Bridging the Methodological Gap Between Inertial Sensors and Optical Motion Capture: Deep Learning as the Path to Accurate Joint Kinematic Modelling Using Inertial Sensors
Abstract
1. Introduction
2. Methods
2.1. Participants
2.2. Data Collection
2.3. Data Pre-Processing
2.4. Sensor Combination
2.5. Model Architecture
2.6. Custom Biomech Loss Function
2.7. Early-Stopping Configuration
2.8. Model Evaluation
2.9. External Validation
3. Results
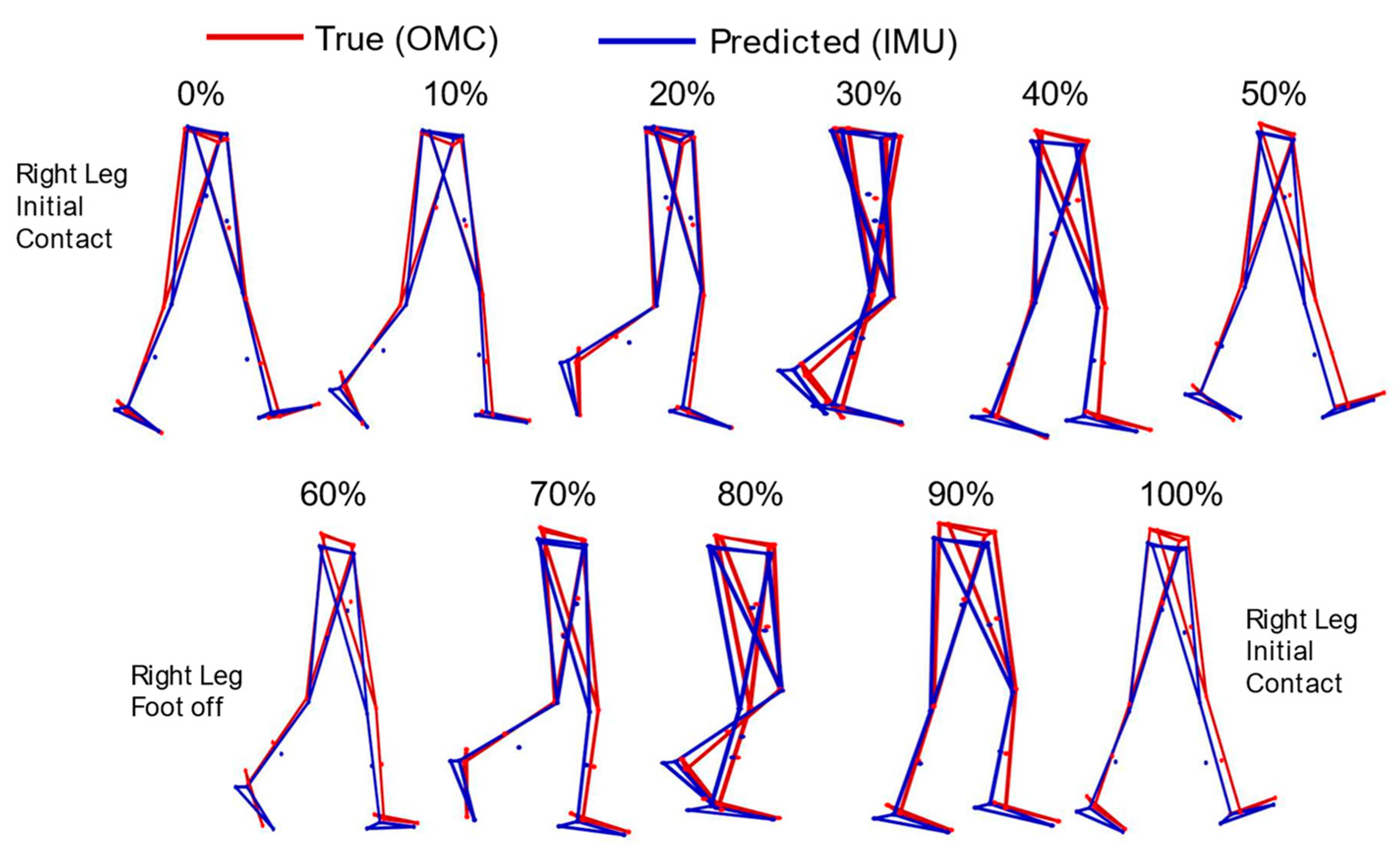
3.1. Marker Positions
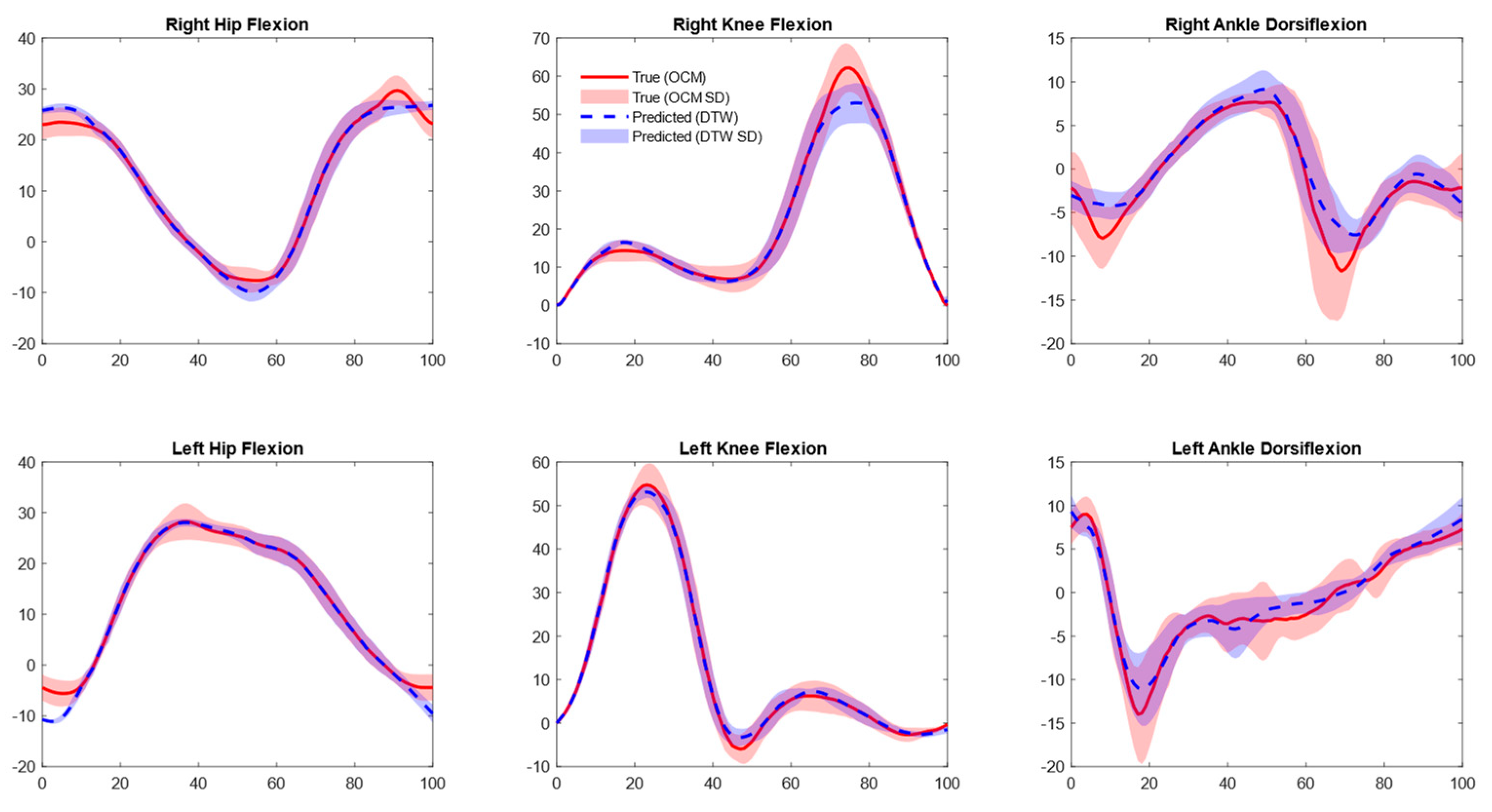
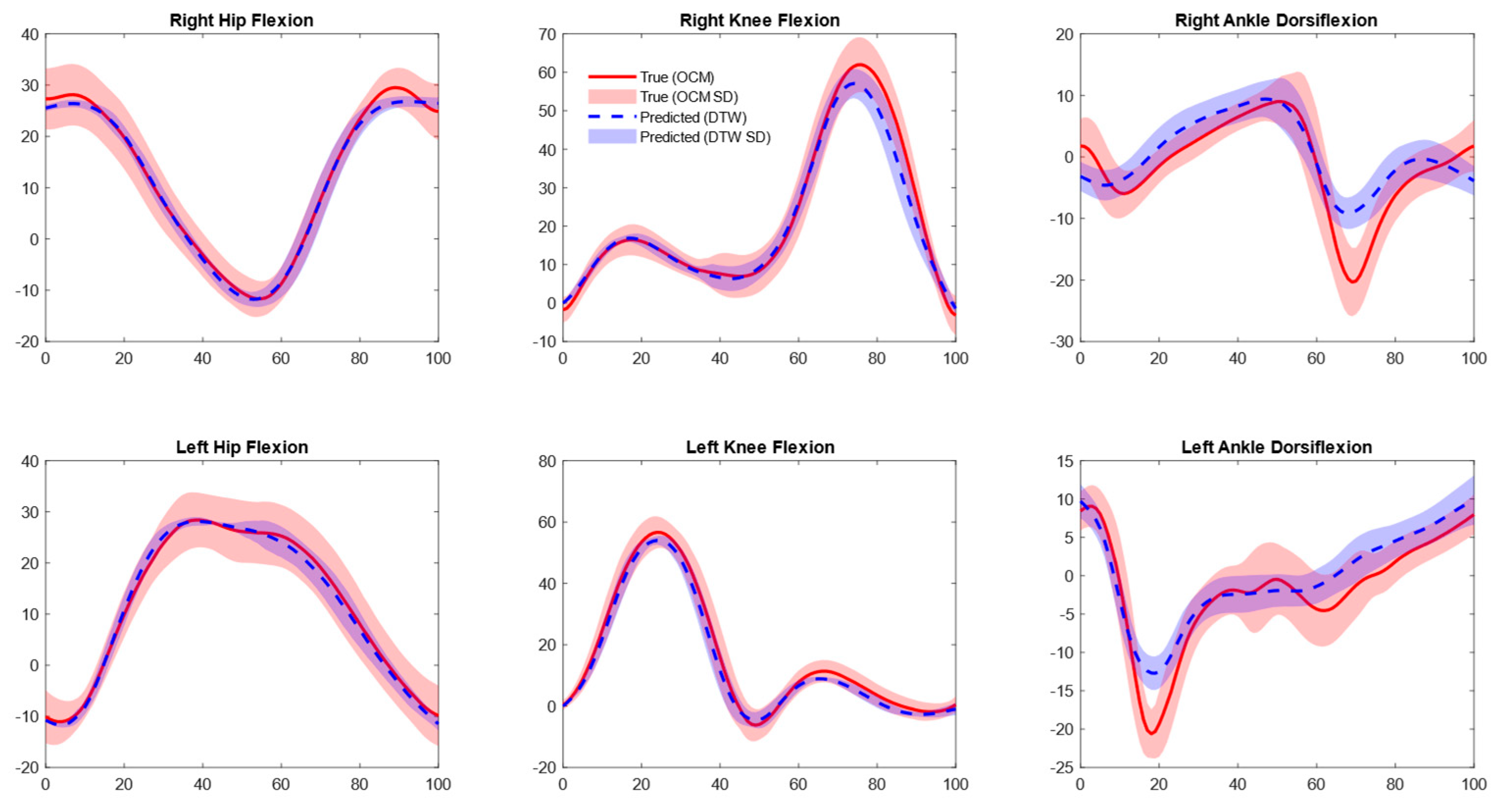
3.2. Joint Kinematics
4. Discussion
4.1. Marker Positions
4.2. OMC Vs. IMU Joint Kinematics
4.3. External Validation and Generalizability
4.4. Conclusion, Limitation and Outlook
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gurchiek, R.D.; Cheney, N.; McGinnis, R.S. McGinnis. Estimating Biomechanical Time-Series with Wearable Sensors: A Systematic Review of Machine Learning Techniques. Sensors 2019, 19, 5227. [Google Scholar] [CrossRef]
- Halilaj, E.; Rajagopal, A.; Fiterau, M.; Hicks, J.L.; Hastie, T.J.; Delp, S.L. Machine learning in human movement biomechanics: Best practices, common pitfalls, and new opportunities. J. Biomech. 2018, 81, 1–11. [Google Scholar] [CrossRef]
- Vienne, A.; Barrois, R.P.; Buffat, S.; Ricard, D.; Vidal, P.-P. Inertial Sensors to Assess Gait Quality in Patients with Neurological Disorders: A Systematic Review of Technical and Analytical Challenges. Front. Psychol. 2017, 8, 817. [Google Scholar] [CrossRef]
- Fullenkamp, A.M.; Tolusso, D.V.; Laurent, C.M.; Campbell, B.M.; Cripps, A.E. A Comparison of Both Motorized and Nonmotorized Treadmill Gait Kinematics to Overground Locomotion. J. Sport Rehabil. 2017, 27, 357–363. [Google Scholar] [CrossRef]
- Renggli, D.; Graf, C.; Tachatos, N.; Singh, N.; Meboldt, M.; Taylor, W.R.; Stieglitz, L.; Daners, M.S. Wearable Inertial Measurement Units for Assessing Gait in Real-World Environments. Front. Physiol. 2020, 11, 90. [Google Scholar] [CrossRef]
- Carcreff, L.; Gerber, C.N.; Paraschiv-Ionescu, A.; De Coulon, G.; Newman, C.J.; Aminian, K.; Armand, S. Comparison of gait characteristics between clinical and daily life settings in children with cerebral palsy. Sci. Rep. 2020, 10, 2091. [Google Scholar] [CrossRef] [PubMed]
- Adesida, Y.; Papi, E.; McGregor, A.H.; McGregor, A.H.; McGregor, A.H. Exploring the Role of Wearable Technology in Sport Kinematics and Kinetics: A Systematic Review. Sensors 2019, 19, 1597. [Google Scholar] [CrossRef] [PubMed]
- Rana, M.; Mittal, V.; Mittal, V.; Mittal, V. Wearable Sensors for Real-Time Kinematics Analysis in Sports: A Review. IEEE Sens. J. 2020, 21, 1187–1207. [Google Scholar] [CrossRef]
- Weygers, I.; Kok, M.; Konings, M.J.; Hallez, H.; De Vroey, H.; Claeys, K. Inertial Sensor-Based Lower Limb Joint Kinematics: A Methodological Systematic Review. Sensors 2020, 20, 673. [Google Scholar] [CrossRef] [PubMed]
- Al Borno, M.; O’dAy, J.; Ibarra, V.; Dunne, J.; Seth, A.; Habib, A.; Ong, C.; Hicks, J.; Uhlrich, S.; Delp, S. OpenSense: An open-source toolbox for inertial-measurement-unit-based measurement of lower extremity kinematics over long durations. J. Neuroeng. Rehabil. 2022, 19, 22. [Google Scholar] [CrossRef]
- Mundt, M.; Thomsen, W.; David, S.; Dupré, T.; Bamer, F.; Potthast, W.; Markert, B. Assessment of the measurement accuracy of inertial sensors during different tasks of daily living. J. Biomech. 2019, 84, 81–86. [Google Scholar] [CrossRef]
- Nüesch, C.; Roos, E.; Pagenstert, G.; Mündermann, A. Measuring joint kinematics of treadmill walking and running: Comparison between an inertial sensor based system and a camera-based system. J. Biomech. 2017, 57, 32–38. [Google Scholar] [CrossRef]
- Robert-Lachaine, X.; Turcotte, R.; Dixon, P.C.; Pearsall, D.J. Impact of hockey skate design on ankle motion and force production. Sports Eng. 2012, 15, 197–206. [Google Scholar] [CrossRef]
- Schepers, M.; Giuberti, M.; Bellusci, G. Xsens MVN: Consistent Tracking of Human Motion Using Inertial Sensing. Xsens Technol 2018, 1, 1–8. [Google Scholar] [CrossRef]
- Shah, V.R.; Dixon, P.C.; Willmott, A.P. Evaluation of lower-body gait kinematics on outdoor surfaces using wearable sensors. J. Biomech. 2024, 177, 112401. [Google Scholar] [CrossRef] [PubMed]
- Dorschky, E.; Nitschke, M.; Martindale, C.F.; van den Bogert, A.J.; Koelewijn, A.D.; Eskofier, B.M. CNN-Based Estimation of Sagittal Plane Walking and Running Biomechanics from Measured and Simulated Inertial Sensor Data. Front. Bioeng. Biotechnol. 2020, 8, 604. [Google Scholar] [CrossRef]
- Mundt, M.; Johnson, W.R.; Potthast, W.; Markert, B.; Mian, A.; Alderson, J. A Comparison of Three Neural Network Approaches for Estimating Joint Angles and Moments from Inertial Measurement Units. Sensors 2021, 21, 4535. [Google Scholar] [CrossRef]
- Shah, V.R.; Dixon, P.C. Gait Speed and Task Specificity in Predicting Lower-Limb Kinematics: A Deep Learning Approach Using Inertial Sensors. Mayo Clin. Proc. Digit. Health 2025, 3, 100183. [Google Scholar] [CrossRef]
- Senanayake, D.; Halgamuge, S.K.; Ackland, D.C. Real-time conversion of inertial measurement unit data to ankle joint angles using deep neural networks. J. Biomech. 2021, 125, 110552. [Google Scholar] [CrossRef]
- Sung, J.; Han, S.; Park, H.; Cho, H.-M.; Hwang, S.; Park, J.W.; Youn, I. Prediction of Lower Extremity Multi-Joint Angles during Overground Walking by Using a Single IMU with a Low Frequency Based on an LSTM Recurrent Neural Network. Sensors 2021, 22, 53. [Google Scholar] [CrossRef]
- Tan, J.-S.; Tippaya, S.; Binnie, T.; Davey, P.; Napier, K.; Caneiro, J.P.; Kent, P.; Smith, A.; O’sullivan, P.; Campbell, A. Predicting Knee Joint Kinematics from Wearable Sensor Data in People with Knee Osteoarthritis and Clinical Considerations for Future Machine Learning Models. Sensors 2022, 22, 446. [Google Scholar] [CrossRef]
- Rapp, E.; Shin, S.; Thomsen, W.; Ferber, R.; Halilaj, E. Estimation of kinematics from inertial measurement units using a combined deep learning and optimization framework. J. Biomech. 2021, 116, 110229. [Google Scholar] [CrossRef]
- Renani, M.S.; Eustace, A.M.; Myers, C.A.; Clary, C.W. The Use of Synthetic IMU Signals in the Training of Deep Learning Models Significantly Improves the Accuracy of Joint Kinematic Predictions. Sensors 2021, 21, 5876. [Google Scholar] [CrossRef] [PubMed]
- Warmerdam, E.; Hansen, C.; Romijnders, R.; Hobert, M.A.; Welzel, J.; Maetzler, W. Full-Body Mobility Data to Validate Inertial Measurement Unit Algorithms in Healthy and Neurological Cohorts. Data 2022, 7, 136. [Google Scholar] [CrossRef]
- Kadaba, M.P.; Ramakrishnan, H.K.; Wootten, M.E. Measurement of lower extremity kinematics during level walking. J. Orthop. Res. 1990, 8, 383–392. [Google Scholar] [CrossRef]
- Ippersiel, P.; Shah, V.; Dixon, P.C. The impact of outdoor walking surfaces on lower-limb coordination and variability during gait in healthy adults. Gait Posture 2022, 91, 7–13. [Google Scholar] [CrossRef]
- Shah, V.; Flood, M.W.; Grimm, B.; Dixon, P.C. Generalizability of deep learning models for predicting outdoor irregular walking surfaces. J. Biomech. 2022, 139, 111159. [Google Scholar] [CrossRef]
- Yu, B.; Gabriel, D.; Noble, L.; An, K.-N. Estimate of the Optimum Cutoff Frequency for the Butterworth Low-Pass Digital Filter. J. Appl. Biomech. 1999, 15, 319–329. [Google Scholar] [CrossRef]
- Google Colab. Available online: https://colab.research.google.com/ (accessed on 12 July 2024).
- tf.keras.callbacks.EarlyStopping, v2.16.1; TensorFlow. Available online: https://www.tensorflow.org/api_docs/python/tf/keras/callbacks/EarlyStopping (accessed on 16 December 2024).
- Dixon, P.C.; Loh, J.J.; Michaud-Paquette, Y.; Pearsall, D.J. Biomechzoo: An open-source toolbox for the processing, analysis, and visualization of biomechanical movement data. Comput. Methods Programs Biomed. 2017, 140, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, J.O.; Silverman, B.W. Functional Data Analysis; Springer Series in Statistics; Springer: New York, NY, USA, 2005. [Google Scholar] [CrossRef]
- Yurtman, A.; Robberechts, P.; Vohl, D.; Ma, E.; Verbruggen, G.; Rossi, M.; Shaikh, M.; Yasirroni, M.; Zieliński, W.; Van Craenendonck, T.; et al. wannesm/dtaidistance; v2.3.5; Zenodo, 2022; Available online: https://zenodo.org/records/5901139 (accessed on 11 September 2025).
- Meert, W. Dtaidistance: Distance Measures for Time Series (Dynamic Time Warping, Fast C Implementation). Python. Available online: https://github.com/wannesm/dtaidistance (accessed on 10 March 2024).
- Chénier, F. Kinetics Toolkit: An Open-Source Python Package to Facilitate Research in Biomechanics. J. Open Source Softw. 2021, 6, 3714. [Google Scholar] [CrossRef]
- Saegner, K.; Romijnders, R.; Hansen, C.; Holder, J.; Warmerdam, E.; Maetzler, W. Inter-joint coordination with and without dopaminergic medication in Parkinson’s disease: A case-control study. J. Neuroeng. Rehabil. 2024, 21, 118. [Google Scholar] [CrossRef]
- Uhlrich, S.D.; Falisse, A.; Kidziński, Ł.; Muccini, J.; Ko, M.; Chaudhari, A.S.; Hicks, J.L.; Delp, S.L.; Marsden, A.L. OpenCap: 3D human movement dynamics from smartphone videos. bioRxiv 2022. [Google Scholar] [CrossRef]
- Kanko, R.M.; Outerleys, J.B.; Laende, E.K.; Selbie, W.S.; Deluzio, K.J. Comparison of Concurrent and Asynchronous Running Kinematics and Kinetics from Marker-Based and Markerless Motion Capture Under Varying Clothing Conditions. J. Appl. Biomech. 2024, 40, 129–137. [Google Scholar] [CrossRef]
- Findlow, A.H.; Goulermas, J.Y.; Nester, C.J.; Howard, D.; Kenney, L. Predicting lower limb joint kinematics using wearable motion sensors. Gait Posture 2008, 28, 120–126. [Google Scholar] [CrossRef]
- Mundt, M.; Koeppe, A.; David, S.; Witter, T.; Bamer, F.; Potthast, W.; Markert, B. Estimation of Gait Mechanics Based on Simulated and Measured IMU Data Using an Artificial Neural Network. Front. Bioeng. Biotechnol. 2020, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Wouda, F.J.; Giuberti, M.; Bellusci, G.; Maartens, E.; Reenalda, J.; van Beijnum, B.-J.F.; Veltink, P.H. Estimation of Vertical Ground Reaction Forces and Sagittal Knee Kinematics During Running Using Three Inertial Sensors. Front. Physiol. 2018, 9, 218. [Google Scholar] [CrossRef] [PubMed]
- McGinley, J.L.; Baker, R.; Wolfe, R.; Morris, M.E. The reliability of three-dimensional kinematic gait measurements: A systematic review. Gait Posture 2009, 29, 360–369. [Google Scholar] [CrossRef] [PubMed]



| Definition | Markers | X (cm) | Y (cm) | Z (cm) |
|---|---|---|---|---|
| Left ASIS | LASI | 2.3 ± 0.8 | 2.4 ± 0.9 | 3.8 ± 2.0 |
| Right ASIS | RASI | 2.1 ± 0.9 | 2.6 ± 0.8 | 3.6 ± 1.8 |
| Left PSIS | LPSI | 2.3 ± 0.5 | 3.0 ± 1.5 | 3.1 ± 1.6 |
| Right PSIS | RPSI | 2.3 ± 0.7 | 3.1 ± 1.4 | 3.2 ± 1.7 |
| Left thigh | LTHI | 2.3 ± 0.6 | 3.2 ± 1.5 | 4.6 ± 2.7 |
| Left knee | LKNE | 2.9 ± 1.2 | 3.4 ± 1.2 | 3.0 ± 1.6 |
| Left tibia | LTIB | 2.7 ± 0.9 | 4.0 ± 1.8 | 3.3 ± 2.2 |
| Left ankle | LANK | 3.0 ± 1.4 | 4.3 ± 2.7 | 2.3 ± 0.8 |
| Left heel | LHEE | 2.5 ± 0.6 | 4.2 ± 2.5 | 2.4 ± 0.9 |
| Left toe | LTOE | 2.9 ± 1.0 | 4.4 ± 3.0 | 1.6 ± 0.7 |
| Right thigh | RTHI | 2.4 ± 0.8 | 3.8 ± 1.1 | 5.9 ± 3.4 |
| Right knee | RKNE | 2.2 ± 0.6 | 3.5 ± 1.5 | 2.7 ± 1.7 |
| Right tibia | RTIB | 2.1 ± 0.6 | 4.4 ± 2.4 | 3.1 ± 1.8 |
| Right ankle | RANK | 2.2 ± 0.5 | 4.5 ± 2.9 | 2.0 ± 1.0 |
| Right heel | RHEE | 2.6 ± 1.1 | 4.6 ± 3.1 | 2.3 ± 1.1 |
| Right toe | RTOE | 2.4 ± 0.9 | 4.9 ± 3.3 | 1.5 ± 0.5 |
| Average All Markers | 2.4 ± 0.8 | 3.8 ± 2.0 | 3.0 ± 1.6 | |
| Datasets | Joints | Right Hip | Right Knee | Right Ankle | Left Hip | Left Knee | Left Ankle |
|---|---|---|---|---|---|---|---|
| Same dataset | Non-DTW | 5.2 ± 1.4 | 5.8 ± 1.3 | 6.8 ± 2.5 | 4.5 ± 1.7 | 5.3 ± 2.2 | 5.4 ± 2.1 |
| DTW | 2.8 ± 1.2 | 2.3 ± 0.8 | 4.0 ± 1.9 | 1.8 ± 0.7 | 2.6 ± 1.1 | 2.1 ± 1.0 | |
| Collected separately | Non-DTW | 3.3 ± 0.6 | 3.6 ± 0.7 | 4.2 ± 0.6 | 2.8 ± 0.5 | 3.2 ± 0.5 | 4.2 ± 0.5 |
| DTW | 1.0 ± 0.2 | 1.4 ± 0.4 | 2.1 ± 0.4 | 0.5 ± 0.1 | 1.1 ± 0.2 | 0.8 ± 0.2 | |
| External Treadmill [24] | Non-DTW | 4.5 ± 1.5 | 7.6 ± 2.2 | 6.4 ± 1.9 | 4.0 ± 1.2 | 5.0 ± 1.4 | 5.4 ± 1.9 |
| DTW | 2.1 ± 0.7 | 3.2 ± 1.1 | 2.8 ± 1.4 | 2.1 ± 0.8 | 2.2 ± 0.8 | 2.0 ± 0.8 | |
| External Overground [24] | Non-DTW | 4.6 ± 2.2 | 7.2 ± 2.2 | 7.1 ± 2.2 | 6.0 ± 2.3 | 5.5 ± 1.9 | 6.0 ± 1.9 |
| DTW | 3.4 ± 1.2 | 4.7 ± 2.5 | 4.6 ± 2.4 | 4.1 ± 1.9 | 3.7 ± 1.8 | 3.9 ± 2.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shah, V.R.; Dixon, P.C. Bridging the Methodological Gap Between Inertial Sensors and Optical Motion Capture: Deep Learning as the Path to Accurate Joint Kinematic Modelling Using Inertial Sensors. Sensors 2025, 25, 5728. https://doi.org/10.3390/s25185728
Shah VR, Dixon PC. Bridging the Methodological Gap Between Inertial Sensors and Optical Motion Capture: Deep Learning as the Path to Accurate Joint Kinematic Modelling Using Inertial Sensors. Sensors. 2025; 25(18):5728. https://doi.org/10.3390/s25185728
Chicago/Turabian StyleShah, Vaibhav R., and Philippe C. Dixon. 2025. "Bridging the Methodological Gap Between Inertial Sensors and Optical Motion Capture: Deep Learning as the Path to Accurate Joint Kinematic Modelling Using Inertial Sensors" Sensors 25, no. 18: 5728. https://doi.org/10.3390/s25185728
APA StyleShah, V. R., & Dixon, P. C. (2025). Bridging the Methodological Gap Between Inertial Sensors and Optical Motion Capture: Deep Learning as the Path to Accurate Joint Kinematic Modelling Using Inertial Sensors. Sensors, 25(18), 5728. https://doi.org/10.3390/s25185728







