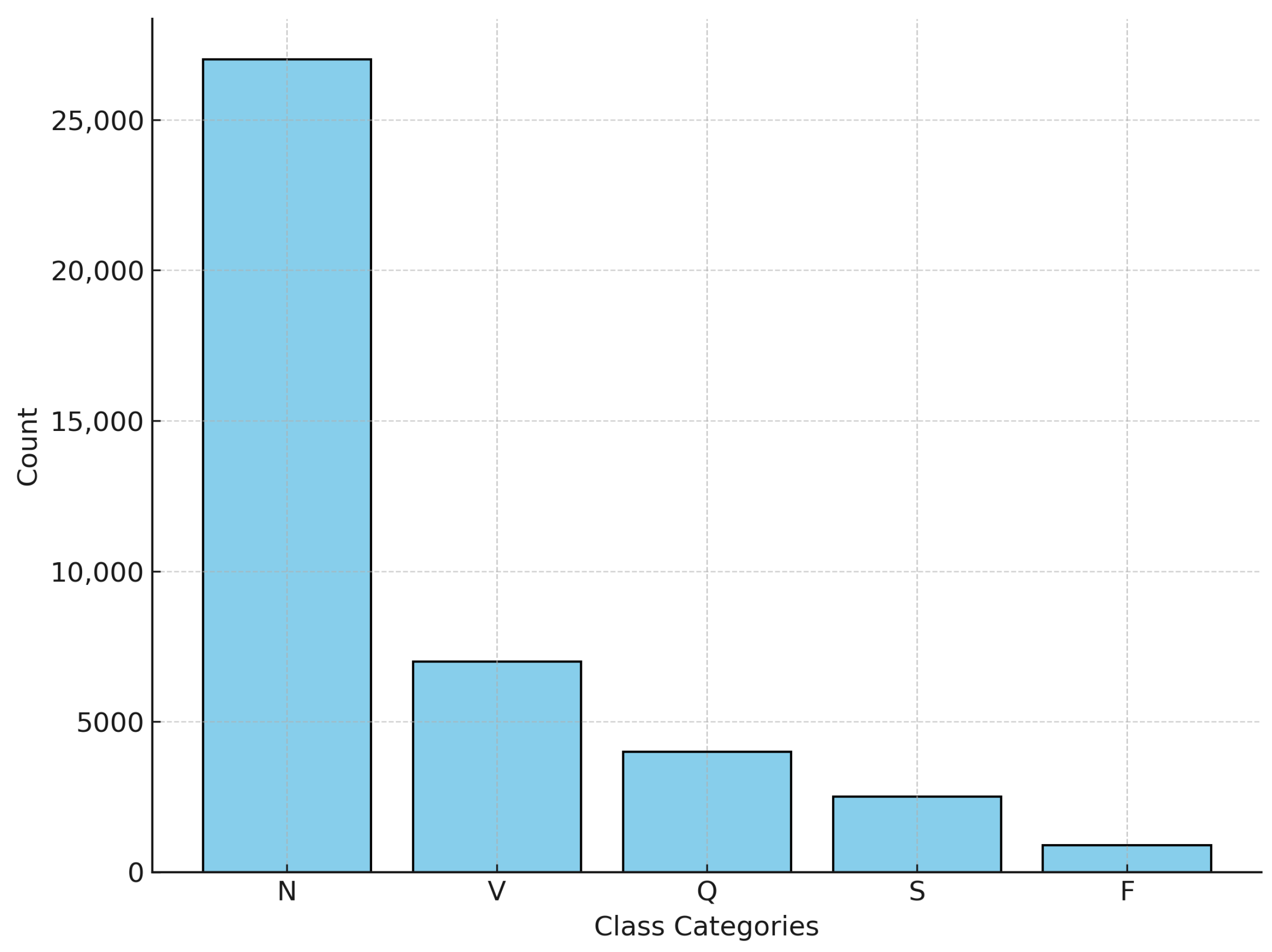
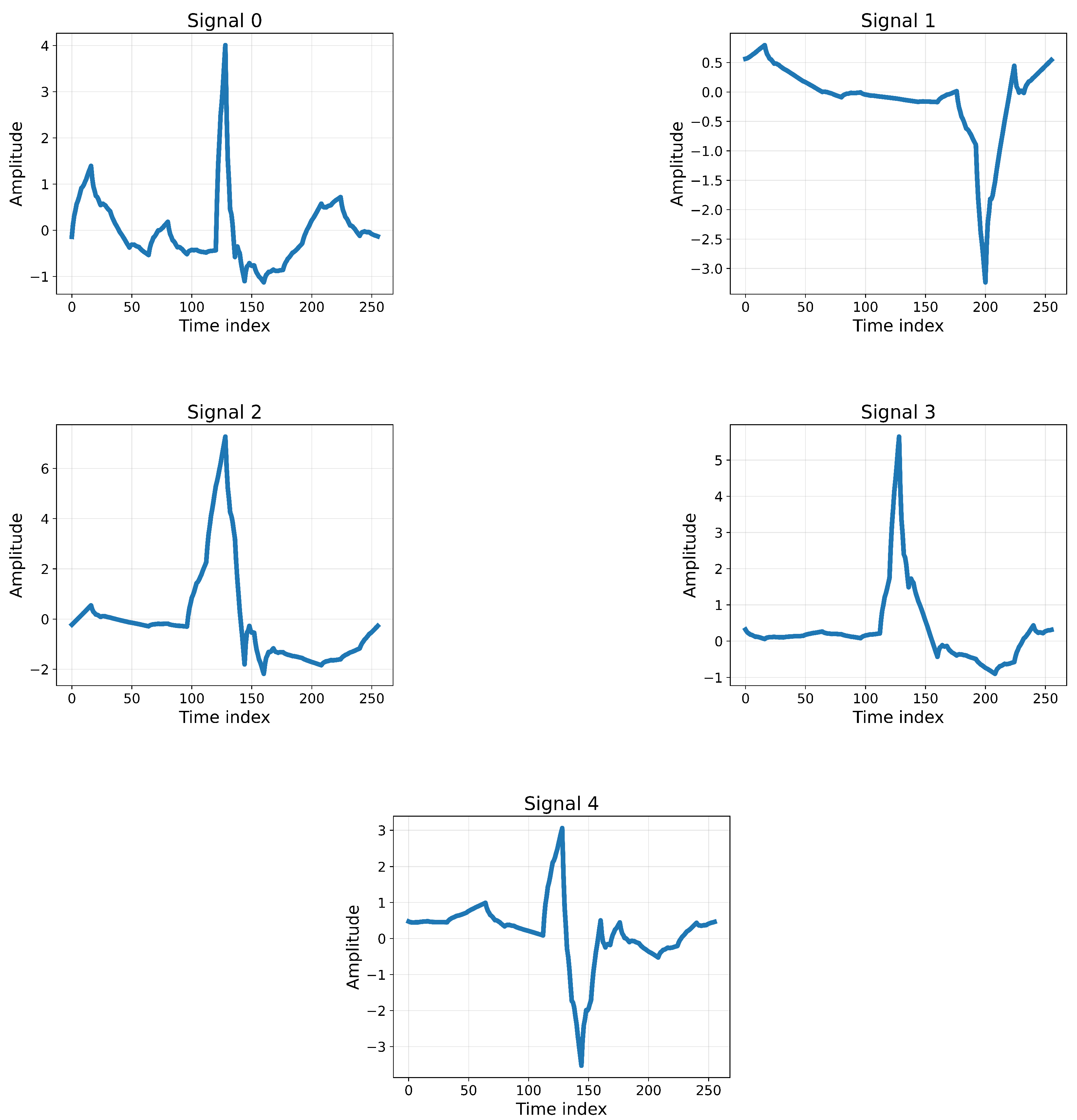
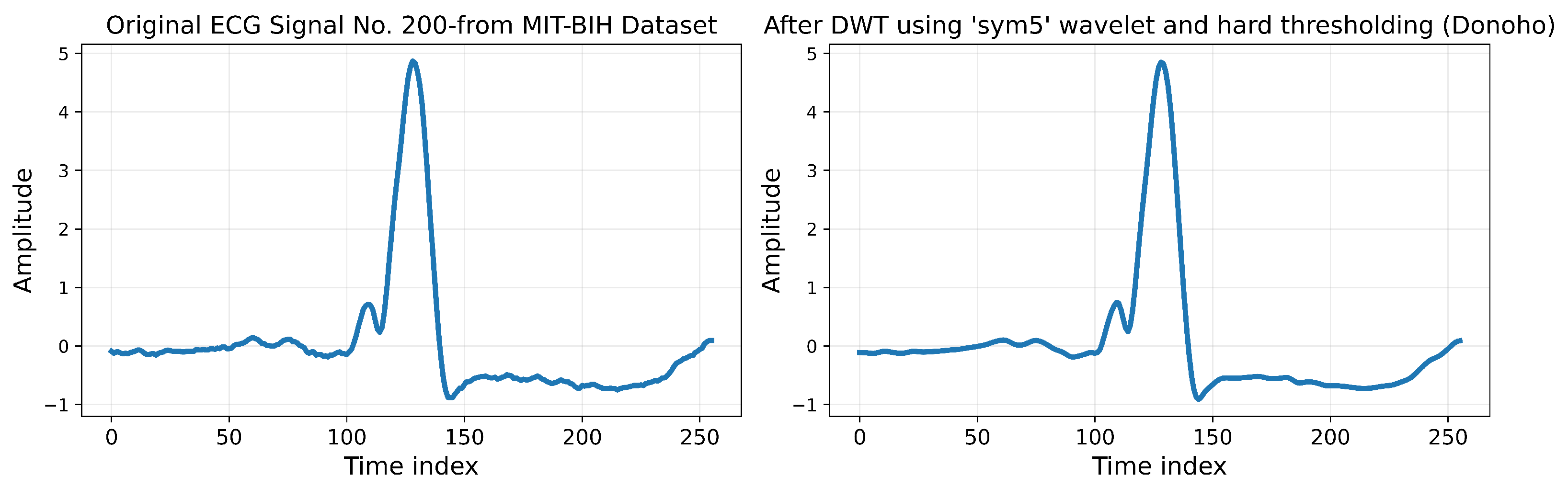
The MIT-BIH Arrhythmia Database was used for this research, which contains approximately 110,000 heartbeat annotations collected from 48 half-hour ECG recordings of 47 subjects. The signals were sampled at 360 Hz with 11-bit resolution over a 10 mV range, ensuring high-quality recordings suitable for detecting subtle arrhythmia patterns. A subset of these signals was carefully selected to ensure a balanced representation of the five arrhythmia classes, as defined by the AAMI standard.
5.1. ML Models
In this experiment, the GBM was trained to iteratively improve classification performance by combining multiple weak learners, typically decision trees, into a strong ensemble. The model was configured using default hyperparameters, which were empirically validated to provide reliable performance on the dataset. The GBM achieved an overall accuracy of 91%, which is considered moderately acceptable; however, it was not among the top-performing models. Given the nature of ECG signals, where diagnostic accuracy is critical, higher performance is expected to ensure clinical reliability. Due to the sequential nature of boosting, training time may increase with larger datasets. As shown in
Figure 10, the confusion matrix for the GBM highlights its classification accuracy across different classes, with notable strengths in well-represented categories and some misclassifications in borderline or underrepresented classes. As detailed in the evaluation matrix shown in
Table 7, the model exhibited high precision and recall for the N and Q classes, while the S and F classes showed moderate performance, suggesting areas for further optimization.
The MLP model, a type of artificial neural network, was configured with one hidden layer of 10 neurons and utilized the ReLU activation function to introduce non-linearity. The model was trained using the Adam optimizer, which dynamically adjusts learning rates during training. With a regularization term
to mitigate overfitting, the MLP achieved an overall accuracy of 95%. As shown in
Figure 11, the confusion matrix reveals that the MLP model performed well overall but exhibited slight misclassifications in certain classes, particularly the F and S categories. As shown in
Table 8, the evaluation matrix corroborates these findings, showing that while the N and Q classes achieved high precision and recall, other classes require improved tuning to enhance model performance.
The confusion matrices and classification reports for both the GBM and MLP models demonstrate their respective strengths and areas for improvement. The GBM achieved an overall accuracy of 91%, while the MLP performed slightly better with an overall accuracy of 95%. These findings underscore the importance of understanding the nuances of each model, including the impact of hyperparameter selection and data preprocessing, to achieve optimal results in arrhythmia classification. Given the critical nature of ECG data, models with higher precision and consistency are favored to ensure reliable diagnostic support.
5.2. DL Models
The 1D-CNN was designed to extract local features from ECG signals through convolutional layers. The model was trained with a learning rate of 0.0002 and the Adam optimizer. The CrossEntropyLoss function was employed, and class weights were applied to address the class imbalance. After training with a batch size of 20, the model achieved an overall accuracy of 92% and demonstrated strong performance across most classes. For class N, it achieved a precision of 97% and a recall of 92%, indicating high precision and effective detection. However, challenges were noted for class F, which had a precision of 47%, suggesting room for improvement. As shown in
Figure 12, the confusion matrix highlights the model’s classification accuracy for each class.
Table 9 presents the evaluation matrix, revealing an F1 score of 95% for class N and 89% for class V, reflecting robust detection capabilities. However, the F1 score for class F was lower at 60%, underscoring difficulties in this category. These results highlight the model’s strengths in accurately classifying common classes but also suggest opportunities to refine its performance for less distinct categories, such as class F. Adjustments to hyperparameters or increased model depth may address these challenges and improve its classification capabilities.
Figure 13 illustrates the training and validation accuracy and loss curves for the 1D-CNN model across 30 epochs.
The LSTM model was implemented to capture the temporal dependencies in ECG signals, which are crucial for arrhythmia classification. Similarly to the 1D-CNN model, the Adam optimizer was used with a learning rate of 0.0002. The model was trained with a batch size of 16, and CrossEntropyLoss was applied. The LSTM model, while effective at capturing temporal patterns, achieved a slightly lower overall accuracy of 75%, likely due to its inability to capture local features, unlike the 1D-CNN. As shown in
Figure 14, the confusion matrix highlights the model’s classification accuracy for each class. The LSTM model shows strong performance in the N and Q classes, achieving high precision and recall metrics. However, it struggles with the S and F classes, as indicated by low precision (35% and 13%, respectively) and moderate recall (56% and 39%, respectively), as shown in
Table 10. These results suggest misclassification issues due to unclear features or insufficient data. The training and validation accuracy and loss curves for the LSTM model over 30 epochs are shown in
Figure 15.
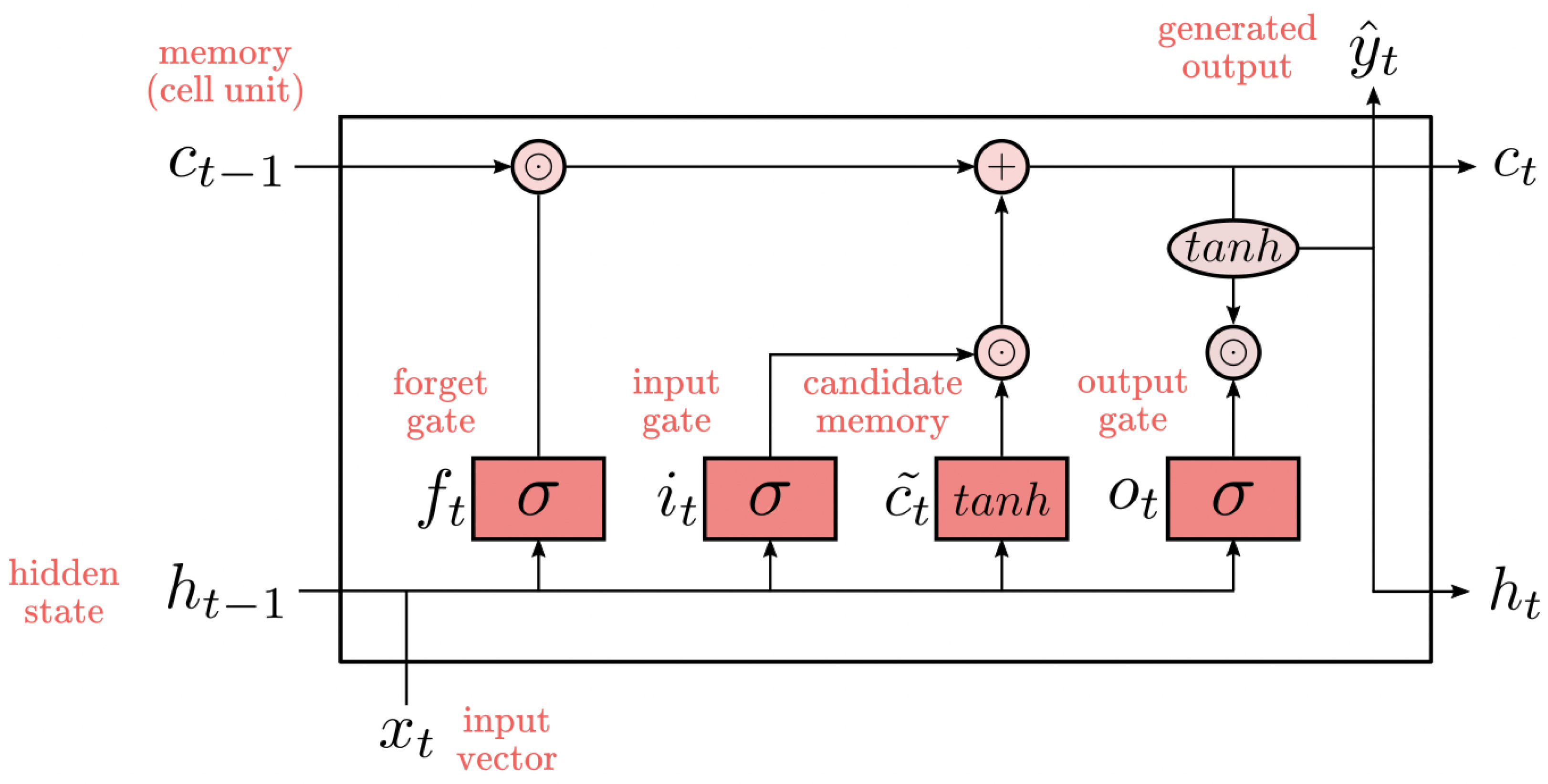
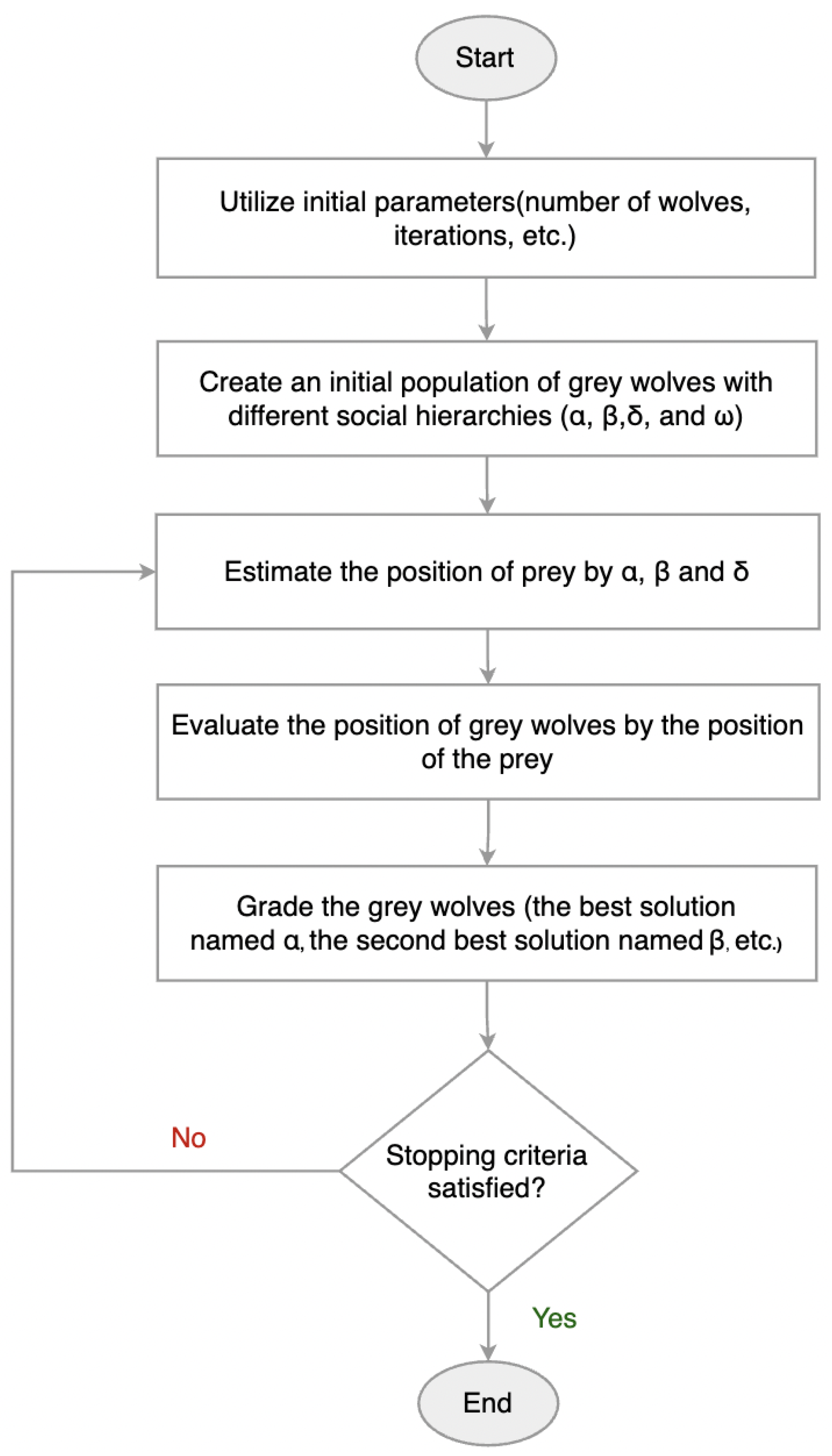
The proposed approach model combines the strengths of both 1D-CNN and LSTM networks, where the 1D-CNN component is responsible for extracting local features from the ECG signals, while the LSTM component captures temporal dependencies within the sequences. The model architecture is based on a 1D-CNN-LSTM hybrid structure.
To optimize the model’s performance, the GWO algorithm was employed to automatically select the best hyperparameters, such as the number of filters in each 1D-CNN layer, kernel sizes, dropout rates, pooling configurations, and LSTM hidden size. This approach replaced the manual tuning process, enabling a more systematic and efficient exploration of the hyperparameter space to enhance model accuracy.
For weight initialization, the Kaiming Normal initialization method was used to ensure efficient gradient flow during training. This technique, also known as He initialization [
44], is particularly suited for layers followed by ReLU activations and helps prevent vanishing or exploding gradients in deep networks. It was applied to all 1D-CNN, linear, and LSTM layers in the model to stabilize training and improve convergence.
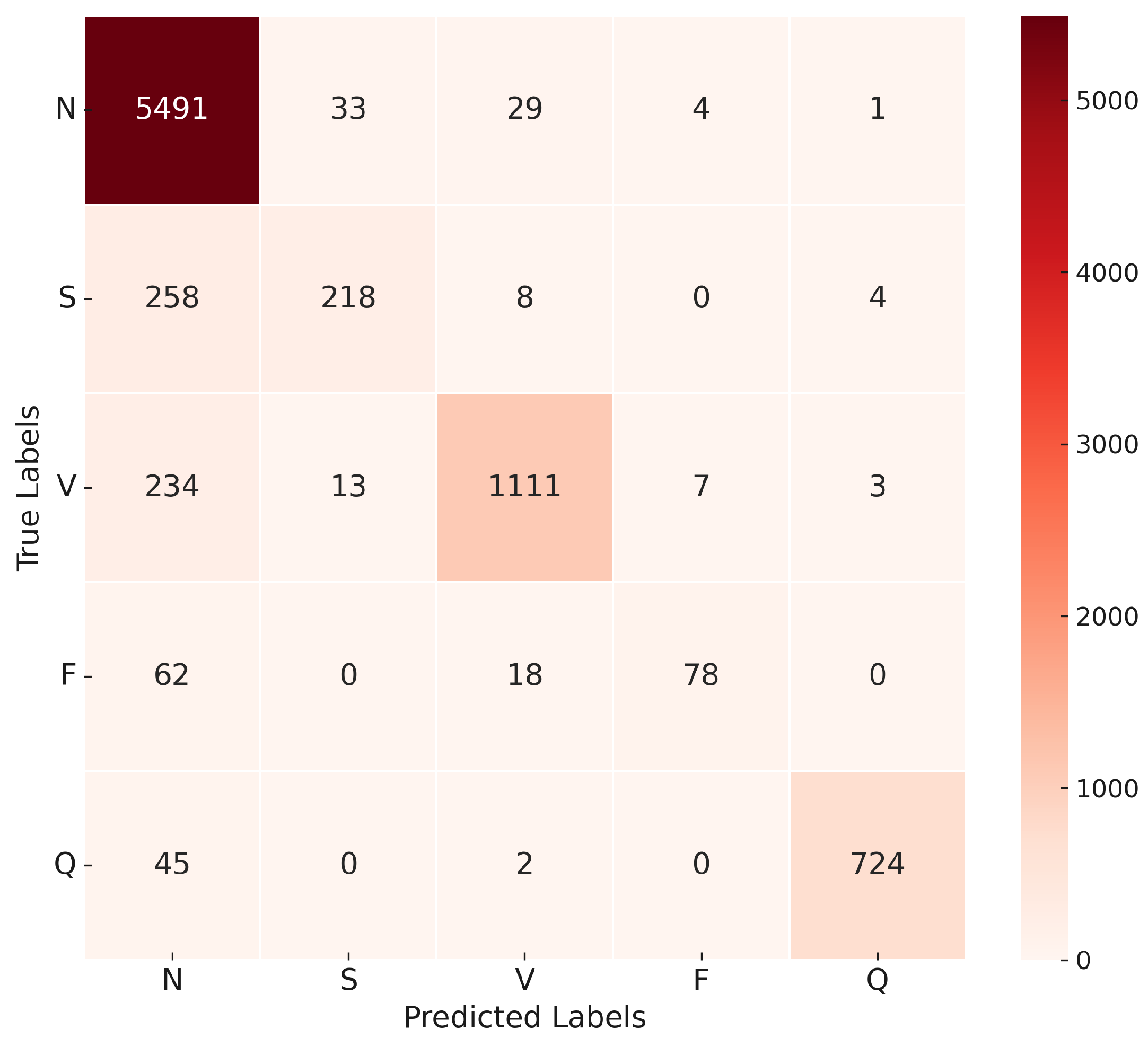
The confusion matrix in
Figure 16 illustrates the model’s ability to distinguish between the five classes of ECG recommended by AAMI. The matrix shows a high true positive rate for the N and V classes, with minor misclassifications occurring in the S and F classes.
The evaluation matrix presented in
Table 11 further supports the effectiveness of the model, with a weighted average precision, recall, and F1 score. Although the model achieved outstanding results in most categories, it showed a relatively lower recall in the F class, suggesting potential areas for future improvement.
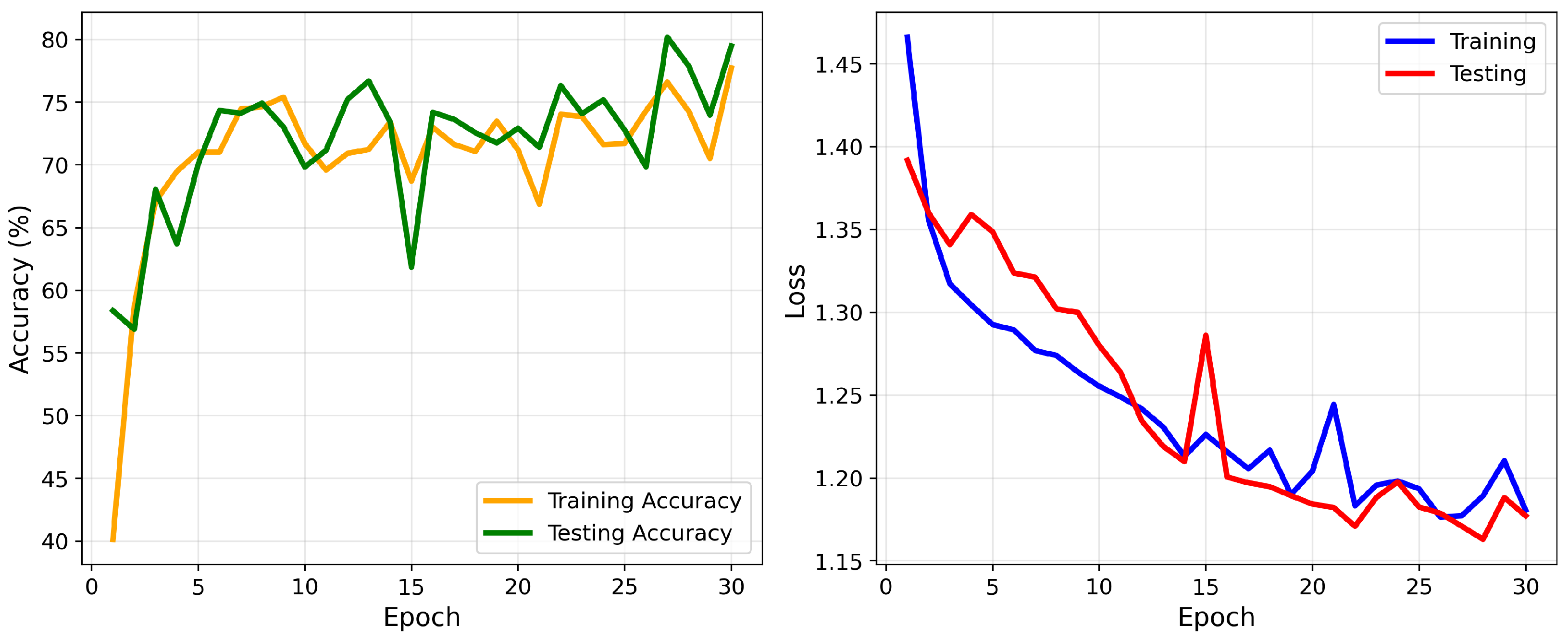
The final model was trained using the Adam optimizer, with the optimal hyperparameters obtained through the GWO. The optimization process achieved a peak validation overall accuracy of 97% and a minimum validation loss of 0.12, reflecting a strong fit to both training and validation datasets. The corresponding convergence of accuracy and loss during training is illustrated in
Figure 17.
The optimized proposed approach, the 1D-CNN–LSTM model, demonstrated computational efficiency, containing only 298,770 trainable parameters with a compact size of 1.14 MB. The training process was completed in approximately 1099 s (18 min) on an NVIDIA Tesla T4 GPU with 14.74 GB of memory. These results indicate that the proposed model is lightweight, resource-efficient, and feasible for deployment in practical healthcare applications.
The novelty of this research lies in the integration of 1D-CNN–LSTM with the GWO, which, unlike prior works, unifies deep feature extraction, temporal sequence learning, and metaheuristic-based hyperparameter tuning within a single framework [
23,
24,
25,
26,
27]. While earlier studies employed 1D-CNN–LSTM without optimization, this work applies the GWO to automatically tune the hyperparameters of the hybrid 1D-CNN–LSTM model. Instead of relying on manual configuration, which is often time-consuming and suboptimal, the GWO offers an intelligent metaheuristic strategy to efficiently search for optimal hyperparameter settings, thereby enhancing the model’s overall accuracy and robustness. Other research has applied the GWO to alternative architectures such as an ANN or R-CNN [
26,
27]; however, this research utilized the complementary strengths of the 1D-CNN, which excels at extracting spatial and morphological features from ECG signals, and LSTM, which effectively captures temporal dependencies in heartbeat sequences. This proposed approach design, optimized through the GWO, enabled the proposed model to achieve superior performance across all arrhythmia classes. The results demonstrate that GWO-driven optimization of 1D-CNN–LSTM significantly enhances classification accuracy, reaching 97%. Furthermore, by benchmarking against both classical ML models (GBM and MLP) and DL baselines models, this research provides clear evidence that the proposed approach delivers superior and robust performance across all five AAMI arrhythmia classes.
To further evaluate the effectiveness of the proposed approach model (1D-CNN-LSTM) enhanced with the GWO, a comparison was performed with recent state-of-the-art studies that have addressed the classification of ECG arrhythmias using either DL or metaheuristic-based optimization methods in the MIT-BIH dataset.
Table 12 summarizes the key characteristics and classification accuracies reported in these studies, in contrast to the results achieved in this work.
As shown in
Table 12, the proposed model outperforms prior studies in terms of classification accuracy on the MIT-BIH dataset. The integration of a 1D-CNN–LSTM hybrid architecture with metaheuristic-based hyperparameter tuning via the GWO contributed to this improvement, underscoring the effectiveness of combining DL with nature-inspired optimization techniques for ECG arrhythmia detection. Furthermore, unlike previous models that often require millions of parameters and substantial storage, the optimized 1D-CNN–LSTM in this research contained only 298,770 trainable parameters (1.14 MB) and completed training in approximately 18 min, demonstrating computational efficiency and practical feasibility for real-world deployment.
Although the proposed approach 1D-CNN–LSTM model with the GWO achieved 97% overall accuracy, it is important to acknowledge that some recent works have reported accuracies above 98% [
21,
24,
25]. However, these approaches often rely on complex attention mechanisms or image-based ECG representations, which increase computational cost. In contrast, our model balances strong accuracy with efficiency and simplicity, making it more practical for clinical deployment.
Several significant limitations should be noted even though the proposed 1D-CNN–LSTM model optimized using the GWO achieved improved performance. First, this research relies solely on the MIT-BIH dataset, which restricts the generalizability of the findings and may not fully capture the diversity of broader clinical populations. Second, class imbalance was addressed only through selective sampling, without comparisons to alternative approaches such as SMOTE, focal loss, or class weighting, which may provide different perspectives on performance. Third, statistical validation techniques, including k-fold cross-validation, confidence intervals, or hypothesis testing, were not performed due to computational constraints, which limits the ability to confirm the robustness of the reported accuracy. Finally, the model does not include explainability analysis. Methods such as Grad-CAM, saliency maps, or attention-based mechanisms could provide useful insights into the decision-making process of the model, thereby improving interpretability and clinical trust.
The proposed approach 1D-CNN–LSTM model optimized with the GWO demonstrates promising results for automated arrhythmia classification. In clinical practice, such a system could be integrated as a decision support tool to assist cardiologists and healthcare providers in the early detection of abnormal heart rhythms. For example, the model could be embedded in hospital ECG monitoring systems to provide real-time alerts for arrhythmic events. This would enable faster diagnosis, reduce the burden on physicians, and potentially improve patient outcomes through timely interventions.
However, several challenges must be addressed before clinical deployment. First, variations in ECG acquisition devices and patient populations may affect model performance, underscoring the need for validation on diverse datasets. Second, the interpretability of DL predictions remains limited, which may hinder trust among clinicians. Third, integration into existing healthcare infrastructure requires consideration of data privacy, regulatory approval, and user training. Addressing these challenges is essential to ensure the safe, reliable, and effective adoption of the model in real-world diagnostic workflows.