A Multidimensional Particle Swarm Optimization-Based Algorithm for Brain MRI Tumor Segmentation
Abstract
1. Introduction
2. Key Contributions
- We propose a multidimensional particle swarm optimization (MDPSO)-based clustering method that dynamically adapts the number of clusters, ensuring generalizability across heterogeneous and non-synthetic MRI data;
- A fitness function has been developed, integrating both pixel intensity and Euclidean distance measures to enhance segmentation accuracy;
- Incorporating features derived from MDPSO-based unsupervised clustering into a supervised learning framework using an RF classifier yields improved performance compared to traditional supervised approaches under equivalent training conditions;
- The proposed methodology bridges the gap between fully supervised and unsupervised methods, offering a more interpretable and resource-efficient alternative for medical image segmentation.
3. Methodology
| Algorithm 1: MDPSO Algorithm |
 |
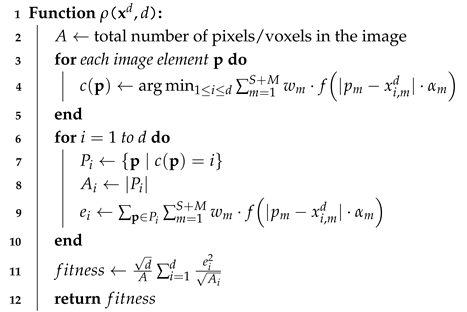
| Algorithm 2: Fitness Function Computation for 2D/3D Images |
 |
3.1. The PSO and MDPSO Algorithms
- N denotes the number of iterations performed during the search process;
- stores the index of the particle that achieves the best result;
- S is the total number of particles in the swarm, and k is the index of the current particle;
- is a vector-valued real loss function that optimizes the solution to the given problem while considering the dimensionality of the search space (see, e.g., Algorithm 2).
3.2. The Proposed Algorithm for 2D and 3D Image Clustering
3.3. Complexity Analysis
3.4. Metrics
- is the set of predicted clusters labeled as lesions;
- denotes a predicted cluster classified as a lesion ();
- is the set of ground truth lesion regions;
- represents the i-th lesion region in the ground truth ().
4. Experiments and Discussion
4.1. Dataset
4.2. Parameter Settings
4.2.1. Clustering Parameters
4.2.2. Random-Forest Feature Extraction and Training
4.3. Results
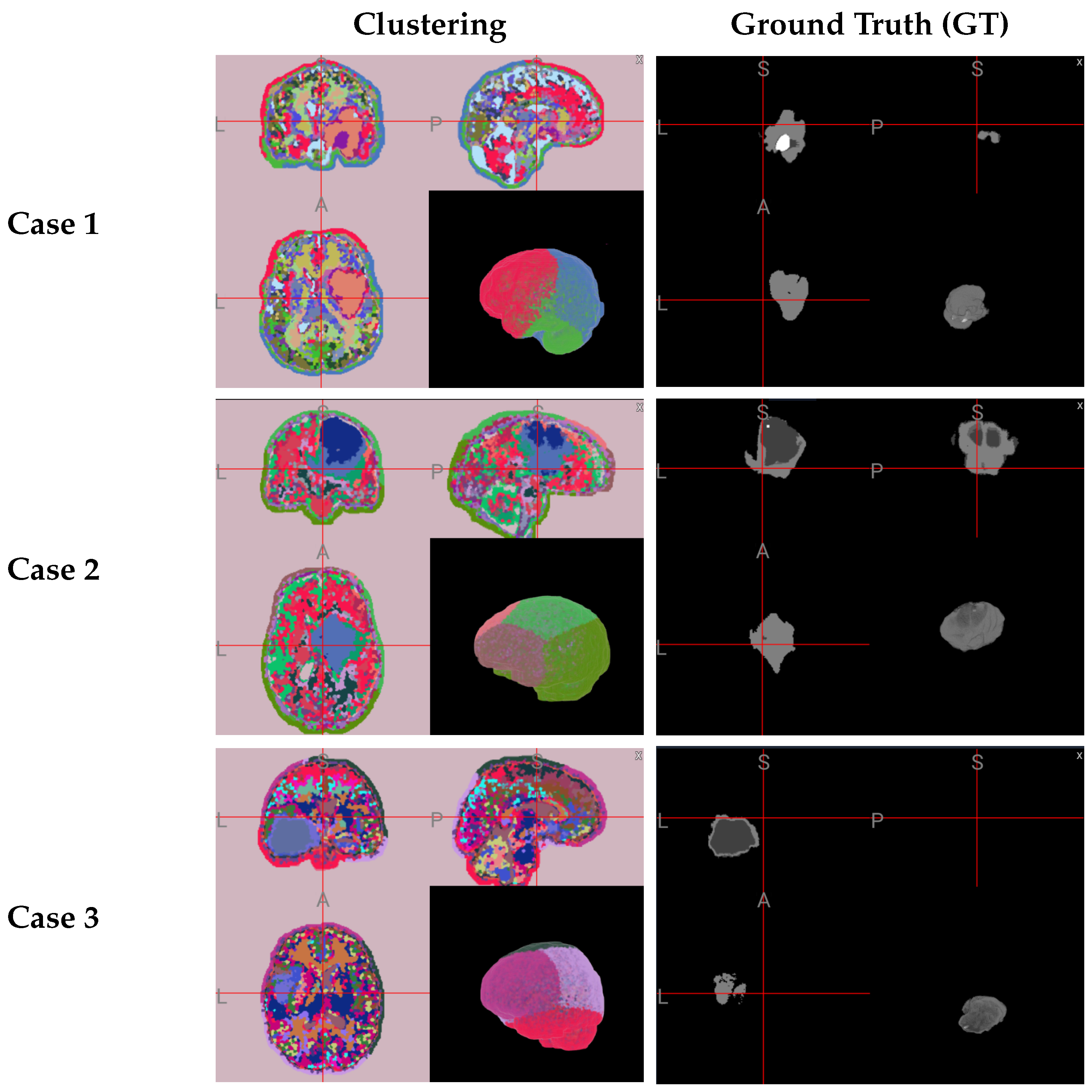
4.3.1. Qualitative Results
4.3.2. Quantitative Results
4.4. Comparison with Other Approaches
5. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dougherty, G. Digital Image Processing for Medical Applications; University Press: Cambridge, UK, 2009. [Google Scholar]
- Westbrook, C.; Talbot, J. MRI in Practice; John Wiley & Sons: Hoboken, NJ, USA, 2018. [Google Scholar]
- Ranjbarzadeh, R.; Caputo, A.; Tirkolaee, E.B.; Ghoushchi, S.J.; Bendechache, M. Brain tumor segmentation of MRI images: A comprehensive review on the application of artificial intelligence tools. Comput. Biol. Med. 2023, 152, 106405. [Google Scholar] [CrossRef] [PubMed]
- McGrath, H.; Li, P.; Dorent, R.; Bradford, R.; Saeed, S.; Bisdas, S.; Ourselin, S.; Shapey, J.; Vercauteren, T. Manual segmentation versus semi-automated segmentation for quantifying vestibular schwannoma volume on MRI. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 1445–1455. [Google Scholar] [CrossRef] [PubMed]
- Veiga-Canuto, D.; Cerdà-Alberich, L.; Sangüesa Nebot, C.; Martínez de las Heras, B.; Pötschger, U.; Gabelloni, M.; Carot Sierra, J.M.; Taschner-Mandl, S.; Düster, V.; Cañete, A.; et al. Comparative multicentric evaluation of inter-observer variability in manual and automatic segmentation of neuroblastic tumors in magnetic resonance images. Cancers 2022, 14, 3648. [Google Scholar] [CrossRef] [PubMed]
- Kelly, P.J. Gliomas: Survival, origin and early detection. Surg. Neurol. Int. 2010, 1, 96. [Google Scholar] [CrossRef]
- Stieb, S.; McDonald, B.; Gronberg, M.; Engeseth, G.M.; He, R.; Fuller, C.D. Imaging for target delineation and treatment planning in radiation oncology: Current and emerging techniques. Hematol. Clin. N. Am. 2019, 33, 963–975. [Google Scholar] [CrossRef]
- Peltonen, J.I.; Mäkelä, T.; Lehmonen, L.; Sofiev, A.; Salli, E. Inter-and intra-scanner variations in four magnetic resonance imaging image quality parameters. J. Med. Imaging 2020, 7, 065501. [Google Scholar] [CrossRef] [PubMed]
- Gyorfi, Á.; Szilágyi, L.; Kovács, L. A fully automatic procedure for brain tumor segmentation from multi-spectral MRI records using ensemble learning and atlas-based data enhancement. Appl. Sci. 2021, 11, 564. [Google Scholar] [CrossRef]
- Luu, H.M.; Park, S.H. Extending nn-UNet for Brain Tumor Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Springer: Cham, Switzerland, 2022; pp. 173–186. [Google Scholar]
- Bareja, R.; Ismail, M.; Martin, D.; Nayate, A.; Yadav, I.; Labbad, M.; Dullur, P.; Garg, S.; Tamrazi, B.; Salloum, R.; et al. nnU-Net–based Segmentation of Tumor Subcompartments in Pediatric Medulloblastoma Using Multiparametric MRI: A Multi-institutional Study. Radiol. Artif. Intell. 2024, 6, e230115. [Google Scholar] [CrossRef]
- Ranjbarzadeh, R.; Bagherian Kasgari, A.; Jafarzadeh Ghoushchi, S.; Anari, S.; Naseri, M.; Bendechache, M. Brain tumor segmentation based on deep learning and an attention mechanism using MRI multi-modalities brain images. Sci. Rep. 2021, 11, 10930. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, P.K.; Soltani, S.; Wong, A.K. A survey of thresholding techniques. Comput. Vision Graph. Image Process. 1988, 41, 233–260. [Google Scholar] [CrossRef]
- Zhang, H.; Fritts, J.E.; Goldman, S.A. Image segmentation evaluation: A survey of unsupervised methods. Comput. Vis. Image Underst. 2008, 110, 260–280. [Google Scholar] [CrossRef]
- Oliva, D.; Abd Elaziz, M.; Hinojosa, S.; Oliva, D.; Abd Elaziz, M.; Hinojosa, S. Multilevel thresholding for image segmentation based on metaheuristic algorithms. In Metaheuristic Algorithms for Image Segmentation: Theory and Applications; Springer: Cham, Switzerland, 2019; pp. 59–69. [Google Scholar]
- Xu, M.; Cao, L.; Lu, D.; Hu, Z.; Yue, Y. Application of swarm intelligence optimization algorithms in image processing: A comprehensive review of analysis, synthesis, and optimization. Biomimetics 2023, 8, 235. [Google Scholar] [CrossRef] [PubMed]
- El Dor, A.; Lepagnot, J.; Nakib, A.; Siarry, P. PSO-2S optimization algorithm for brain MRI segmentation. In Genetic and Evolutionary Computing, Proceedings of the Seventh International Conference on Genetic and Evolutionary Computing, ICGEC 2013, Prague, Czech Republic, 25–27 August 2013; Springer: Cham, Switzerland, 2014; pp. 13–22. [Google Scholar]
- Krishna, K.; Murty, M.N. Genetic K-means algorithm. IEEE Trans. Syst. Man Cybern. Part B (Cybern.) 1999, 29, 433–439. [Google Scholar] [CrossRef]
- Cheng, Y. Mean shift, mode seeking, and clustering. IEEE Trans. Pattern Anal. Mach. Intell. 1995, 17, 790–799. [Google Scholar] [CrossRef]
- Cheng, B.; Schwing, A.; Kirillov, A. Per-pixel classification is not all you need for semantic segmentation. Adv. Neural Inf. Process. Syst. 2021, 34, 17864–17875. [Google Scholar]
- Kennedy, J.; Eberhart, R. Particle swarm optimization. In Proceedings of the ICNN’95—International Conference on Neural Networks, Perth, WA, Australia, 27 November–1 December 1995; Volume 4, pp. 1942–1948. [Google Scholar]
- Kiranyaz, S.; Pulkkinen, J.; Gabbouj, M. Multi-dimensional particle swarm optimization in dynamic environments. Expert Syst. Appl. 2011, 38, 2212–2223. [Google Scholar] [CrossRef]
- Acharya, U.K.; Kumar, S. Particle swarm optimized texture based histogram equalization (PSOTHE) for MRI brain image enhancement. Optik 2020, 224, 165760. [Google Scholar] [CrossRef]
- Kiranyaz, S.; Ince, T.; Gabbouj, M. Multidimensional Particle Swarm Optimization for Machine Learning and Pattern Recognition; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar]
- Roy, S.; Mazumdar, N.; Pamula, R. An energy optimized and QoS concerned data gathering protocol for wireless sensor network using variable dimensional PSO. Ad Hoc Netw. 2021, 123, 102669. [Google Scholar] [CrossRef]
- Kovács, P.; Fridli, S.; Schipp, F. Generalized rational variable projection with application in ECG compression. IEEE Trans. Signal Process. 2019, 68, 478–492. [Google Scholar] [CrossRef]
- Kovács, P.; Kiranyaz, S.; Gabbouj, M. Hyperbolic particle swarm optimization with application in rational identification. In Proceedings of the 21st European Signal Processing Conference (EUSIPCO 2013), Marrakech, Morocco, 9–13 September 2013; pp. 1–5. [Google Scholar]
- Shi, Y.; Eberhart, R. A modified particle swarm optimizer. In Proceedings of the 1998 IEEE International Conference on Evolutionary Computation Proceedings. IEEE World Congress on Computational Intelligence (Cat. No. 98TH8360), Anchorage, AK, USA, 4–9 May 1998; pp. 69–73. [Google Scholar]
- Liu, J.; Yang, Y.H. Multiresolution color image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 1994, 16, 689–700. [Google Scholar]
- Van den Bergh, M.; Boix, X.; Roig, G.; De Capitani, B.; Van Gool, L. Seeds: Superpixels extracted via energy-driven sampling. In Computer Vision—ECCV 2012: 12th European Conference on Computer Vision, Florence, Italy, 7–13 October 2012, Proceedings, Part VII; Springer: Cham, Switzerland, 2012; pp. 13–26. [Google Scholar]
- Amendola, D.; Basile, A.; Castellano, G.; Vessio, G.; Zaza, G. From Voxels to Insights: Exploring the Effectiveness and Transparency of Graph Neural Networks in Brain Tumor Segmentation. In Proceedings of the 2024 International Joint Conference on Neural Networks (IJCNN), Yokohama, Japan, 30 June–5 July 2024; pp. 1–7. [Google Scholar]
- Baid, U.; Ghodasara, S.; Mohan, S.; Bilello, M.; Calabrese, E.; Colak, E.; Farahani, K.; Kalpathy-Cramer, J.; Kitamura, F.C.; Pati, S.; et al. The rsna-asnr-miccai brats 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv 2021, arXiv:2107.02314. [Google Scholar]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef]
- Szilágyi, L.; Iclanzan, D.; Kapás, Z.; Szabó, Z.; Gyorfi, A.; Lefkovits, L. Low and high grade glioma segmentation in multispectral brain MRI data. Acta Univ. Sapientiae Inform. 2018, 10, 110–132. [Google Scholar] [CrossRef]
- Piotrowski, A.P.; Napiorkowski, J.J.; Piotrowska, A.E. Population size in particle swarm optimization. Swarm Evol. Comput. 2020, 58, 100718. [Google Scholar] [CrossRef]
- Karun, B.; Thiyagarajan, A.; Murugan, P.R.; Jeyaprakash, N.; Ramaraj, K.; Makreri, R. Advanced Hybrid Brain Tumor Segmentation in MRI: Elephant Herding Optimization Combined with Entropy-Guided Fuzzy Clustering. Math. Comput. Appl. 2025, 30, 1. [Google Scholar] [CrossRef]
- Houssein, E.H.; Mohamed, G.M.; Djenouri, Y.; Wazery, Y.M.; Ibrahim, I.A. Nature inspired optimization algorithms for medical image segmentation: A comprehensive review. Clust. Comput. 2024, 27, 14745–14766. [Google Scholar] [CrossRef]


| Method | Statistic | Dice (%) | Precision (%) | Sensitivity (%) | Accuracy (%) | Specificity (%) | Hausdorff95 (mm) |
|---|---|---|---|---|---|---|---|
| GF+ RFC (2-D) | Mean | 81.92 | 88.89 | 77.20 | 95.02 | 99.20 | 19.34 |
| Median | 85.32 | 90.50 | 81.11 | 97.84 | 99.32 | 7.00 | |
| IQR | 22.01 | 12.37 | 26.88 | 2.39 | 0.92 | 11.01 | |
| GF+ MDPSO+ RFC (2-D) | Mean | 84.94 | 91.87 | 81.22 | 97.72 | 99.31 | 10.41 |
| Median | 87.64 | 93.81 | 85.37 | 98.16 | 99.41 | 6.21 | |
| IQR | 14.88 | 8.17 | 20.23 | 1.69 | 0.81 | 7.87 | |
| GF+ RFC (3-D) | Mean | 82.55 | 87.62 | 78.43 | 96.21 | 98.94 | 23.68 |
| Median | 84.81 | 89.05 | 80.02 | 97.02 | 99.08 | 9.55 | |
| IQR | 19.34 | 11.48 | 22.77 | 2.12 | 1.12 | 13.73 | |
| GF+ MDPSO+ RFC (3-D) | Mean | 84.37 | 90.42 | 80.51 | 97.11 | 99.12 | 15.28 |
| Median | 86.72 | 91.63 | 83.77 | 97.88 | 99.27 | 8.33 | |
| IQR | 15.21 | 9.84 | 19.59 | 1.88 | 0.97 | 9.44 |
| Method (Type) | Dataset (Train & Test Size, Dimensionality) | Dice Score (%) | Train & Inference Time | Hardware | Year |
|---|---|---|---|---|---|
| Binary Decision Trees Ensemble [9] | BraTS2019 (76 LGG, 259 HGG, 5:1 split, 3D MRI) | LGG: 84.79 HGG: 85.16 | Train: N/R Test: 58 s/vol. | CPU: Intel Core i7, 16 GB RAM | 2021 |
| nnU-Net (3D U-Net with modifications) [10] | BraTS2021 (1251/219, 3D MRI) | ET: 84.51 TC: 87.81 WT: 92.75 | N/R | GPU: NVIDIA RTX 3090(24 GB) | 2021 |
| Cascade CNN + Distance-Wise Attention [12] | BRATS2018 (75 LGG, 210 HGG, 9:1 split, 3D MRI) | WT: 92.03 ET: 91.13 TC: 87.26 | Train: 13 h, Test: 7 s/vol. | CPU: Intel Core i7, 32 GB RAM, GPU: NVIDIA GeForce GTX 1080Ti | 2021 |
| nnU-Net–based Medulloblastoma Segmentation [11] | Local data from 3 inst. (78 cases, leave-one-institution-out split strategy, 2D MRI) | 84.33 | N/R | N/R | 2024 |
| EHO-EnFCM [37] | BraTS (20 images, 3D MRI) | 80.07 | Train: N/R Test: 26.57 s/img. | CPU: Intel Core i7, 8 GB RAM | 2025 |
| MDPSO + RFC for 2D images (Ours) | BraTS2021 (235/100, 2D MRI) | 84.94 | Train: 1.5 h, Test: 22 s/img. | GPU: NVIDIA GeForce RTX 3060 (12 GB) | 2025 |
| MDPSO + RFC for 3D data (Ours) | BraTS2019 (235/100, 3D MRI) | 84.37 | Train: 3 h, Test: 37 s/img. | GPU: NVIDIA GeForce RTX 3060 (12 GB) | 2025 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boga, Z.; Sándor, C.; Kovács, P. A Multidimensional Particle Swarm Optimization-Based Algorithm for Brain MRI Tumor Segmentation. Sensors 2025, 25, 2800. https://doi.org/10.3390/s25092800
Boga Z, Sándor C, Kovács P. A Multidimensional Particle Swarm Optimization-Based Algorithm for Brain MRI Tumor Segmentation. Sensors. 2025; 25(9):2800. https://doi.org/10.3390/s25092800
Chicago/Turabian StyleBoga, Zsombor, Csanád Sándor, and Péter Kovács. 2025. "A Multidimensional Particle Swarm Optimization-Based Algorithm for Brain MRI Tumor Segmentation" Sensors 25, no. 9: 2800. https://doi.org/10.3390/s25092800
APA StyleBoga, Z., Sándor, C., & Kovács, P. (2025). A Multidimensional Particle Swarm Optimization-Based Algorithm for Brain MRI Tumor Segmentation. Sensors, 25(9), 2800. https://doi.org/10.3390/s25092800






