STCCA: Spatial–Temporal Coupled Cross-Attention Through Hierarchical Network for EEG-Based Speech Recognition
Abstract
1. Introduction
- We propose a novel CCA module, which applies attention mechanisms bidirectionally to two input features. To the best of our knowledge, this is the first CCA fusion method designed to fuse temporal and spatial features in EEG-based speech recognition.
- We propose an innovative STCCA network that hierarchically captures local and global characteristics of EEG signals. The LFEM is employed to extract local temporal and spatial features, the CCA module fuses these features by capturing their coupled interactions, and the GFEM extracts global dependencies.
- STCCA achieves accuracy improvements of 1.95%, 3.98%, and 1.98% on three EEG-based speech datasets with 22 subjects. Ablation experiments further validate the superior performance of the CCA compared to other commonly used fusion methods, and highlight the essential roles of LFEM and GFEM in enhancing overall performance.
2. Method
2.1. Problem Definition
2.2. Overview of STCCA
- (1)
- LFEM. LFEM extracts local fine-grained temporal and spatial features from EEG signals through convolutional layers along the time and channel dimensions, enabling effective representation of both temporal dynamics and spatial distributions.
- (2)
- CCA. CCA leverages a bidirectional attention mechanism to dynamically assign weights, enabling the modeling of complex interactions between temporal and spatial features. By capturing joint spatial–temporal dependencies, CCA effectively overcomes the limitations of traditional feature fusion approaches.
- (3)
- GFEM. GFEM employs multi-head self-attention layers to model long-range dependencies, thereby enhancing the representation of global spatial–temporal patterns. The final classification results are then obtained through the fully connected layers.
| Algorithm 1: Pseudo-code of STCCA |
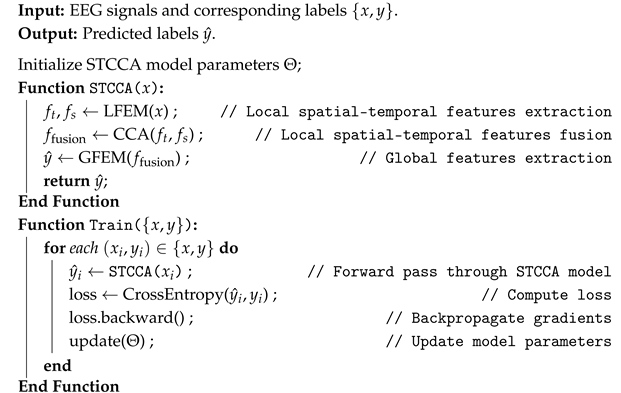 |
2.3. Local Feature Extraction Module
2.4. Coupled Cross-Attention Fusion Module
2.5. Global Feature Extraction Module
3. Experiment and Results
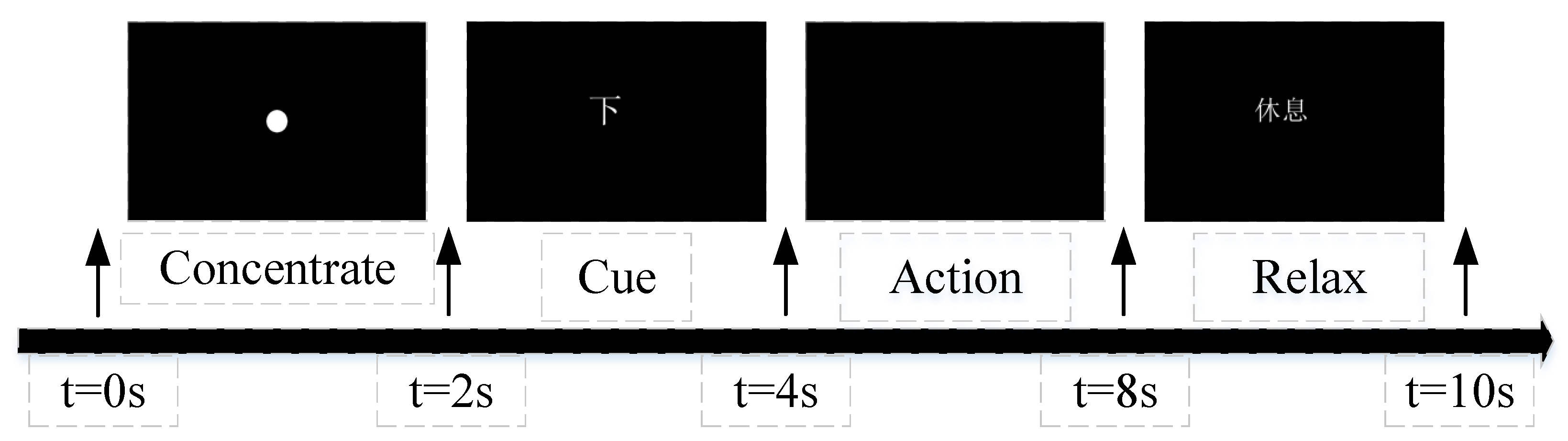
3.1. EEG Data and Preprocessing
3.2. Experimental Setting
3.3. Experimental Results and Analyses
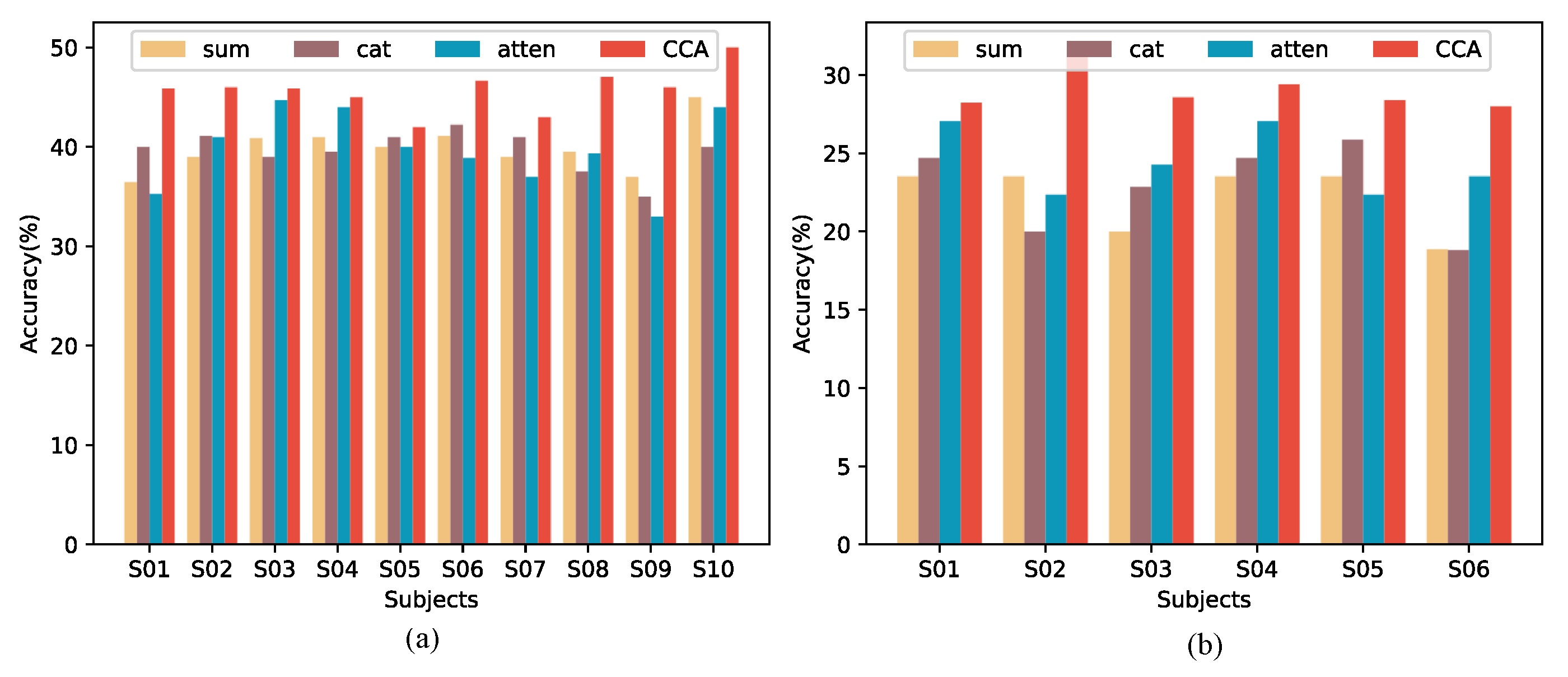
3.4. Ablation Studies
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EEG | electroencephalogram |
| BCI | brain–computer interface |
| DL | deep learning |
| CNN | convolutional neural network |
| LFEM | local feature extraction module |
| CCA | coupled cross-attention |
| GFEM | global feature extraction module |
| Conv | convolutional layers |
| AG | average pooling layers |
| BN | batch normalization layers |
| LN | layer normalization |
| MHA | multi-head self-attention |
Appendix A
Appendix A.1
| S01 | S02 | S03 | S04 | S05 | S06 | S07 | S08 | S09 | S10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Class0 | 125 | 150 | 125 | 150 | 150 | 135 | 150 | 125 | 150 | 150 |
| Class1 | 125 | 150 | 125 | 150 | 150 | 135 | 150 | 125 | 150 | 150 |
| Class2 | 125 | 150 | 125 | 150 | 150 | 135 | 150 | 125 | 150 | 150 |
| Class3 | 125 | 150 | 125 | 150 | 150 | 135 | 150 | 125 | 150 | 150 |
| S01 | S02 | S03 | S04 | S05 | S06 | |
|---|---|---|---|---|---|---|
| Class0 | 12 | 12 | 12 | 12 | 15 | 12 |
| Class1 | 12 | 12 | 12 | 12 | 15 | 12 |
| Class2 | 12 | 12 | 12 | 12 | 15 | 12 |
| Class3 | 12 | 12 | 12 | 12 | 15 | 12 |
| Class4 | 12 | 12 | 12 | 12 | 15 | 12 |
| Class5 | 12 | 12 | 12 | 12 | 15 | 12 |
| Class6 | 12 | 12 | 12 | 12 | 15 | 12 |
| Class7 | 12 | 12 | 12 | 12 | 15 | 12 |
| Class8 | 12 | 12 | 12 | 12 | 15 | 12 |
| Class9 | 12 | 12 | 12 | 12 | 15 | 12 |
| Class10 | 12 | 12 | 12 | 12 | 15 | 12 |
| S01 | S02 | S03 | S04 | S05 | Sub6 | |
|---|---|---|---|---|---|---|
| Class0 | 50 | 40 | 50 | 40 | 50 | 40 |
| Class1 | 50 | 40 | 50 | 40 | 50 | 40 |
| Class2 | 50 | 40 | 50 | 40 | 50 | 40 |
| Class3 | 50 | 40 | 50 | 40 | 50 | 40 |
| Class4 | 50 | 40 | 50 | 40 | 50 | 40 |
| Class5 | 50 | 40 | 50 | 40 | 50 | 40 |
| Class6 | 50 | 40 | 50 | 40 | 50 | 40 |
| Class7 | 50 | 40 | 50 | 40 | 50 | 40 |
| Class8 | 50 | 40 | 50 | 40 | 50 | 40 |
| Class9 | 50 | 40 | 50 | 40 | 50 | 40 |
Appendix A.2
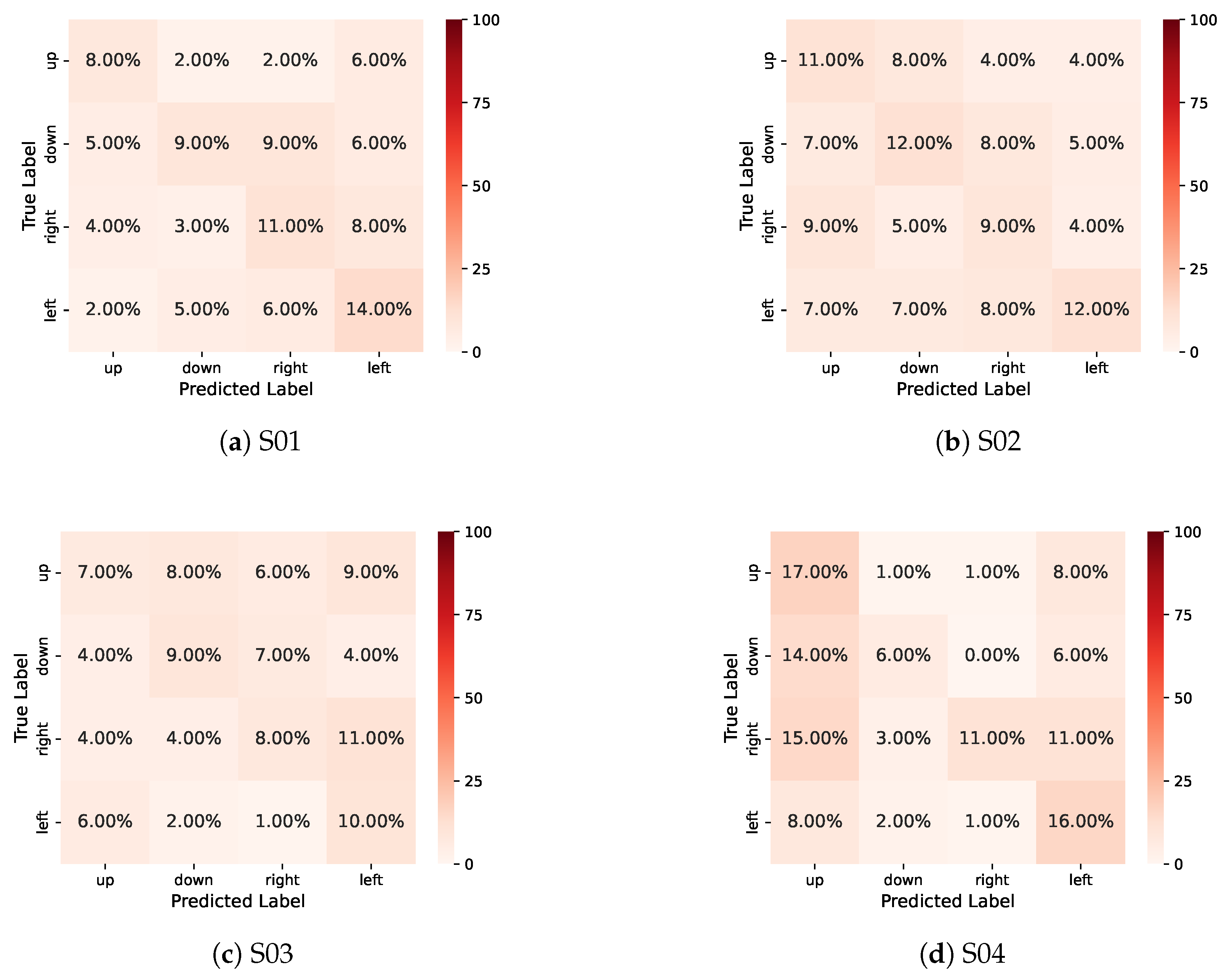
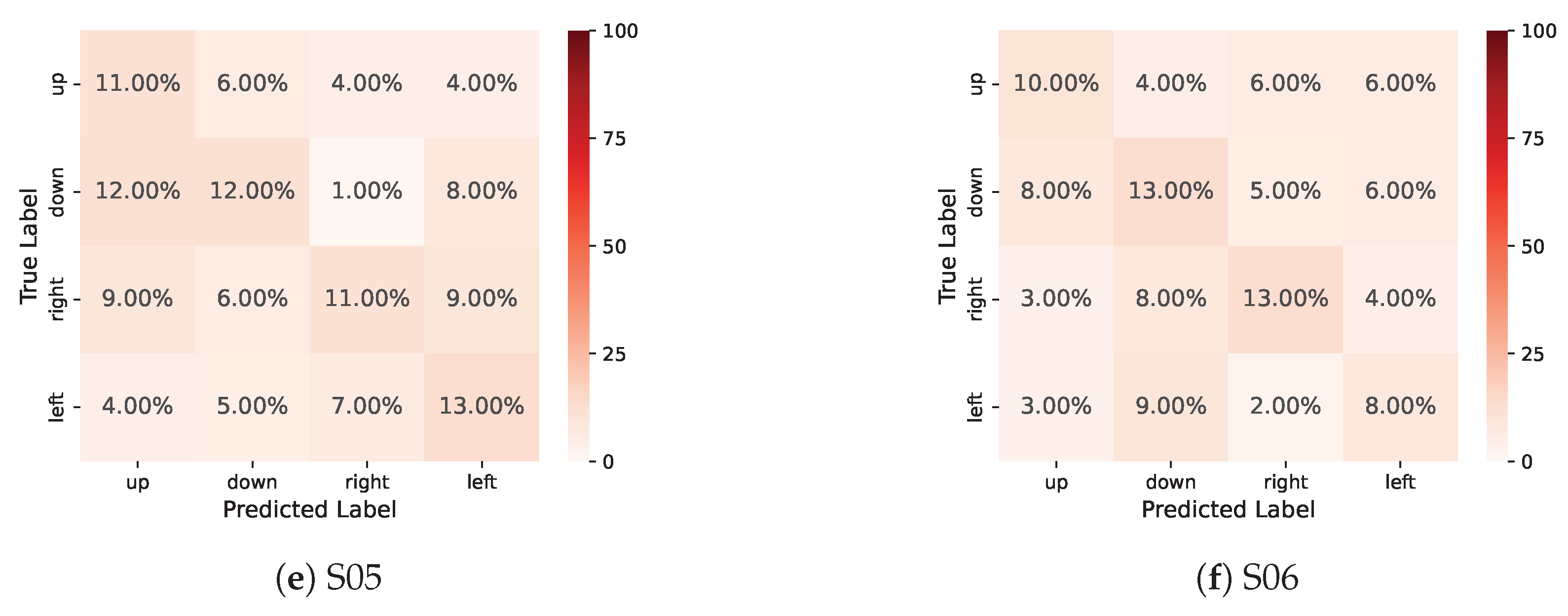
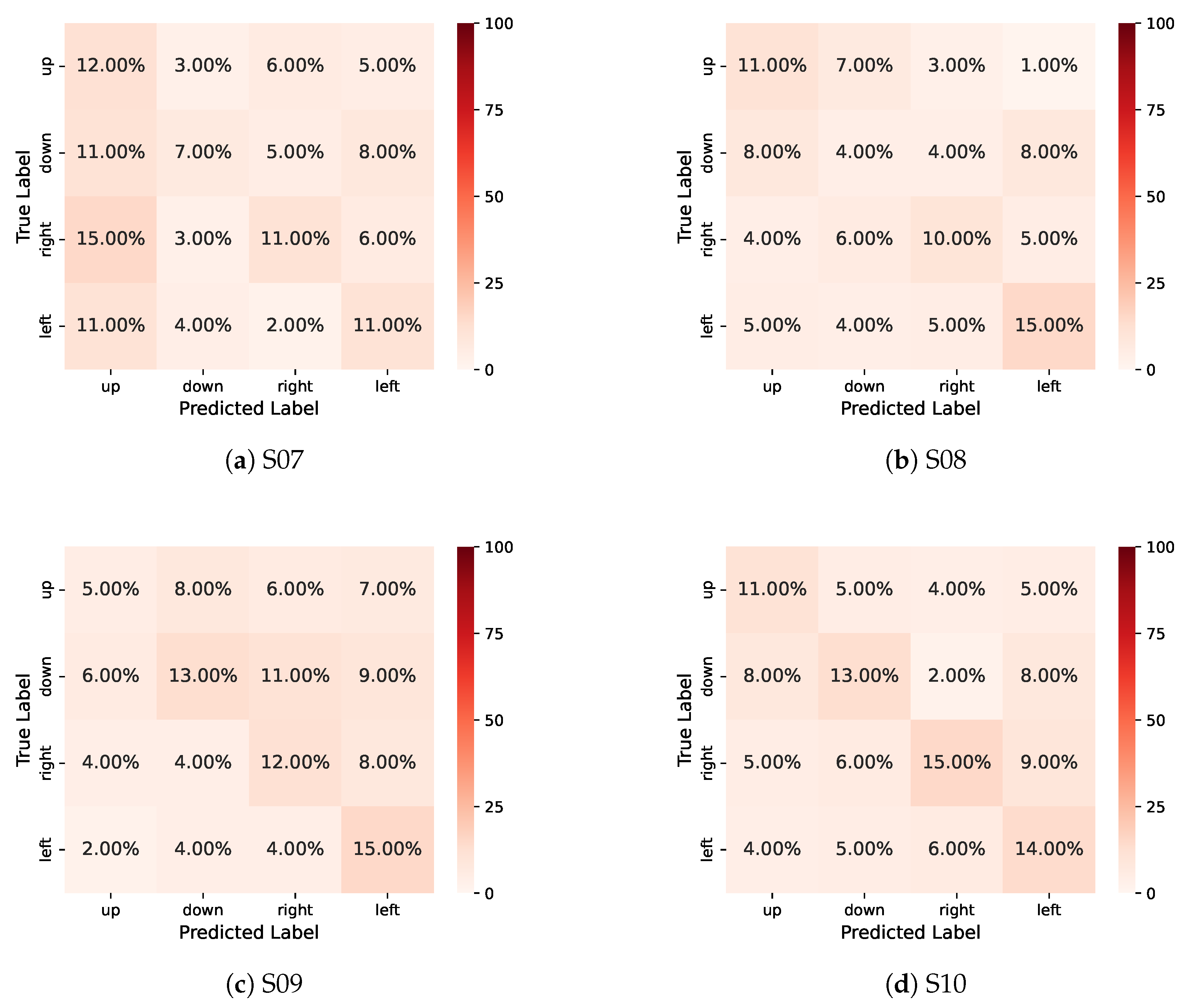
References
- Zhang, L.; Zhou, Y.; Gong, P.; Zhang, D. Speech imagery decoding using EEG signals and deep learning: A survey. IEEE Trans. Cogn. Dev. Syst. 2024, 17, 22–39. [Google Scholar] [CrossRef]
- Guetschel, P.; Ahmadi, S.; Tangermann, M. Review of deep representation learning techniques for brain–computer interfaces. J. Neural Eng. 2024, 21, 061002. [Google Scholar] [CrossRef]
- Musso, M.; Hübner, D.; Schwarzkopf, S.; Bernodusson, M.; LeVan, P.; Weiller, C.; Tangermann, M. Aphasia recovery by language training using a brain–computer interface: A proof-of-concept study. Brain Commun. 2022, 4, fcac008. [Google Scholar] [CrossRef]
- Lopez-Bernal, D.; Balderas, D.; Ponce, P.; Molina, A. Exploring inter-trial coherence for inner speech classification in EEG-based brain–computer interface. J. Neural Eng. 2024, 21, 026048. [Google Scholar] [CrossRef]
- Kamble, A.; Ghare, P.H.; Kumar, V. Optimized rational dilation wavelet transform for automatic imagined speech recognition. IEEE Trans. Instrum. Meas. 2023, 72, 4002210. [Google Scholar] [CrossRef]
- Rahman, N.; Khan, D.M.; Masroor, K.; Arshad, M.; Rafiq, A.; Fahim, S.M. Advances in brain-computer interface for decoding speech imagery from EEG signals: A systematic review. Cogn. Neurodyn. 2024, 18, 3565–3583. [Google Scholar] [CrossRef]
- Cai, Z.; Luo, T.j.; Cao, X. Multi-branch spatial-temporal-spectral convolutional neural networks for multi-task motor imagery EEG classification. Biomed. Signal Process. Control 2024, 93, 106156. [Google Scholar] [CrossRef]
- Li, X.; Tang, J.; Li, X.; Yang, Y. CWSTR-Net: A Channel-Weighted Spatial–Temporal Residual Network based on nonsmooth nonnegative matrix factorization for fatigue detection using EEG signals. Biomed. Signal Process. Control 2024, 97, 106685. [Google Scholar] [CrossRef]
- Nieto, N.; Peterson, V.; Rufiner, H.L.; Kamienkowski, J.E.; Spies, R. Thinking out loud, an open-access EEG-based BCI dataset for inner speech recognition. Sci. Data 2022, 9, 52. [Google Scholar] [CrossRef] [PubMed]
- Kamble, A.; Ghare, P.H.; Kumar, V. Deep-learning-based BCI for automatic imagined speech recognition using SPWVD. IEEE Trans. Instrum. Meas. 2022, 72, 4001110. [Google Scholar] [CrossRef]
- Li, C.; Wang, H.; Liu, Y.; Zhu, X.; Song, L. Silent EEG classification using cross-fusion adaptive graph convolution network for multilingual neurolinguistic signal decoding. Biomed. Signal Process. Control 2024, 87, 105524. [Google Scholar] [CrossRef]
- Li, C.; Liu, Y.; Li, J.; Miao, Y.; Liu, J.; Song, L. Decoding Bilingual EEG Signals With Complex Semantics Using Adaptive Graph Attention Convolutional Network. IEEE Trans. Neural Syst. Rehabil. Eng. 2024, 32, 249–258. [Google Scholar] [CrossRef]
- Liu, K.; Yang, M.; Yu, Z.; Wang, G.; Wu, W. FBMSNet: A filter-bank multi-scale convolutional neural network for EEG-based motor imagery decoding. IEEE Trans. Biomed. Eng. 2022, 70, 436–445. [Google Scholar] [CrossRef]
- Cao, L.; Yu, B.; Dong, Y.; Liu, T.; Li, J. Convolution spatial-temporal attention network for EEG emotion recognition. Physiol. Meas. 2024, 45, 125003. [Google Scholar] [CrossRef]
- Zhao, W.; Jiang, X.; Zhang, B.; Xiao, S.; Weng, S. CTNet: A convolutional transformer network for EEG-based motor imagery classification. Sci. Rep. 2024, 14, 20237. [Google Scholar] [CrossRef] [PubMed]
- Park, D.; Park, H.; Kim, S.; Choo, S.; Lee, S.; Nam, C.S.; Jung, J.Y. Spatio-temporal explanation of 3D-EEGNet for motor imagery EEG classification using permutation and saliency. IEEE Trans. Neural Syst. Rehabil. Eng. 2023. [Google Scholar] [CrossRef]
- Lawhern, V.J.; Solon, A.J.; Waytowich, N.R.; Gordon, S.M.; Hung, C.P.; Lance, B.J. EEGNet: A compact convolutional neural network for EEG-based brain–computer interfaces. J. Neural Eng. 2018, 15, 056013. [Google Scholar] [CrossRef]
- Ding, Y.; Robinson, N.; Zhang, S.; Zeng, Q.; Guan, C. TSception: Capturing temporal dynamics and spatial asymmetry from EEG for emotion recognition. IEEE Trans. Affect. Comput. 2022, 14, 2238–2250. [Google Scholar] [CrossRef]
- Chang, Y.; Zheng, X.; Chen, Y.; Li, X.; Miao, Q. Spatiotemporal Gated Graph Transformer for EEG-based Emotion Recognition. IEEE Signal Process. Lett. 2024. [Google Scholar] [CrossRef]
- Chen, W.; Wang, C.; Xu, K.; Yuan, Y.; Bai, Y.; Zhang, D. D-FaST: Cognitive Signal Decoding with Disentangled Frequency-Spatial-Temporal Attention. IEEE Trans. Cogn. Dev. Syst. 2024. [Google Scholar] [CrossRef]
- Ding, Y.; Li, Y.; Sun, H.; Liu, R.; Tong, C.; Liu, C.; Zhou, X.; Guan, C. EEG-Deformer: A dense convolutional transformer for brain-computer interfaces. IEEE J. Biomed. Health Inform. 2024, 29, 1909–1918. [Google Scholar] [CrossRef]
- Song, Y.; Zheng, Q.; Liu, B.; Gao, X. EEG conformer: Convolutional transformer for EEG decoding and visualization. IEEE Trans. Neural Syst. Rehabil. Eng. 2022, 31, 710–719. [Google Scholar] [CrossRef]
- Zhang, X.; Cheng, X. A transformer convolutional network with the method of image segmentation for EEG-based emotion recognition. IEEE Signal Process. Lett. 2024, 31, 401–405. [Google Scholar] [CrossRef]
- Vaswani, A. Attention is all you need. In Proceedings of the Advances in Neural Information Processing Systems 30, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Zhao, S.; Rudzicz, F. Classifying phonological categories in imagined and articulated speech. In Proceedings of the 2015 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), South Brisbane, Australia, 19–24 April 2015; pp. 992–996. [Google Scholar]
- Wong, T.T.; Yeh, P.Y. Reliable Accuracy Estimates from k-Fold Cross Validation. IEEE Trans. Knowl. Data Eng. 2020, 32, 1586–1594. [Google Scholar] [CrossRef]
- Schirrmeister, R.T.; Springenberg, J.T.; Fiederer, L.D.J.; Glasstetter, M.; Eggensperger, K.; Tangermann, M.; Hutter, F.; Burgard, W.; Ball, T. Deep learning with convolutional neural networks for EEG decoding and visualization. Hum. Brain Mapp. 2017, 38, 5391–5420. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, X.; Zhang, X.; Yu, C. MCMTNet: Advanced network architectures for EEG-based motor imagery classification. Neurocomputing 2024, 620, 129255. [Google Scholar] [CrossRef]
- Moctezuma, L.A.; Suzuki, Y.; Furuki, J.; Molinas, M.; Abe, T. GRU-powered sleep stage classification with permutation-based EEG channel selection. Sci. Rep. 2024, 14, 17952. [Google Scholar] [CrossRef]
- Fang, Z.; Wang, W.; Ren, S.; Wang, J.; Shi, W.; Liang, X.; Fan, C.C.; Hou, Z.G. Learning Regional Attention Convolutional Neural Network for Motion Intention Recognition Based on EEG Data. In Proceedings of the Twenty-Ninth International Conference on International Joint Conferences on Artificial Intelligence, Yokohama, Japan, 11–17 July 2020; pp. 1570–1576. [Google Scholar]
- Palazzo, S.; Spampinato, C.; Kavasidis, I.; Giordano, D.; Schmidt, J.; Shah, M. Decoding brain representations by multimodal learning of neural activity and visual features. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 43, 3833–3849. [Google Scholar] [CrossRef] [PubMed]
- Miao, Z.; Zhao, M.; Zhang, X.; Ming, D. LMDA-Net: A lightweight multi-dimensional attention network for general EEG-based brain-computer interfaces and interpretability. NeuroImage 2023, 276, 120209. [Google Scholar] [CrossRef]
- Kamble, A.; Ghare, P.H.; Kumar, V.; Kothari, A.; Keskar, A.G. Spectral analysis of EEG signals for automatic imagined speech recognition. IEEE Trans. Instrum. Meas. 2023, 72, 4009409. [Google Scholar] [CrossRef]




| Layer Name | Layer Function | In | Out | Kernel | Stride |
|---|---|---|---|---|---|
| Conv1 | Conv2d | 1 | k | (1, fs/10) | (1, 1) |
| Conv2 | Conv2d | k | k | (ch/2, 1) | (1, 1) |
| Conv3 | Conv2d | 1 | k | (ch/2, 1) | (1, 1) |
| Conv4 | Conv2d | k | k | (1, fs/10) | (1, 1) |
| BN | Batchnorm2d | - | - | k | - |
| AG | Avgpool2d | - | - | (4, 4) | - |
| Dataset I | Dataset II | Dataset III | |
|---|---|---|---|
| Number of subjects | 10 | 6 | 6 |
| Number of channels | 128 | 62 | 32 |
| Length of time samples | 1000 | 1000 | 1000 |
| Frequency of samples | 256 | 250 | 250 |
| Number of classes | 4 | 11 | 10 |
| Size of train dataset | 400 | 105 | 400 |
| Size of test dataset | 100 | 27 | 100 |
| Method | S01 | S02 | S03 | S04 | S05 | S06 | S07 | S08 | S09 | S10 | Avg |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ShallowConvNet [27,28] | 33.23 ± 4.15 | 32.33 ± 3.46 | 36.00 ± 2.00 | 33.33 ± 3.33 | 32.67 ± 5.35 | 31.48 ± 3.93 | 30.67 ± 1.90 | 34.80 ± 6.42 | 29.67 ± 3.80 | 32.67 ± 3.65 | 32.69 ± 3.80 |
| DeepConvNet [27,28] | 34.45 ± 2.97 | 35.00 ± 6.77 | 36.45 ± 6.07 | 33.67 ± 5.19 | 32.67 ± 3.46 | 37.41 ± 4.42 | 36.33 ± 3.42 | 37.60 ± 4.56 | 34.00 ± 1.90 | 37.00 ± 5.45 | 35.46 ± 4.42 |
| EEGNet [17,29] | 29.67 ± 3.85 | 28.33 ± 4.41 | 32.40 ± 2.97 | 37.00 ± 2.17 | 30.33 ± 5.94 | 28.52 ± 3.10 | 29.00 ± 6.08 | 32.00 ± 8.37 | 26.33 ± 5.94 | 29.00 ± 4.65 | 30.26 ± 4.75 |
| RACNN [30] | 29.23 ± 3.03 | 23.00 ± 2.98 | 26.45 ± 2.19 | 26.00 ± 2.24 | 25.67 ± 2.53 | 25.19 ± 3.10 | 28.00 ± 1.39 | 22.89 ± 3.35 | 24.00 ± 3.46 | 27.67 ± 6.52 | 25.81 ± 3.08 |
| EEG-ChannelNet [31] | 35.67 ± 4.34 | 36.00 ± 2.79 | 32.00 ± 3.16 | 31.00 ± 2.24 | 32.00 ± 4.15 | 34.07 ± 1.01 | 31.33 ± 2.17 | 35.23 ± 3.35 | 30.67 ± 0.91 | 36.33 ± 4.31 | 33.43 ± 2.84 |
| Conformer [22] | 39.67 ± 6.07 | 33.67 ± 5.06 | 38.89 ± 7.56 | 36.33 ± 6.60 | 36.67 ± 4.08 | 38.52 ± 3.56 | 37.00 ± 6.71 | 40.00 ± 4.69 | 33.00 ± 5.70 | 42.67 ± 5.96 | 37.64 ± 5.60 |
| LMDA-Net [32] | 36.89 ± 5.93 | 35.00 ± 3.54 | 37.23 ± 7.01 | 29.00 ± 2.53 | 32.67 ± 4.50 | 33.33 ± 4.72 | 30.33 ± 2.74 | 32.45 ± 4.98 | 30.67 ± 6.73 | 28.67 ± 2.98 | 32.62 ± 4.57 |
| AISR (SPWVD+CNN) [33] | 30.08 ± 0.44 | 31.92 ± 2.32 | 33.34 ± 1.05 | 33.05 ± 3.81 | 37.72 ± 1.37 | 29.17 ± 3.98 | 28.46 ± 0.93 | 35.37 ± 1.96 | 33.59 ± 3.28 | 34.65 ± 2.58 | 32.74 ± 2.17 |
| EEG-Deformer [21] | 42.41 ± 11.47 | 47.00 ± 2.74 | 40.59 ± 3.22 | 43.00 ± 9.08 | 41.00 ± 11.18 | 46.27 ± 7.45 | 41.00 ± 3.25 | 42.94 ± 8.32 | 42.00 ± 8.37 | 48.75 ± 2.55 | 43.50 ± 6.76 |
| D-FaST [20] | 35.67 ± 2.97 | 34.67 ± 2.98 | 37.23 ± 4.15 | 30.33 ± 3.80 | 31.67 ± 4.25 | 31.48 ± 5.24 | 34.00 ± 9.17 | 34.89 ± 4.60 | 30.00 ± 2.64 | 40.67 ± 7.87 | 34.06 ± 4.77 |
| STCCA (Proposed) | 45.88 ± 4.92 | 46.00 ± 8.22 | 45.88 ± 6.44 | 44.00 ± 5.48 | 40.00 ± 3.54 | 46.67 ± 6.33 | 43.00 ± 6.71 | 47.06 ± 4.16 | 46.00 ± 9.62 | 50.00 ± 9.35 | 45.45 ± 6.48 |
| Method | S01 | S02 | S03 | S04 | S05 | S06 | Avg |
|---|---|---|---|---|---|---|---|
| ShallowConvNet [27,28] | 11.98 ± 4.19 | 8.90 ± 3.02 | 13.30 ± 5.79 | 24.07 ± 8.83 | 13.52 ± 3.59 | 16.47 ± 11.31 | 14.71 ± 6.12 |
| DeepConvNet [27,28] | 14.97 ± 6.52 | 17.91 ± 4.03 | 19.34 ± 8.52 | 25.38 ± 4.05 | 26.81 ± 3.53 | 21.18 ± 5.26 | 20.93 ± 5.32 |
| EEGNet [17,29] | 10.44 ± 4.04 | 16.37 ± 2.87 | 16.59 ± 6.71 | 15.05 ± 5.65 | 25.38 ± 4.05 | 14.12 ± 5.26 | 16.33 ± 4.76 |
| RACNN [30] | 13.52 ± 3.59 | 19.45 ± 4.28 | 22.42 ± 5.13 | 19.45 ± 4.28 | 20.88 ± 6.30 | 15.29 ± 3.22 | 18.50 ± 4.47 |
| EEG-ChannelNet [31] | 10.33 ± 6.21 | 17.80 ± 6.37 | 11.76 ± 6.11 | 23.85 ± 6.09 | 32.75 ± 2.71 | 23.74 ± 5.42 | 20.04 ± 5.49 |
| Conformer [22] | 10.55 ± 4.42 | 16.26 ± 7.59 | 14.97 ± 6.52 | 19.56 ± 10.58 | 19.01 ± 12.65 | 20.00 ± 3.22 | 16.73 ± 7.50 |
| LMDA-Net [32] | 16.26 ± 5.66 | 13.30 ± 5.79 | 14.70 ± 5.63 | 25.60 ± 7.51 | 16.48 ± 6.28 | 18.82 ± 4.92 | 17.53 ± 5.97 |
| AISR (SPWVD+CNN) [33] | 12.69 ± 1.06 | 12.15 ± 0.91 | 13.71 ± 2.51 | 12.53 ± 0.08 | 13.23 ± 3.95 | 11.11 ± 0.60 | 12.57 ± 1.52 |
| EEG-Deformer [21] | 20.00 ± 11.18 | 21.00 ± 7.68 | 22.02 ± 5.68 | 25.00 ± 1.20 | 20.00 ± 2.92 | 23.56 ± 5.95 | 21.93 ± 5.77 |
| D-FaST [20] | 13.41 ± 6.00 | 15.05 ± 5.65 | 16.37 ± 6.15 | 28.24 ± 7.65 | 22.42 ± 7.48 | 20.00 ± 3.22 | 19.25 ± 6.03 |
| STCCA (Proposed) | 18.23 ± 3.57 | 22.23 ± 5.04 | 23.50 ± 2.50 | 31.25 ± 5.70 | 35.50 ± 7.43 | 24.72 ± 4.82 | 25.91 ± 4.84 |
| Method | S01 | S02 | S03 | S04 | S05 | S06 | Avg |
|---|---|---|---|---|---|---|---|
| ShallowConvNet [27,28] | 18.82 ± 9.67 | 19.12 ± 5.63 | 17.86 ± 7.14 | 21.18 ± 9.84 | 21.18 ± 6.71 | 20.00 ± 3.22 | 19.69 ± 7.04 |
| DeepConvNet [27,28] | 12.94 ± 12.75 | 15.29 ± 5.26 | 18.57 ± 3.91 | 12.94 ± 7.67 | 14.12 ± 9.84 | 20.00 ± 7.89 | 15.64 ± 7.89 |
| EEGNet [17,29] | 20.59 ± 3.42 | 14.12 ± 3.22 | 22.86 ± 9.31 | 17.65 ± 4.80 | 20.00 ± 8.92 | 18.82 ± 10.52 | 19.01 ± 6.70 |
| RACNN [30] | 27.06 ± 3.22 | 24.71 ± 4.92 | 21.43 ± 2.04 | 24.71 ± 6.44 | 16.47 ± 4.92 | 23.53 ± 8.32 | 22.99 ± 4.98 |
| EEG-ChannelNet [31] | 20.00 ± 7.89 | 24.71 ± 6.44 | 14.29 ± 3.50 | 21.18 ± 3.22 | 25.88 ± 5.26 | 16.47 ± 4.92 | 20.42 ± 5.21 |
| Conformer [22] | 22.35 ± 2.63 | 21.18 ± 5.26 | 21.43 ± 7.14 | 25.88 ± 6.71 | 23.53 ± 3.06 | 23.53 ± 4.16 | 22.98 ± 4.83 |
| LMDA-Net [32] | 20.00 ± 6.71 | 15.29 ± 7.89 | 22.86 ± 13.74 | 22.35 ± 7.67 | 18.82 ± 7.67 | 23.53 ± 4.16 | 20.48 ± 7.97 |
| AISR(SPWVD+CNN) [33] | 11.88 ± 0.97 | 12.44 ± 1.18 | 11.34 ± 0.57 | 12.04 ± 0.67 | 12.10 ± 0.23 | 11.08 ± 1.44 | 11.81 ± 0.84 |
| EEG-Deformer [21] | 27.41 ± 4.16 | 27.06 ± 5.26 | 25.71 ± 3.91 | 28.24 ± 4.92 | 30.59 ± 6.44 | 23.53 ± 4.16 | 27.09 ± 4.81 |
| D-FaST [20] | 19.40 ± 9.09 | 20.00 ± 8.92 | 18.82 ± 2.63 | 18.57 ± 3.91 | 26.00 ± 15.17 | 22.35 ± 2.63 | 20.86 ± 7.06 |
| STCCA (Proposed) | 28.24 ± 2.63 | 31.76 ± 5.26 | 28.57 ± 8.75 | 29.41 ± 4.16 | 28.41 ± 5.88 | 28.00 ± 5.95 | 29.07 ± 5.44 |
| S01 | S02 | S03 | S04 | S05 | S06 | S07 | S08 | S09 | S10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| F1_score | 0.4125 | 0.3996 | 0.3570 | 0.3852 | 0.4132 | 0.4094 | 0.3594 | 0.4452 | 0.3764 | 0.4554 |
| Acc_5 | 30.00 | 33.33 | 27.00 | 31.67 | 35.00 | 30.56 | 25.83 | 34.00 | 30.83 | 35.83 |
| Acc_10 | 32.00 | 29.17 | 27.00 | 32.50 | 33.33 | 30.56 | 26.67 | 34.00 | 30.00 | 33.33 |
| Method | Parameters (M) | FLOPs (M) | Inference Time (ms) |
|---|---|---|---|
| EEG-ChannelNet | 36.682 | 13,547.434 | 12.126 |
| Conformer | 31.155 | 2012.787 | 16.094 |
| EEG-Deformer | 1.096 | 98.146 | 6.067 |
| D-FaST | 6.386 | 7252.466 | 99.338 |
| STCCA (Proposed) | 7.155 | 2581.882 | 4.597 |
| Method | S01 | S02 | S03 | S04 | S05 | S06 | S07 | S08 | S09 | S10 |
|---|---|---|---|---|---|---|---|---|---|---|
| ShallowConvNet | 27.18 | 25.50 | 28.17 | 27.67 | 25.33 | 27.04 | 27.00 | 27.38 | 26.33 | 26.17 |
| DeepConvNet | 29.56 | 28.67 | 28.97 | 27.00 | 29.17 | 29.26 | 26.00 | 30.36 | 28.50 | 25.50 |
| EEGNet | 27.78 | 27.17 | 30.36 | 30.00 | 29.17 | 24.63 | 26.83 | 26.98 | 25.83 | 27.17 |
| EEG-ChannelNet | 29.37 | 26.83 | 27.18 | 26.00 | 27.33 | 27.96 | 28.33 | 29.37 | 28.83 | 27.83 |
| LMDA | 29.17 | 29.33 | 28.17 | 24.83 | 28.00 | 27.78 | 26.83 | 26.19 | 25.17 | 26.17 |
| Deformer | 28.57 | 25.50 | 29.17 | 26.33 | 27.50 | 26.48 | 26.00 | 27.98 | 25.33 | 25.33 |
| D-FaST | 30.00 | 29.00 | 26.98 | 28.83 | 25.83 | 27.22 | 27.00 | 29.54 | 28.67 | 27.83 |
| STCCA (Proposed) | 30.36 | 29.83 | 29.37 | 30.17 | 29.33 | 29.30 | 27.67 | 30.37 | 27.50 | 27.83 |
| Method | S01 | S02 | S03 | S04 | S05 | S06 | S07 | S08 | S09 | S10 | Avg |
|---|---|---|---|---|---|---|---|---|---|---|---|
| w/o LFEM | 40.06 ± 7.20 | 42.00 ± 4.47 | 44.71 ± 5.26 | 42.76 ± 7.42 | 38.70 ± 9.08 | 44.44 ± 3.93 | 40.05 ± 7.42 | 42.35 ± 9.67 | 42.00 ± 10.95 | 40.00 ± 7.07 | 41.71 ± 7.25 |
| w/o GFEM | 41.88 ± 9.67 | 43.00 ± 8.37 | 43.41 ± 5.26 | 40.36 ± 4.18 | 39.45 ± 4.58 | 43.36 ± 2.74 | 39.48 ± 4.92 | 40.59 ± 6.71 | 45.00 ± 3.54 | 43.63 ± 5.84 | 42.02 ± 5.58 |
| STCCA (Proposed) | 45.88 ± 4.92 | 46.00 ± 8.22 | 45.88 ± 6.44 | 44.00 ± 5.48 | 40.00 ± 3.54 | 46.67 ± 6.33 | 43.00 ± 6.71 | 47.06 ± 4.16 | 46.00 ± 9.62 | 50.00 ± 9.35 | 45.45 ± 6.48 |
| Method | S01 | S02 | S03 | S04 | S05 | S06 | Avg |
|---|---|---|---|---|---|---|---|
| w/o LFEM | 15.23 ± 2.62 | 20.50 ± 5.02 | 20.65 ± 3.69 | 15.00 ± 13.69 | 31.00 ± 3.69 | 8.00 ± 10.95 | 18.40 ± 4.94 |
| w/o GFEM | 17.04 ± 3.69 | 13.69 ± 4.13 | 19.89 ± 2.92 | 30.00 ± 20.92 | 30.00 ± 1.18 | 12.00 ± 10.92 | 20.44 ± 7.29 |
| STCCA (Proposed) | 18.23 ± 3.57 | 22.23 ± 5.04 | 23.50 ± 2.50 | 31.25 ± 5.70 | 35.50 ± 7.43 | 24.72 ± 4.82 | 25.91 ± 4.84 |
| Method | S01 | S02 | S03 | S04 | S05 | S06 | Avg |
|---|---|---|---|---|---|---|---|
| w/o LFEM | 22.35 ± 7.67 | 27.06 ± 6.71 | 22.86 ± 7.83 | 22.76 ± 9.84 | 24.71 ± 6.44 | 23.53 ± 8.32 | 23.88 ± 7.80 |
| w/o GFEM | 23.41 ± 4.16 | 29.41 ± 7.2 | 25.71 ± 6.39 | 20.59 ± 6.44 | 21.76 ± 6.71 | 25.49 ± 6.79 | 24.40 ± 6.28 |
| STCCA (Proposed) | 28.24 ± 2.63 | 31.76 ± 5.26 | 28.57 ± 8.75 | 29.41 ± 4.16 | 28.41 ± 5.88 | 28.00 ± 5.95 | 29.07 ± 5.44 |
| Parameters (M) | FLOPs (M) | Inference Time (ms) | |
|---|---|---|---|
| sum | 7.155 | 2547.787 | 4.250 |
| cat | 7.155 | 2567.283 | 5.473 |
| atten | 7.155 | 2548.901 | 4.308 |
| CCA | 7.155 | 2581.882 | 4.597 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, L.; Shao, H.; Zhang, L.; Li, L. STCCA: Spatial–Temporal Coupled Cross-Attention Through Hierarchical Network for EEG-Based Speech Recognition. Sensors 2025, 25, 6541. https://doi.org/10.3390/s25216541
Dong L, Shao H, Zhang L, Li L. STCCA: Spatial–Temporal Coupled Cross-Attention Through Hierarchical Network for EEG-Based Speech Recognition. Sensors. 2025; 25(21):6541. https://doi.org/10.3390/s25216541
Chicago/Turabian StyleDong, Liang, Hengyi Shao, Lin Zhang, and Lei Li. 2025. "STCCA: Spatial–Temporal Coupled Cross-Attention Through Hierarchical Network for EEG-Based Speech Recognition" Sensors 25, no. 21: 6541. https://doi.org/10.3390/s25216541
APA StyleDong, L., Shao, H., Zhang, L., & Li, L. (2025). STCCA: Spatial–Temporal Coupled Cross-Attention Through Hierarchical Network for EEG-Based Speech Recognition. Sensors, 25(21), 6541. https://doi.org/10.3390/s25216541








