huan, X.; Sienko, K. Exploring Random Forest Machine Learning for Fetal Movement Detection Using Abdominal Acceleration and Angular Rate Data. In Proceedings of the 2024 46th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 15 July 2024; IEEE: New York, NY, USA, 2024; pp. 1–4. [Google Scholar] [CrossRef]
- Shan, G. Monte Carlo Cross-Validation for a Study with Binary Outcome and Limited Sample Size. BMC Med. Inform. Decis. Mak. 2022, 22, 270. [Google Scholar] [CrossRef]
- Fadili, Y.; El Yamani, Y.; Kilani, J.; El Kamoun, N.; Baddi, Y.; Bensalah, F. An Enhancing Timeseries Anomaly Detection Using LSTM and Bi-LSTM Architectures. In Proceedings of the 2024 11th International Conference on Wireless Networks and Mobile Communications (WINCOM), Leeds, UK, 23 July 2024; IEEE: New York, NY, USA, 2024; pp. 1–6. [Google Scholar]
- Graves, A.; Fernández, S.; Schmidhuber, J. Bidirectional LSTM Networks for Improved Phoneme Classification and Recognition. In Proceedings of the International Conference on Artificial Neural Networks (ICANN), Warsaw, Poland, 11 September 2005; Springer: Berlin/Heidelberg, Germany, 2005; pp. 799–804. [Google Scholar]
- Addison, P.; Walker, J.; Guido, R. Time—Frequency Analysis of Biosignals. In IEEE Engineering in Medicine and Biology Magazine; IEEE: New York, NY, USA, 2009; Volume 28, pp. 14–29. [Google Scholar]
- David, M.; Hirsch, M.; Karin, J.; Toledo, E.; Akselrod, S.; An, A.S. An Estimate of Fetal Autonomic State by Time-Frequency Analysis of Fetal Heart Rate Variability. J. Appl. Physiol. 2007, 102, 1057–1064. [Google Scholar] [CrossRef]
- Chauhan, R.; Ghanshala, K.K.; Joshi, R.C. Convolutional Neural Network (CNN) for Image Detection and Recognition. In Proceedings of the 2018 First International Conference on Secure Cyber Computing and Communication (ICSCCC), Jalandhar, India, 15 December 2018; IEEE: New York, NY, USA, 2018; pp. 278–282. [Google Scholar]
- Šimundić, A.-M. Measures of Diagnostic Accuracy: Basic Definitions. eJIFCC 2009, 19, 203. [Google Scholar]
- Rudin, C. Stop Explaining Black Box Machine Learning Models for High Stakes Decisions and Use Interpretable Models Instead. Nat. Mach. Intell. 2019, 1, 206–215. [Google Scholar] [CrossRef]
- Tonekaboni, S.; Joshi, S.; Mccradden, M.D.; Goldenberg, A. What Clinicians Want: Contextualizing Explainable Machine Learning for Clinical End Use. In Proceedings of the Machine Learning for Healthcare Conference, Ann Arbor, MI, USA, 9–10 August 2019; pp. 359–380. [Google Scholar]
- Stiglic, G.; Kocbek, P.; Fijacko, N.; Zitnik, M.; Verbert, K.; Cilar, L. Interpretability of Machine Learning-Based Prediction Models in Healthcare. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2020, 10, e1379. [Google Scholar] [CrossRef]
- Nazir, Z.; Zarymkanov, T.; Park, J.-G. A Machine Learning Model Selection Considering Tradeoffs between Accuracy and Interpretability. In Proceedings of the 2021 13th International Conference on Information Technology and Electrical Engineering (ICITEE), Virtual, 14 October 2021; IEEE: New York, NY, USA, 2021; pp. 63–68. [Google Scholar]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of Deep Learning: Concepts, CNN Architectures, Challenges, Applications, Future Directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef]
- Rodriguez-Villegas, E.; Iranmanesh, S.; Imtiaz, S.A. Wearable Medical Devices: High-Level System Design Considerations and Tradeoffs. IEEE Solid-State Circuits Mag. 2018, 10, 43–52. [Google Scholar] [CrossRef]
- Peahl, A.F.; Gourevitch, R.A.; Luo, E.M.; Fryer, K.E.; Moniz, M.H.; Dalton, V.K.; Fendrick, A.M.; Shah, N. Right-Sizing Prenatal Care to Meet Patients’ Needs and Improve Maternity Care Value. Obstet. Gynecol. 2020, 135, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Carter, E.B.; Tuuli, M.G.; Caughey, A.B.; Odibo, A.O.; Macones, G.A.; Cahill, A.G. Number of Prenatal Visits and Pregnancy Outcomes in Low-Risk Women. J. Perinatol. 2016, 36, 178–181. [Google Scholar] [CrossRef] [PubMed]
- Besinger, R.E.; Johnson, T.R.B. Doppler Recordings of Fetal Movement: Clinical Correlation with Real-Time Ultrasound. Obstet. Gynecol. 1989, 74, 277–280. [Google Scholar]
- Raynes-Greenow, C.H.; Gordon, A.; Li, Q.; Hyett, J.A. A Cross-Sectional Study of Maternal Perception of Fetal Movements and Antenatal Advice in a General Pregnant Population, Using a Qualitative Framework. BMC Pregnancy Childbirth 2013, 13, 32. [Google Scholar] [CrossRef]
- Mangesi, L.; Hofmeyr, G.J.; Smith, V.; Smyth, R.M.D. Fetal Movement Counting for Assessment of Fetal Wellbeing. Cochrane Database Syst. Rev. 2015, 10, CD004909. [Google Scholar] [CrossRef]
- Ozkaya, E.; Baser, E.; Cinar, M.; Korkmaz, V.; Kucukozkan, T. Does Diurnal Rhythm Have an Impact on Fetal Biophysical Profile? J. Matern.-Fetal Neonatal Med. 2012, 25, 335–338. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spicher, L.; Bell, C.; Sienko, K.H.; Huan, X. Comparative Analysis of Machine Learning Approaches for Fetal Movement Detection with Linear Acceleration and Angular Rate Signals. Sensors 2025, 25, 2944. https://doi.org/10.3390/s25092944
Spicher L, Bell C, Sienko KH, Huan X. Comparative Analysis of Machine Learning Approaches for Fetal Movement Detection with Linear Acceleration and Angular Rate Signals. Sensors. 2025; 25(9):2944. https://doi.org/10.3390/s25092944
Chicago/Turabian StyleSpicher, Lucy, Carrie Bell, Kathleen H. Sienko, and Xun Huan. 2025. "Comparative Analysis of Machine Learning Approaches for Fetal Movement Detection with Linear Acceleration and Angular Rate Signals" Sensors 25, no. 9: 2944. https://doi.org/10.3390/s25092944
APA StyleSpicher, L., Bell, C., Sienko, K. H., & Huan, X. (2025). Comparative Analysis of Machine Learning Approaches for Fetal Movement Detection with Linear Acceleration and Angular Rate Signals. Sensors, 25(9), 2944. https://doi.org/10.3390/s25092944








