CLCFM3: A 3D Reconstruction Algorithm Based on Photogrammetry for High-Precision Whole Plant Sensing Using All-Around Images
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Cultivation
2.2. CLCFM3: 3D Reconstruction Algorithm
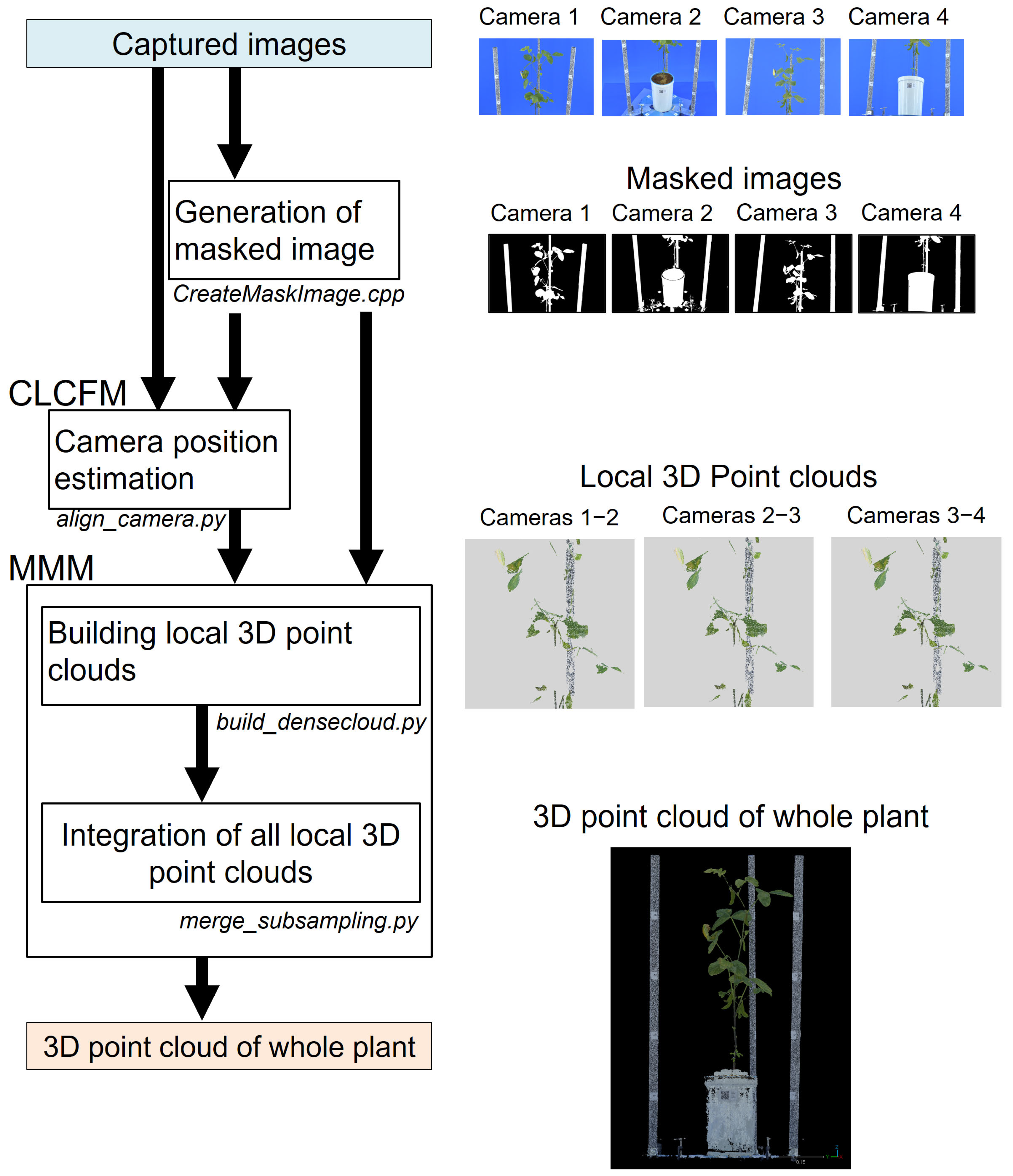
2.2.1. MMM: Multi-Masked Matching
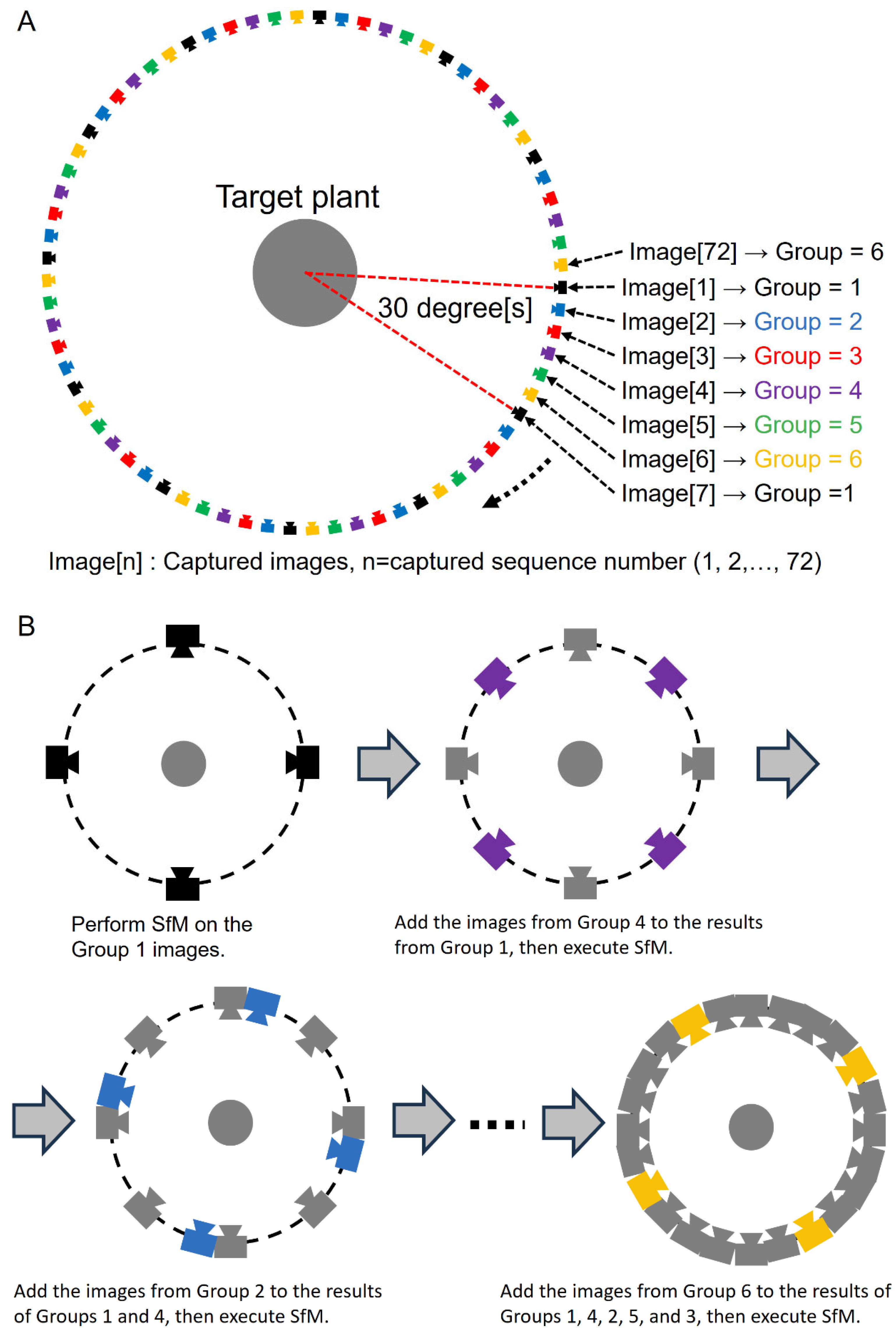
2.2.2. CLCFM: Closed-Loop Coarse-to-Fine Method for SfM
2.3. Three-Dimensional Point Cloud Reconstruction Pipeline Program
- CreateMask.cpp
- 2.
- align_camera.py
- 3.
- build_densecloud.py
- matchPhotos(): The images were processed at 100% scale with accuracy set to ‘high’.
- buildDepthMaps(): To evaluate the effectiveness of MMM, the images were processed at 1/4 scale (1500 × 1000 pixels) with quality set to ‘medium’.
- 4.
- merge_subsampling.py
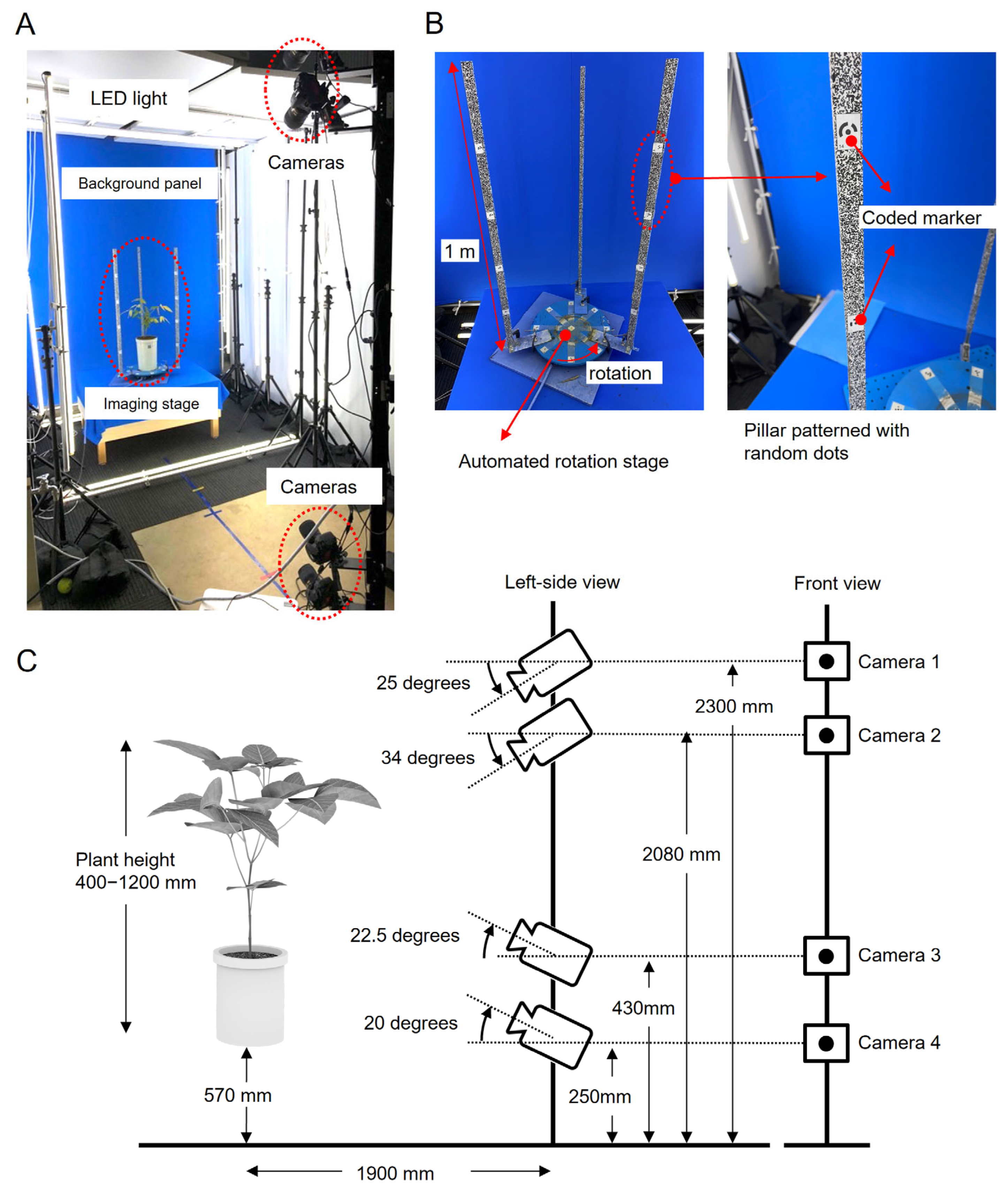
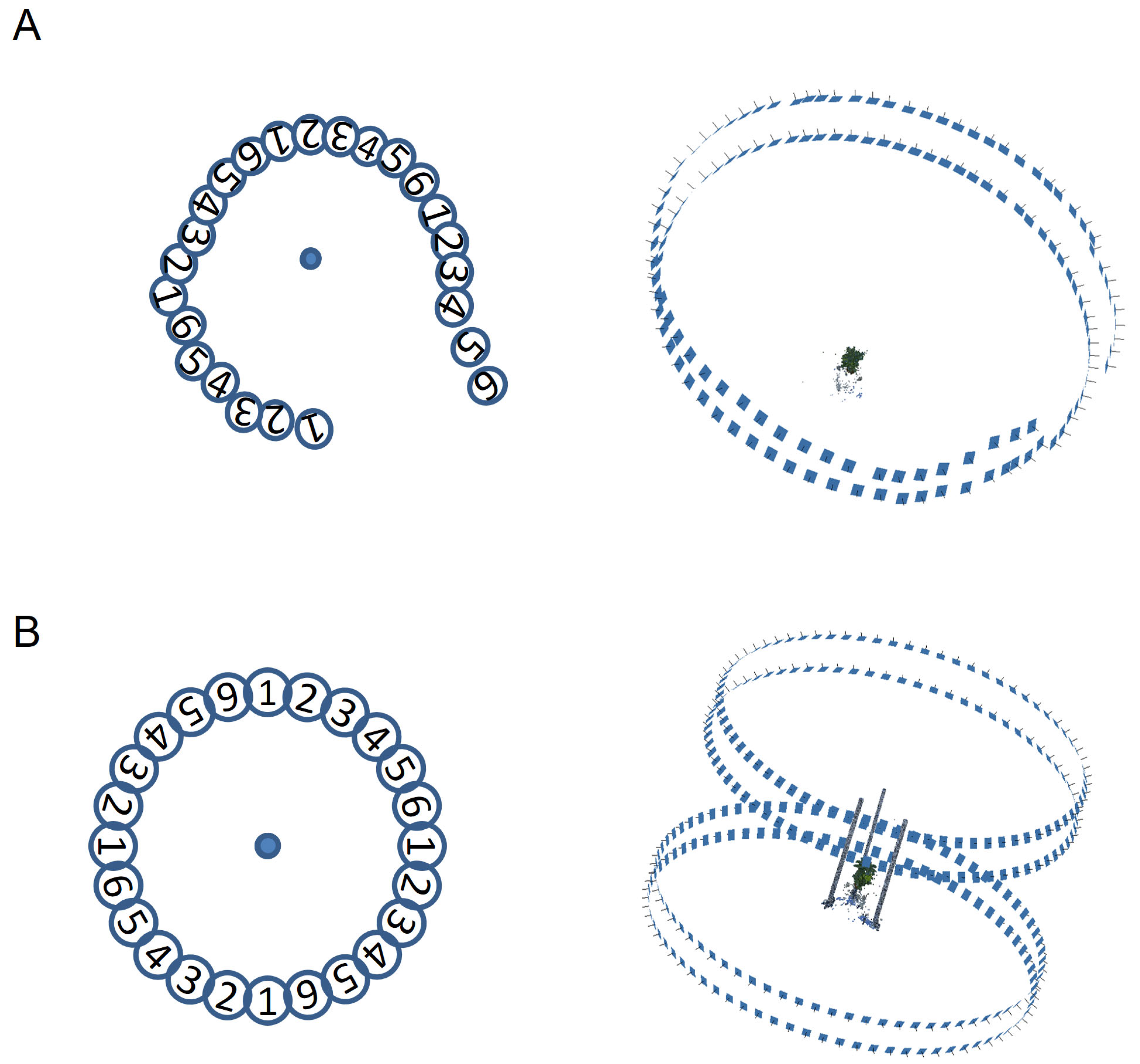
2.4. Imaging
3. Results
3.1. ‘baseCloud’ Construction Process in MMM
3.2. Viewing Angle of the Mask Image Used in the MMM Point Removal Process
3.3. Three-Dimensional Point Cloud Reconstruction of Soybean Plants
3.4. Effects of the Point Removal Processes in MMM
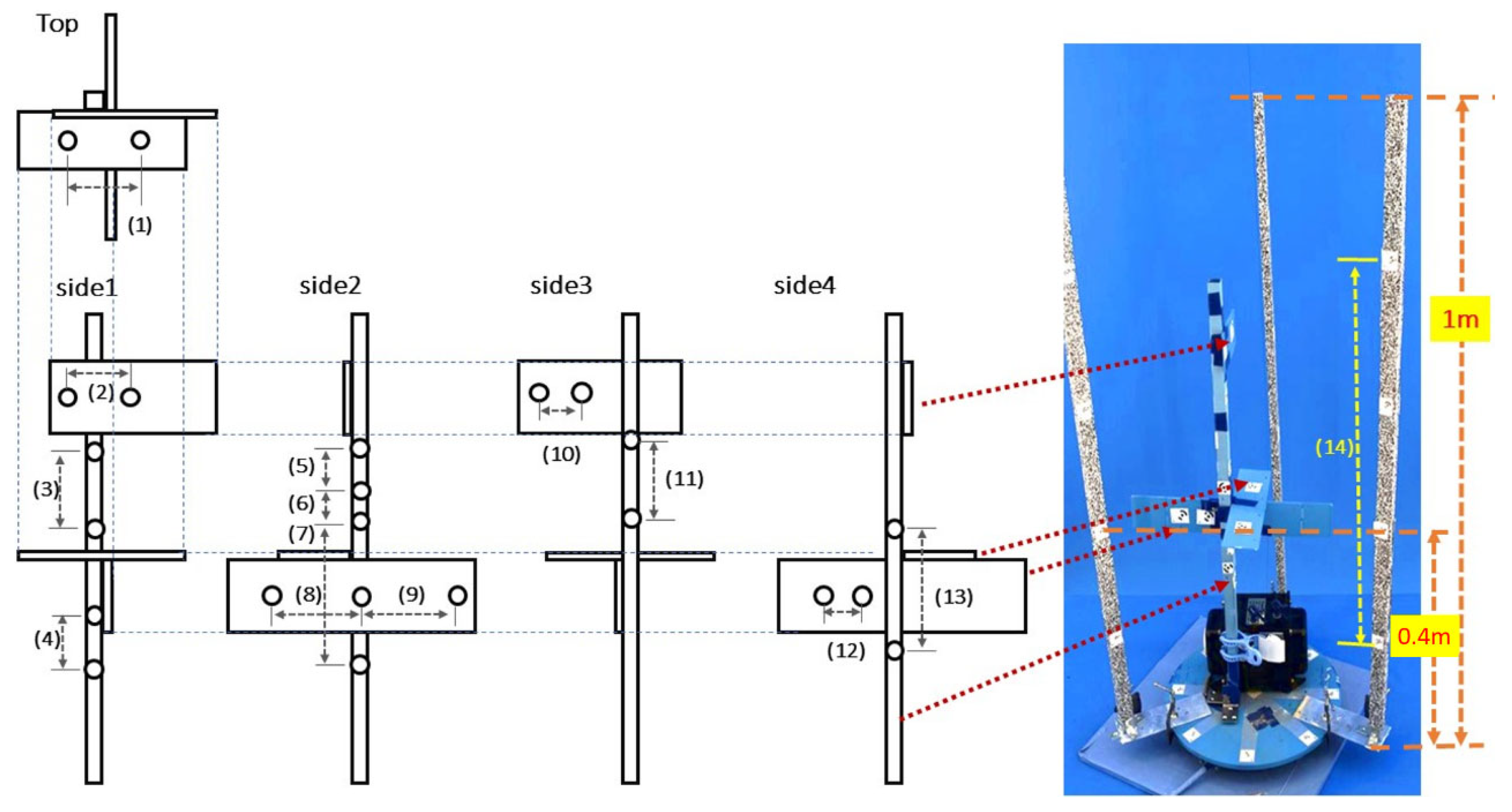
3.5. Validation of Position Accuracy in the Reconstructed Point Cloud
4. Discussion
4.1. Generating Local 3D Points and Removing Noise with Mask Images
4.2. Enhancing Camera Position Accuracy for All-Around Images
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Z.; Guo, R.; Li, M.; Chen, Y.; Li, G. A review of computer vision technologies for plant phenotyping. Comput. Electron. Agric. 2020, 176, 105672. [Google Scholar] [CrossRef]
- Yang, W.; Feng, H.; Zhang, X.; Zhang, J.; Doonan, J.H.; Batchelor, W.D.; Xiong, L.; Yan, J. Crop Phenomics and High-Throughput Phenotyping: Past Decades, Current Challenges, and Future Perspectives. Mol. Plant 2020, 13, 187–214. [Google Scholar] [CrossRef]
- Okura, F. 3D modeling and reconstruction of plants and trees: A cross-cutting review across computer graphics, vision, and plant phenotyping. Breed. Sci. 2022, 72, 31–47. [Google Scholar] [CrossRef]
- Zhang, Y.; Teng, P.; Aono, M.; Shimizu, Y.; Hosoi, F.; Omasa, K. 3D monitoring for plant growth parameters in field with a single camera by multi-view approach. J. Agric. Meteorol. 2018, 74, 129–139. [Google Scholar] [CrossRef]
- Kunita, I.; Morita, M.T.; Toda, M.; Higaki, T. A Three-Dimensional Scanning System for Digital Archiving and Quantitative Evaluation of Arabidopsis Plant Architectures. Plant Cell Physiol. 2021, 62, 1975–1982. [Google Scholar] [CrossRef]
- Akhtar, M.S.; Zafar, Z.; Nawaz, R.; Fraz, M.M. Unlocking plant secrets: A systematic review of 3D imaging in plant phenotyping techniques. Comput. Electron. Agric. 2024, 222, 109033. [Google Scholar] [CrossRef]
- Li, S.; Cui, Z.; Yang, J.; Wang, B. A Review of Optical-Based Three-Dimensional Reconstruction and Multi-Source Fusion for Plant Phenotyping. Sensors 2025, 25, 3401. [Google Scholar] [CrossRef]
- Paulus, S. Measuring crops in 3D: Using geometry for plant phenotyping. Plant Methods 2019, 15, 103. [Google Scholar] [CrossRef] [PubMed]
- Kochi, N.; Hayashi, A.; Shinohara, Y.; Tanabata, T.; Kodama, K.; Isobe, S. All-around 3D plant modeling system using multiple images and its composition. Breed. Sci. 2022, 72, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Salter, W.T.; Shrestha, A.; Barbour, M.M. Open source 3D phenotyping of chickpea plant architecture across plant development. Plant Methods 2021, 17, 95. [Google Scholar] [CrossRef]
- Snavely, N. Bundler: Structure from Motion (sfm) for Unordered Image Collections. Available online: https://www.cs.cornell.edu/~snavely/bundler/ (accessed on 2 April 2025).
- Wu, C. Towards Linear-Time Incremental Structure from Motion. In Proceedings of the 2013 International Conference on 3D Vision, Seattle, WA, USA, 29 June–1 July 2013. [Google Scholar] [CrossRef]
- Furukawa, Y.; Ponce, J. Accurate, Dense, and Robust Multiview Stereopsis. IEEE Trans. Pattern Anal. Mach. Intell. 2010, 32, 1362–1376. [Google Scholar] [CrossRef]
- Townsend, A.J.; Retkute, R.; Chinnathambi, K.; Randall, J.W.P.; Foulkes, J.; Carmo-Silva, E.; Murchie, E.H. Suboptimal Acclimation of Photosynthesis to Light in Wheat Canopies. Plant Physiol. 2018, 176, 1233–1246. [Google Scholar] [CrossRef]
- Santos, T.T.; Oliveira, A.A.d. Image-based 3D digitizing for plant architecture analysis and phenotyping. In Proceedings of the Workshop on Industry Applications (WGARI) in SIBGRAPI 2012 (XXV Conference on Graphics, Patterns and Images), Ouro Preto, Barzil, 22–25 August 2012. [Google Scholar] [CrossRef]
- Gibbs, J.A.; Pound, M.P.; French, A.P.; Wells, D.M.; Murchie, E.H.; Pridmore, T.P. Active Vision and Surface Reconstruction for 3D Plant Shoot Modelling. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 17, 1907–1917. [Google Scholar] [CrossRef] [PubMed]
- Barazzetti, L.; Forlani, G.; Remondino, F.; Roncella, R.; Scaioni, M. Experiences and achievements in automated image sequence orientation for close-range photogrammetric projects. In Proceedings of the Videometrics, Range Imaging, and Applications XI, Munich, Germany, 25–26 May 2011; p. 8085. [Google Scholar] [CrossRef]
- Anai, T.; Kochi, N.; Yamada, M.; Sasaki, T.; Otani, H.; Sasaki, D.; Nishimura, S.; Kimoto, K.; Yasui, N. Examination about influence for precision of 3D image measurement from the ground control point measurement and surface matching. In Proceedings of the Joint Workshop with ISPRS WG IV/7 and WG V/4, Tokyo, Japan, 21–22 May 2015; pp. 165–170. [Google Scholar] [CrossRef]
- Sattler, T.; Torii, A.; Sivic, J.; Pollefeys, M.; Taira, H.; Okutomi, M.; Pajdla, T. Are Large-Scale 3D Models Really Necessary for Accurate Visual Localization? In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1637–1646. [Google Scholar]
- Scaramuzza, D.; Fraundorfer, F.; Pollefeys, M. Closing the loop in appearance-guided omnidirectional visual odometry by using vocabulary trees. Robot. Auton. Syst. 2010, 58, 820–827. [Google Scholar] [CrossRef]
- Havlena, M.; Torii, A.; Pajdla, T. Efficient Structure from Motion by Graph Optimization. In Proceedings of the 11th European Conference on Computer Vision, Crete, Greece, 5–11 September 2010; pp. 110–113. [Google Scholar] [CrossRef]
- Sweeney, C.; Sattler, T.; Höllerer, T.; Turk, M.; Pollefeys, M. Optimizing the Viewing Graph for Structure-from-Motion. In Proceedings of the 2015 IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 801–809. [Google Scholar]
- Schönberger, J.L.; Frahm, J. Structure-from-Motion Revisited. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 4104–4113. [Google Scholar] [CrossRef]
- Zhang, Y.; Teng, P.; Shimizu, Y.; Hosoi, F.; Omasa, K. Estimating 3D leaf and stem shape of nursery paprika plants by a novel multi-camera photography system. Sensors 2016, 16, 874. [Google Scholar] [CrossRef] [PubMed]
- Sabov, A.; Krüger, J. Identification and correction of flying pixels in range camera data. In Proceedings of the 24th Spring Conference on Computer Graphics, Budmerice Castle, Slovakia, 21–23 April 2008; pp. 135–142. [Google Scholar]
- Harandi, N.; Vandenberghe, B.; Vankerschaver, J.; Depuydt, S.; Van Messem, A. How to make sense of 3D representations for plant phenotyping: A compendium of processing and analysis techniques. Plant Methods 2023, 19, 60. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, J.; Zhang, B.; Wang, Y.; Yao, J.; Zhang, X.; Fan, B.; Li, X.; Hai, Y.; Fan, X. Three-dimensional reconstruction and phenotype measurement of maize seedlings based on multi-view image sequences. Front. Plant Sci. 2022, 13, 974339. [Google Scholar] [CrossRef]
- Mildenhall, B.; Srinivasan, P.P.; Tancik, M.; Barron, J.T.; Ramamoorthi, R.; Ng, R. NeRF: Representing Scenes as Neural Radiance Fields for View Synthesis. arXiv 2020, arXiv:2003.08934. [Google Scholar] [CrossRef]
- Hu, K.; Ying, W.; Pan, Y.; Kang, H.; Chen, C. High-fidelity 3D reconstruction of plants using Neural Radiance Fields. Comput. Electron. Agric. 2024, 220, 108848. [Google Scholar] [CrossRef]
- Croce, V.; Billi, D.; Caroti, G.; Piemonte, A.; De Luca, L.; Véron, P. Comparative Assessment of Neural Radiance Fields and Photogrammetry in Digital Heritage: Impact of Varying Image Conditions on 3D Reconstruction. Remote Sens. 2024, 16, 301. [Google Scholar] [CrossRef]
- Tsukamoto, Y.; Hayashi, A.; Tokuda, K.; Kochi, N. Comparative Verification of 3D Reconstructed Point Cloud Data: Comparison of AI Estimation and Multi-View Stereo Methods using Nerfstudio and Metashape. In Proceedings of the SICE Festival 2024 with Annual Conference, Kochi, Japan, 27–30 August 2024. FrAT3.2. [Google Scholar]
- Kochi, N.; Isobe, S.; Hayashi, A.; Kodama, K.; Tanabata, T. Introduction of All-Around 3D Modeling Methods for Investigation of Plants. Int. J. Autom. Technol. 2021, 15, 301–312. [Google Scholar] [CrossRef]
- KUROHIRA. Available online: https://www.gene.affrc.go.jp/databases-plant_search_detail.php?jp=27863 (accessed on 7 July 2025).
- FUKUYUTAKA. Available online: https://www.gene.affrc.go.jp/databases-plant_search_detail.php?jp=29668 (accessed on 7 July 2025).
- HOUJAKU. Available online: https://www.gene.affrc.go.jp/databases-plant_search_detail.php?jp=28770 (accessed on 7 July 2025).
- Misuzudaizu. Available online: https://legumebase.nbrp.jp/glycine/rilParentStrainDetailAction.do?&strainId=0 (accessed on 7 July 2025).
- Agisoft Metashape. Available online: https://www.agisoft.com (accessed on 7 July 2025).
- Tanabata, T.; Yamada, T.; Shimizu, Y.; Shinozaki, Y.; Kanekatsu, M.; Takano, M. Development of automatic segmentation software for efficient measurement of area on the digital images of plant organs. Hortic. Res. 2010, 9, 501–506. [Google Scholar] [CrossRef][Green Version]
- Tanabata, T.; Shibaya, T.; Hori, K.; Ebana, K.; Yano, M. SmartGrain: High-throughput phenotyping software for measuring seed shape through image analysis. Plant Physiol. 2012, 160, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Yang, H.; Liu, D.; Wang, X. Research on automatic 3D reconstruction of plant phenotype based on Multi-View images. Comput. Electron. Agric. 2024, 220, 108866. [Google Scholar] [CrossRef]
- Zhu, R.; Sun, K.; Yan, Z.; Yan, X.; Yu, J.; Shi, J.; Hu, Z.; Jiang, H.; Xin, D.; Zhang, Z.; et al. Analysing the phenotype development of soybean plants using low-cost 3D reconstruction. Sci. Rep. 2020, 10, 7055. [Google Scholar] [CrossRef] [PubMed]
- Leménager, M.; Burkiewicz, J.; Schoen, D.J.; Joly, S. Studying flowers in 3D using photogrammetry. New Phytol. 2022, 237, 1922–1933. [Google Scholar] [CrossRef]
- Karara, H.M. Camera-Calibration in Non-Topographic Photogrammetry; American Society for Photogrammetry and Remote Sensing: Bethesda, MD, USA, 1989; Chapter 5. [Google Scholar]




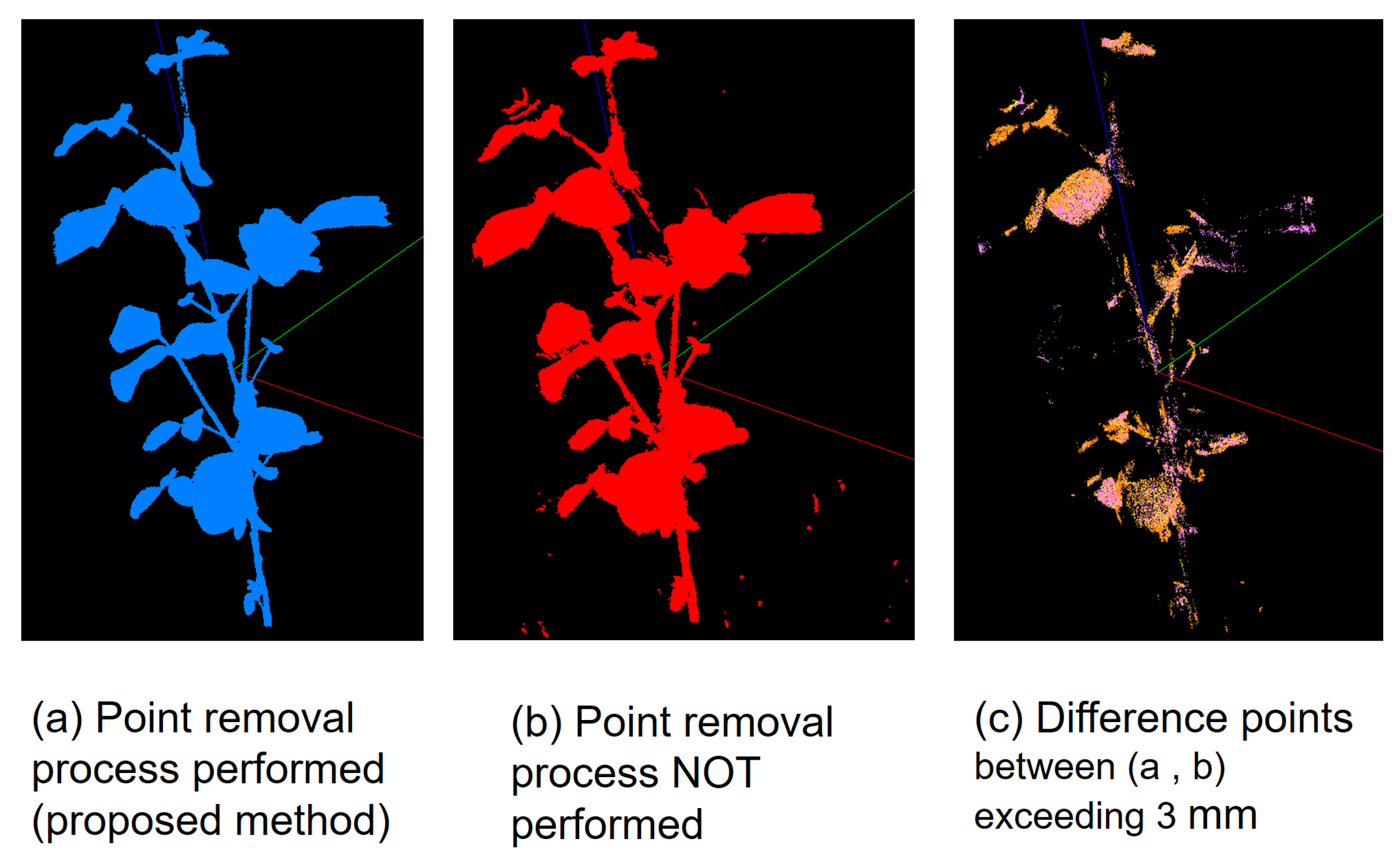

| Range (mm) | Number of Points | % |
|---|---|---|
| 0–3 | 1,732,066 | 95.7 |
| 3– | 78,637 | 4.3 |
| Total | 1,810,703 | 100.0 |
| Measurement Position | Caliper (mm) | CLCFM3 | Distance (mm) | ||
|---|---|---|---|---|---|
| Number | Direction | Avg(mm) | SD | ||
| (1) | width | 124.24 | 124.352 | 0.0000314 | −0.112 |
| (2) | width | 84.51 | 84.651 | 0.0000346 | −0.141 |
| (3) | height | 120.05 | 120.104 | 0.0000271 | −0.054 |
| (4) | height | 69.15 | 69.160 | 0.0000068 | −0.010 |
| (5) | height | 63.11 | 63.151 | 0.0000115 | −0.041 |
| (6) | height | 42.45 | 42.356 | 0.0000110 | 0.094 |
| (7) | height | 156.36 | 156.357 | 0.0000260 | 0.003 |
| (8) | width | 131.41 | 131.130 | 0.0000225 | 0.280 |
| (9) | width | 133.6 | 133.509 | 0.0000176 | 0.091 |
| (10) | width | 44.27 | 44.356 | 0.0000099 | −0.086 |
| (11) | height | 101.05 | 101.183 | 0.0000201 | −0.133 |
| (12) | width | 47.6 | 47.793 | 0.0000067 | −0.193 |
| (13) | height | 144.38 | 144.579 | 0.0000191 | −0.199 |
| (14) | height | 593 | 593.970 | 0.0001225 | −0.970 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hayashi, A.; Kochi, N.; Kodama, K.; Isobe, S.; Tanabata, T. CLCFM3: A 3D Reconstruction Algorithm Based on Photogrammetry for High-Precision Whole Plant Sensing Using All-Around Images. Sensors 2025, 25, 5829. https://doi.org/10.3390/s25185829
Hayashi A, Kochi N, Kodama K, Isobe S, Tanabata T. CLCFM3: A 3D Reconstruction Algorithm Based on Photogrammetry for High-Precision Whole Plant Sensing Using All-Around Images. Sensors. 2025; 25(18):5829. https://doi.org/10.3390/s25185829
Chicago/Turabian StyleHayashi, Atsushi, Nobuo Kochi, Kunihiro Kodama, Sachiko Isobe, and Takanari Tanabata. 2025. "CLCFM3: A 3D Reconstruction Algorithm Based on Photogrammetry for High-Precision Whole Plant Sensing Using All-Around Images" Sensors 25, no. 18: 5829. https://doi.org/10.3390/s25185829
APA StyleHayashi, A., Kochi, N., Kodama, K., Isobe, S., & Tanabata, T. (2025). CLCFM3: A 3D Reconstruction Algorithm Based on Photogrammetry for High-Precision Whole Plant Sensing Using All-Around Images. Sensors, 25(18), 5829. https://doi.org/10.3390/s25185829







