MS-UNet: A Hybrid Network with a Multi-Scale Vision Transformer and Attention Learning Confusion Regions for Soybean Rust Fungus
Abstract
1. Introduction
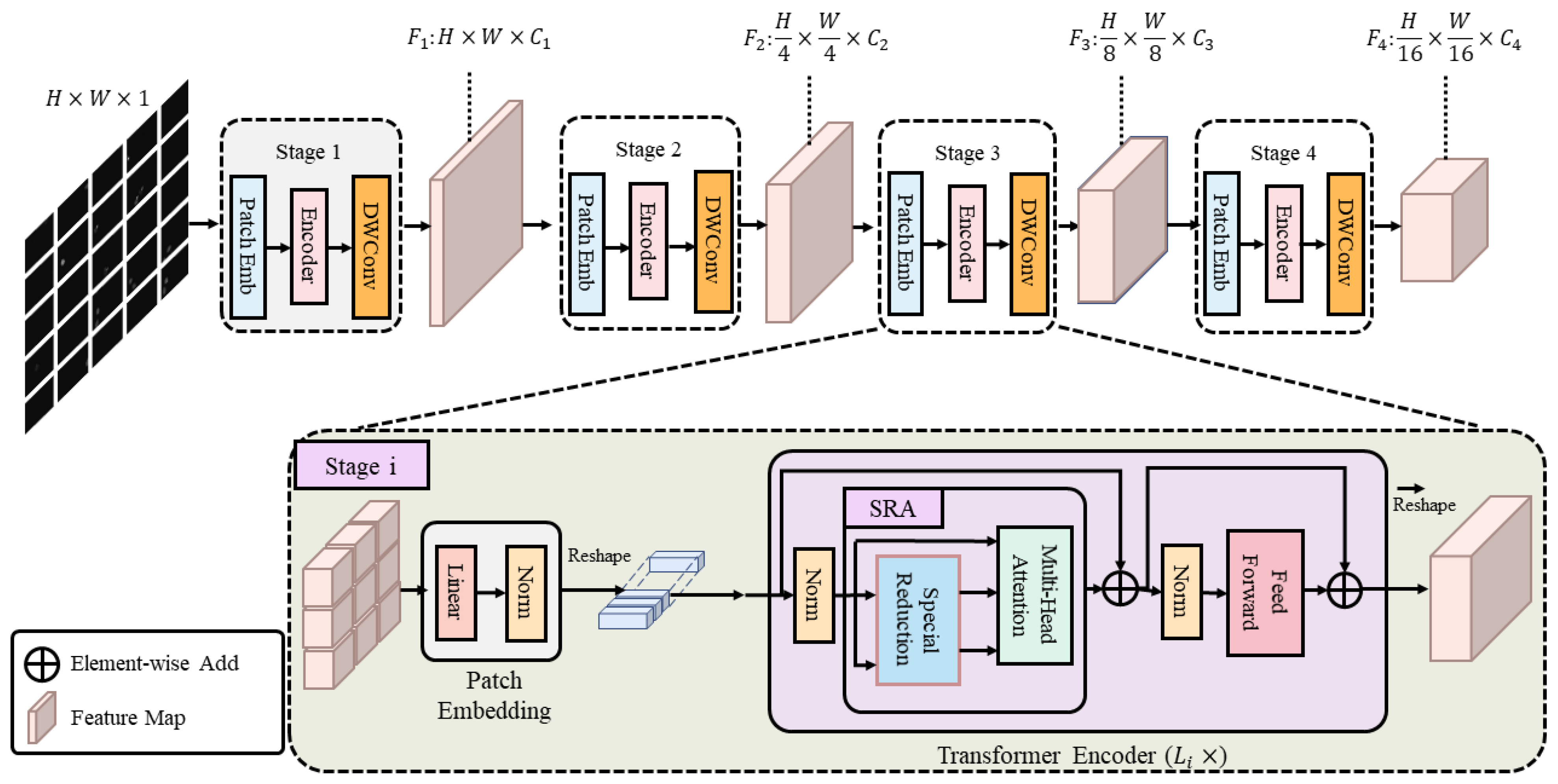
- We design a lightweight progressive hierarchical ViT encoder capable of learning multi-scale and high-resolution features;
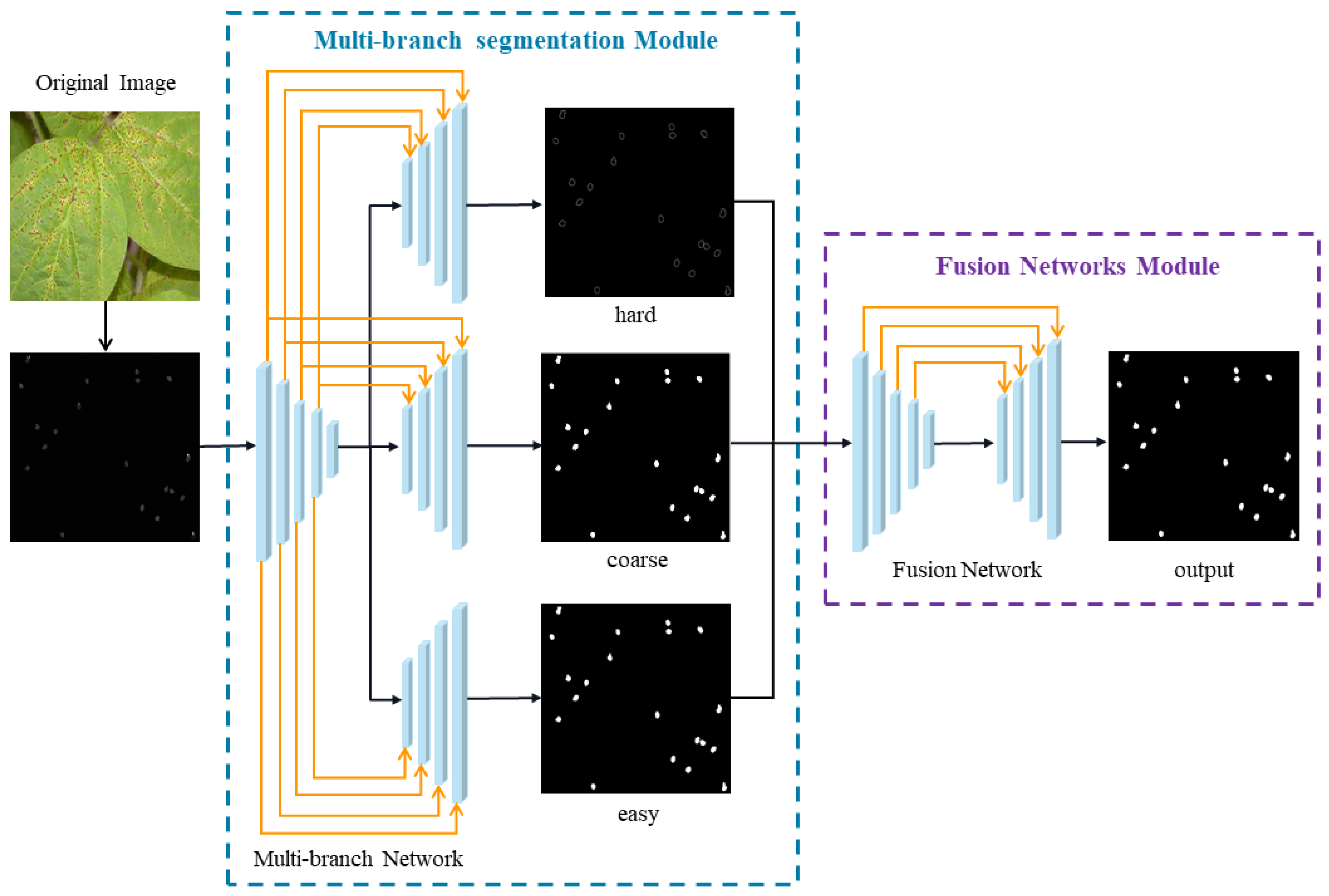
- We develop a multi-branch decoder that leverages low-confidence pixels in the output to compel the network to focus on challenging regions that are difficult to segment;
- The proposed MS-UNet is tailored to the unique characteristics of soybean rust imagery and achieves satisfactory results on the PPS task.
2. Related Work
2.1. Semantic Segmentation
2.2. Vision Transformers
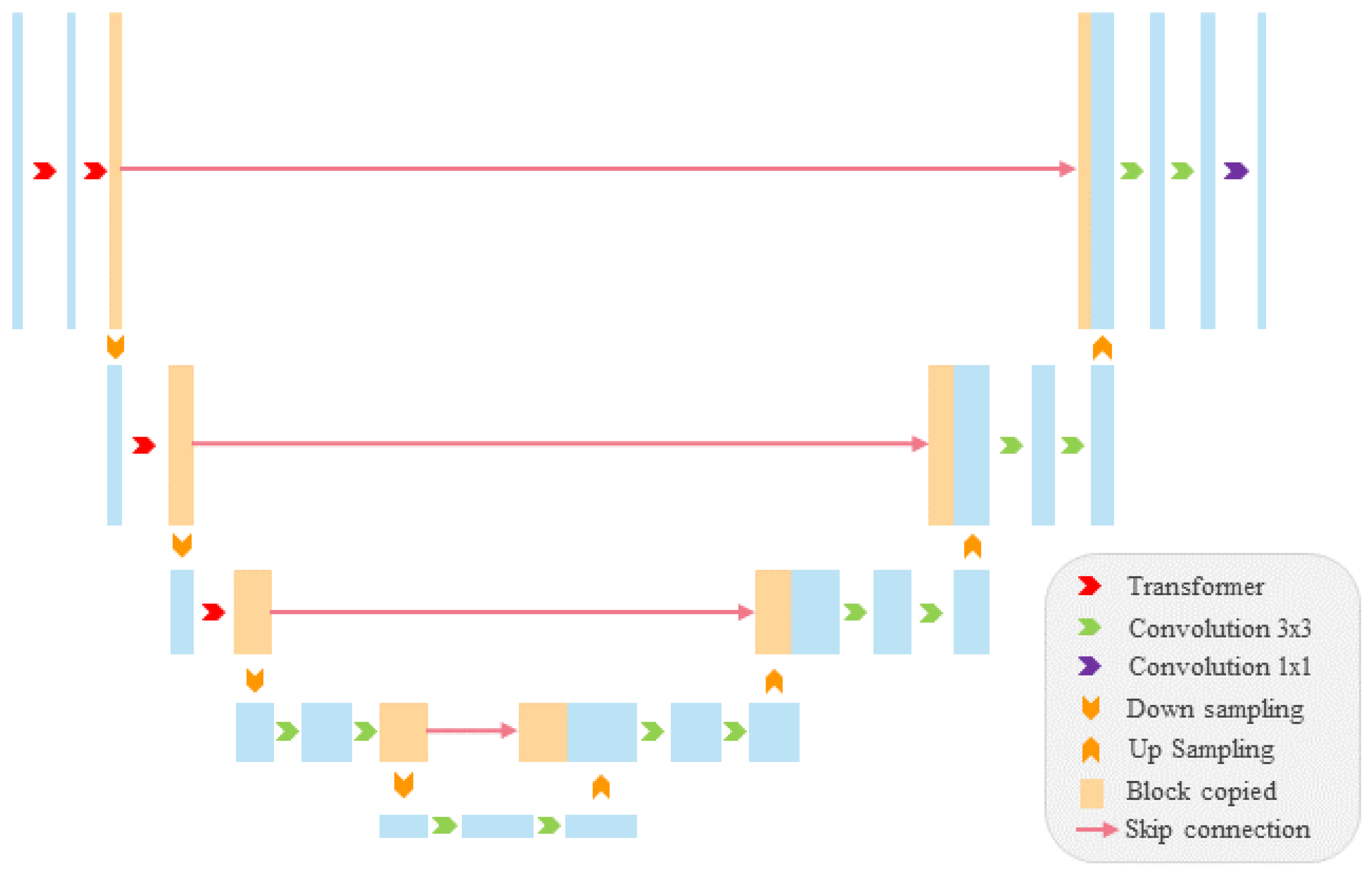
3. The Method
3.1. The Progressive Hierarchical VIT Encoder
3.2. The Multi-Branch Decoder
3.3. The Loss Function
4. Experiments
4.1. Databases
4.2. Evaluation Metrics
4.3. Implementation Details
5. Results and Discussion
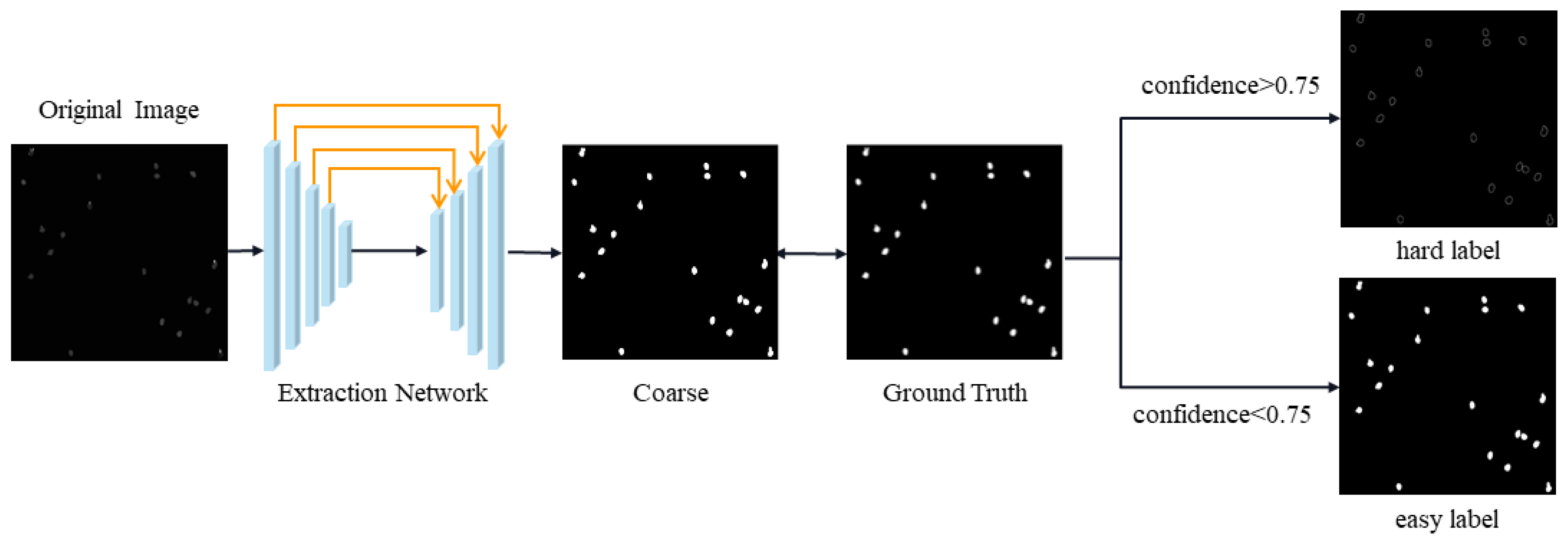
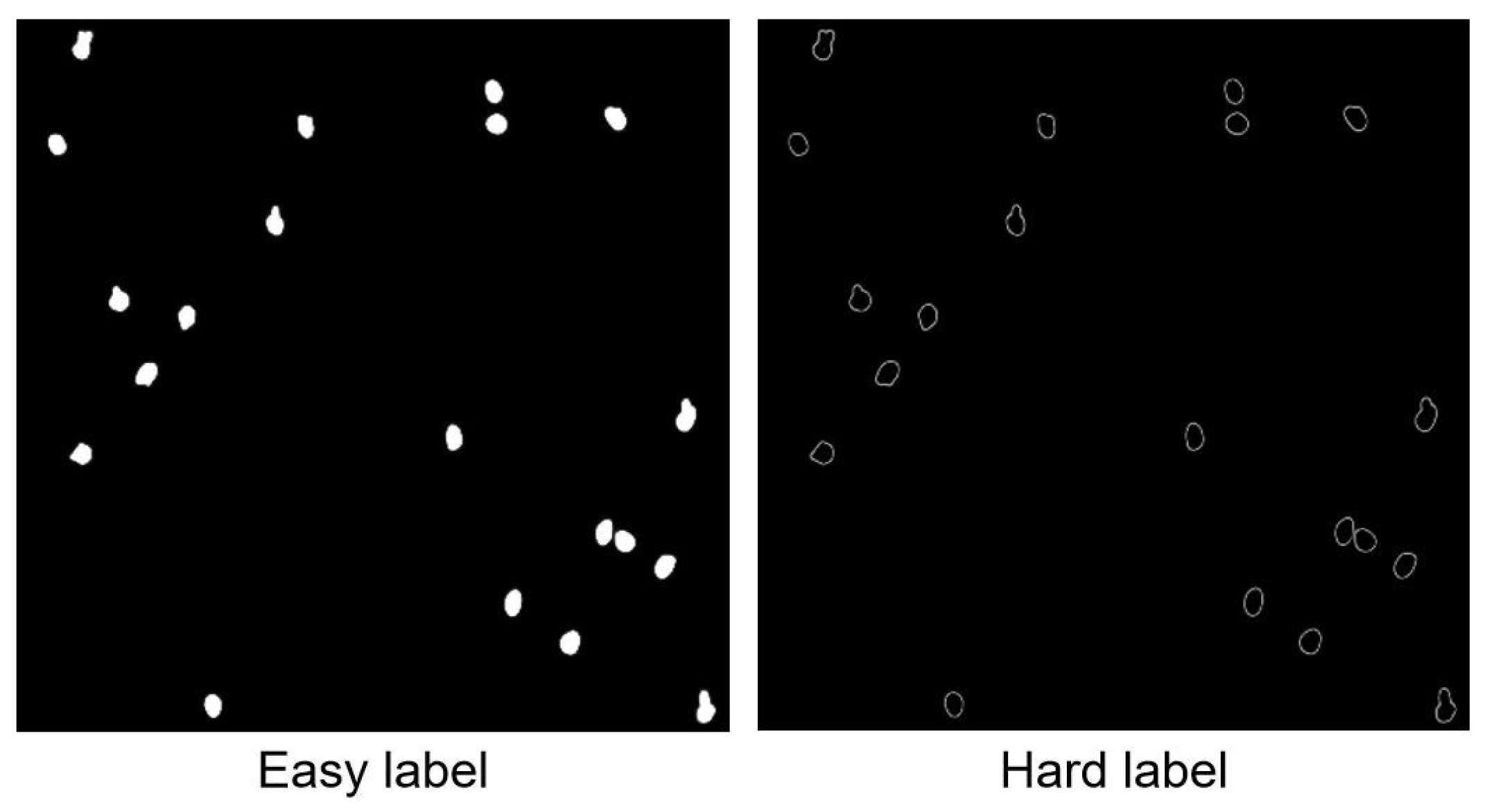
5.1. Experiments with Automatic Label Generation
5.2. Comparison with Other Methods
5.3. An Ablation Study on Individual Components
5.4. Ablation Experiments on the Threshold
5.5. Computational Analysis
6. Conclusions
- A segmentation dataset of Phakopsora pachyrhizi was produced manually for the related research.
- A multi-branch segmentation network was proposed to improve the segmentation accuracy in PPS by accurately segmenting difficult- and easy-to-segment regions from images of Phakopsora pachyrhizi.
- To ensure the segmentation performance of the multi-branch segmentation network, an upgraded U-Net network was proposed as the base segmentation network. Specifically, a combination of a transformer and convolution was used to create a progressive hierarchical ViT encoder capable of learning multi-scale and high-resolution features, resulting in a segmentation network suitable for the PPS problem.
- A fusion network was employed to solve the issue of overlapping regions in images of Phakopsora pachyrhizi by fusing two distinct sections of a multi-branch segmentation network.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cui, D.; Zhang, Q.; Li, M.; Hartman, G.L.; Zhao, Y. Image processing methods for quantitatively detecting soybean rust from multispectral images. Biosyst. Eng. 2010, 107, 186–193. [Google Scholar] [CrossRef]
- Beyer, S.F.; Beesley, A.; Rohmann, P.F.W.; Schultheiss, H.; Conrath, U.; Langenbach, C.J.G. The Arabidopsis non-host defence-associated coumarin scopoletin protects soybean from Asian soybean rust. Plant J. 2019, 99, 397–413. [Google Scholar] [CrossRef]
- Cavanagh, H.; Mosbach, A.; Scalliet, G.; Lind, R.; Endres, R.G. Physics-informed deep learning characterizes morphodynamics of Asian soybean rust disease. Nat. Commun. 2021, 12, 6424. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; IEEE: Washington, DC, USA, 2015; pp. 3431–3440. [Google Scholar]
- Yuan, Y.; Fu, R.; Huang, L.; Lin, W.; Zhang, C.; Chen, X.; Wang, J. Hrformer: High-resolution vision transformer for dense predict. Adv. Neural Inf. Process. Syst. 2021, 34, 7281–7293. [Google Scholar]
- Chen, L.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the Neural Information Processing Systems (NIPS), Long Beach, CA, USA, 4–9 December 2017; Volume 30. [Google Scholar]
- Guo, Y.; Liu, Y.; Georgiou, T.; Lew, M.S. A review of semantic segmentation using deep neural networks. Int. J. Multimed. Inf. Retr. 2018, 7, 87–93. [Google Scholar] [CrossRef]
- Zhao, Q.; Cao, J.; Ge, J.; Zhu, Q.; Chen, X.; Liu, W. Multi-UNet: An effective Multi-U convolutional networks for semantic segmentation. Knowl. Based Syst. 2025, 309, 11. [Google Scholar] [CrossRef]
- Tang, M.; He, Y.; Aslam, M.; Akpokodje, E.; Jilani, S.F. Enhanced U-Net for Improved Semantic Segmentation in Landslide Detection. Sensors 2025, 25, 2670. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Yang, Z.; Tan, L.; Wang, Y.; Sun, W.; Sun, M.; Tang, Y. Methods and datasets on semantic segmentation: A review. Neurocomputing 2018, 304, 82–103. [Google Scholar] [CrossRef]
- Farabet, C.; Couprie, C.; Najman, L.; LeCun, Y. Learning hierarchical features for scene labeling. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 35, 1915–1929. [Google Scholar] [CrossRef]
- Ning, F.; Delhomme, D.; LeCun, Y.; Piano, F.; Bottou, L.; Barbano, P.E. Toward automatic phenotyping of developing embryos from videos. IEEE Trans. Image Process. 2005, 14, 1360–1371. [Google Scholar] [CrossRef]
- Yuan, Y.; Chen, X.; Wang, J. Object-contextual representations for semantic segmentation. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2020; pp. 173–190. [Google Scholar]
- Siam, M.; Elkerdawy, S.; Jagersand, M.; Yogamani, S. Deep semantic segmentation for automated driving: Taxonomy, roadmap and challenges. In Proceedings of the 2017 IEEE 20th International Conference on Intelligent Transportation Systems (ITSC), Yokohama, Japan, 16–19 October 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 1–8. [Google Scholar]
- Sun, K.; Xiao, B.; Liu, D.; Wang, J. Deep high-resolution representation learning for human pose estimation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; IEEE: Washington, DC, USA, 2019; pp. 5693–5703. [Google Scholar]
- Xie, E.; Wang, W.; Yu, Z.; An kumar, A.; Alvarez, J.M.; Luo, P. SegFormer: Simple and efficient design for semantic segmentation with transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 12077–12090. [Google Scholar]
- Yurtkulu, S.C.; Şahin, Y.H.; Unal, G. Semantic segmentation with extended DeepLabv3 architecture. In Proceedings of the 2019 27th Signal Processing and Communications Applications Conference (SIU), Sivas, Turkey, 24–26 April 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–4. [Google Scholar]
- Chen, L.-C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; Springer: Cham, Switzerland, 2018; pp. 801–818. [Google Scholar]
- Vaswani, A.; Ramachandran, P.; Srinivas, A.; Parmar, N.; Hechtman, B.; Shlens, J. Scaling local self-attention for parameter efficient visual backbones. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; IEEE: Washington, DC, USA, 2021; pp. 12894–12904. [Google Scholar]
- Ramachandran, P.; Parmar, N.; Vaswani, A.; Bello, I.; Levskaya, A.; Shlens, J. Stand-alone self-attention in vision models. In Proceedings of the 33rd International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019; Volume 32. [Google Scholar]
- Chen, Y.; Kalantidis, Y.; Li, J.; Yan, S.; Feng, J. A2-nets: Double attention networks. In Proceedings of the 32nd International Conference on Neural Information Processing Systems, Montréal, DC, Canada, 3–8 December 2018; Volume 31. [Google Scholar]
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-end object detection with transformers. In European Conference on Computer Vision; Springer: Cham, Switzerland, 2020; pp. 213–229. [Google Scholar]
- Sun, M.; Zhao, R.; Yin, X.; Xu, L.; Ruan, C.; Jia, W. FBoT-Net: Focal bottleneck transformer network for small green apple detection. Comput. Electron. Agric. 2023, 205, 107609. [Google Scholar] [CrossRef]
- Li, X.; Xiang, Y.; Li, S. Combining convolutional and vision transformer structures for sheep face recognition. Comput. Electron. Agric. 2023, 205, 107651. [Google Scholar] [CrossRef]
- Jiang, J.; He, X.; Zhu, X.; Wang, W.; Liu, J. CGViT: Cross-image GroupViT for zero-shot semantic segmentation. Pattern Recognit. 2025, 164, 111505. [Google Scholar] [CrossRef]
- Touazi, F.; Gaceb, D.; Boudissa, N.; Assas, S. Enhancing Breast Mass Cancer Detection Through Hybrid ViT-Based Image Segmentation Model. In International Conference on Computing Systems and Applications; Springer: Cham, Switzerland, 2024. [Google Scholar] [CrossRef]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.W.; Heng, P.A. H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation From CT Volumes. IEEE Trans. Med. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef] [PubMed]
- Bosma, M.; Dushatskiy, A.; Grewal, M.; Alderliesten, T.; Bosman, P. Mixed-Block Neural Architecture Search for Medical Image Segmentation. arXiv 2022, arXiv:2202.11401. [Google Scholar] [CrossRef]
- Wang, C.; MacGillivray, T.; Macnaught, G.; Yang, G.; Newby, D. A two-stage 3D Unet framework for multi-class segmentation on full resolution image. arXiv 2018, arXiv:1804.04341. [Google Scholar]
- Guan, S.; Khan, A.A.; Sikdar, S.; Chitnis, P.V. Fully dense UNet for 2-D sparse photoacoustic tomography artifact removal. IEEE J. Biomed. Health Inform. 2019, 24, 568–576. [Google Scholar] [CrossRef]
- Xiao, X.; Lian, S.; Luo, Z.; Li, S. Weighted res-unet for high-quality retina vessel segmentation. In Proceedings of the 2018 9th International Conference on Information Technology in Medicine and Education (ITME), Hangzhou, China, 19–21 October 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 327–331. [Google Scholar]
- Guo, X.; Chen, C.; Lu, Y.; Meng, K.; Chen, H.; Zhou, K.; Wang, Z.; Xiao, R. Retinal vessel segmentation combined with generative adversarial networks and dense U-Net. IEEE Access 2020, 8, 194551–194560. [Google Scholar] [CrossRef]
- de Melo, M.J.; Gonçalves, D.N.; Gomes, M.d.N.B.; Faria, G.; de Andrade Silva, J.; Ramos, A.P.M.; Osco, L.P.; Furuya, M.T.G.; Junior, J.M.; Gonxcxalves, W.N. Automatic segmentation of cattle rib-eye area in ultrasound images using the UNet++ deep neural network. Comput. Electron. Agric. 2022, 195, 106818. [Google Scholar] [CrossRef]
- Xiao, L.; Pan, Z.; Du, X.; Chen, W.; Qu, W.; Bai, Y.; Xu, T. Weighted skip-connection feature fusion: A method for augmenting UAV oriented rice panicle image segmentation. Comput. Electron. Agric. 2023, 207, 107754. [Google Scholar] [CrossRef]
- Yogalakshmi, G.; Rani, B.S. Enhanced BT segmentation with modified U-Net architecture: A hybrid optimization approach using CFO-SFO algorithm. Int. J. Syst. Assur. Eng. Manag. 2025, 16, 1451–1467. [Google Scholar] [CrossRef]
- Fan, Y.; Song, J.; Lu, Y.; Fu, X.; Huang, X.; Yuan, L. DPUSegDiff: A Dual-Path U-Net Segmentation Diffusion model for medical image segmentation. Electron. Res. Arch. 2025, 33, 2947–2971. [Google Scholar] [CrossRef]
- Fan, Y.; Song, J.; Lu, Y.; Fu, X.; Huang, X.; Yuan, L. Shape-intensity-guided U-net for medical image segmentation. Neurocomputing 2024, 610, 128534. [Google Scholar] [CrossRef]
- Wang, W.; Xie, E.; Li, X.; Fan, D.P.; Shao, L. Pyramid Vision Transformer: A Versatile Backbone for Dense Prediction without Convolutions. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021. [Google Scholar]
- Chu, X.; Tian, Z.; Zhang, B.; Wang, X.; Wei, X.; Xia, H.; Shen, C. Conditional Positional Encodings for Vision Transformers. arXiv 2021, arXiv:2102.10882. [Google Scholar]
- Yang, L.; Wang, H.; Zeng, Q.; Liu, Y.; Bian, G. A Hybrid Deep Segmentation Network for Fundus Vessels via Deep-Learning Framework. Neurocomputing 2021, 448, 168–178. [Google Scholar] [CrossRef]
- Gao, Y.; Zhou, M.; Metaxas, D.N. UTNet: A hybrid transformer architecture for medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; Springer: Cham, Switzerland, 2021; pp. 61–71. [Google Scholar]
- Xie, E.; Wang, W.; Yu, Z.; An kumar, A.; Álvarez, J.M.; Luo, P. SegFormer: Simple and efficient design for semantic segmentation with transformers. In Proceedings of the Neural Information Processing Systems, Online, 6–14 December 2021; Curran Associates, Inc.: Red Hook, NY, USA, 2021; pp. 1–13. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-Unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the ECCV Workshops, Tel Aviv, Israel, 23 October 2022; Springer: Cham, Switzerland, 2021; pp. 205–218. [Google Scholar]
- Mehta, S.; Rastegari, M. MobileViT: Light-weight, general-purpose, and mobile-friendly vision transformer. Sensors 2021, 21, 9210. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
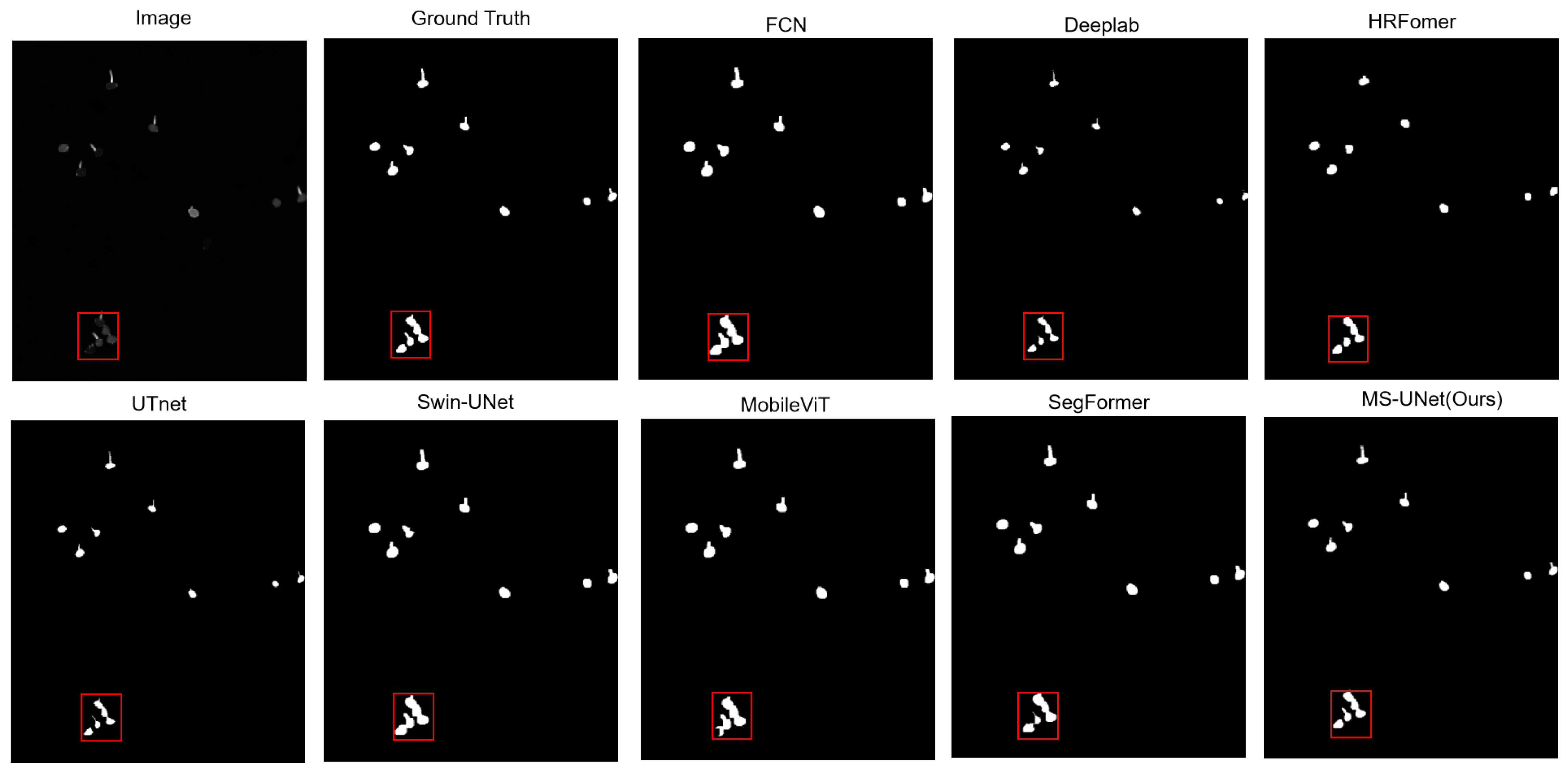
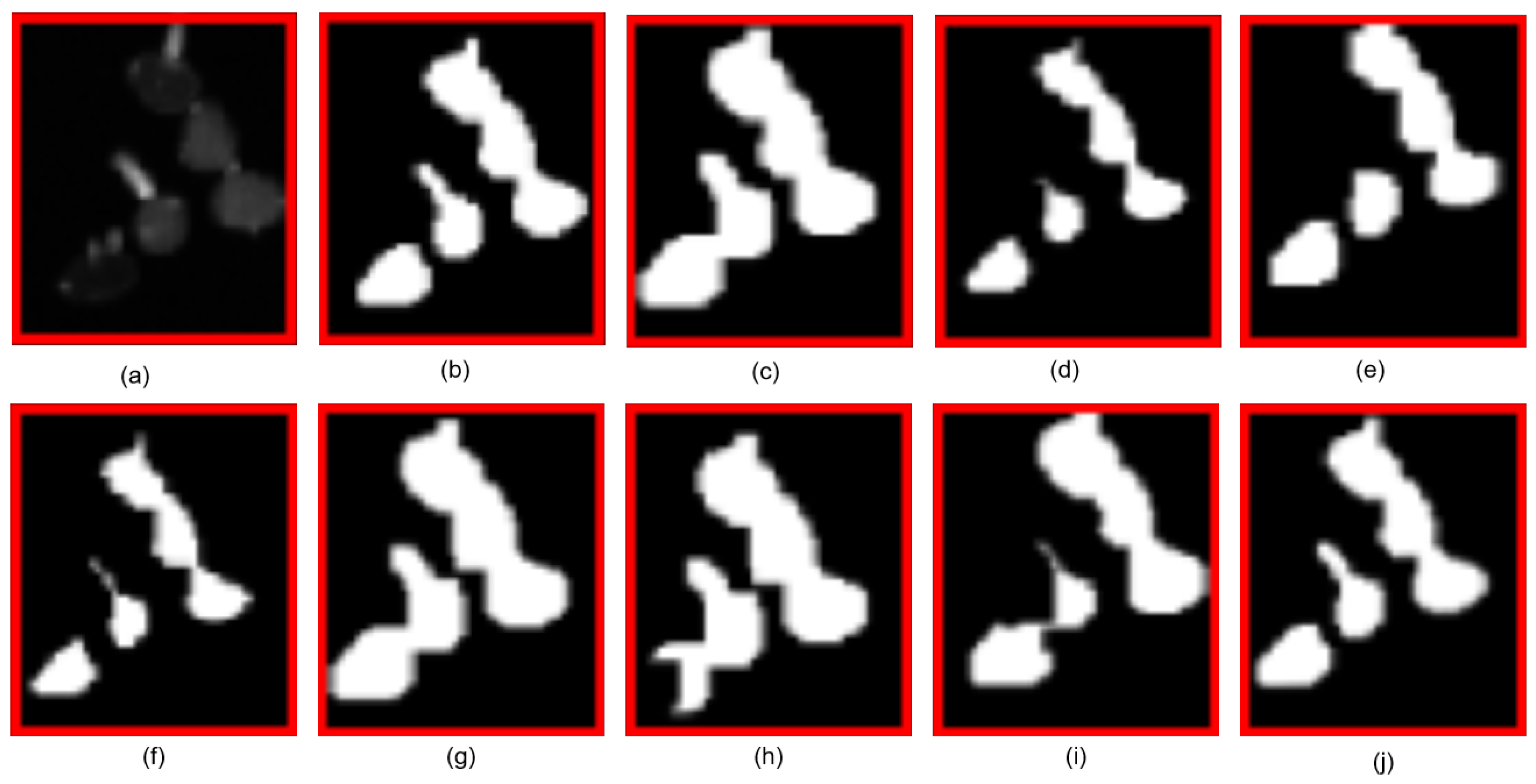
| Methods | Pre | Re | MIoU | F1 | Boundary IoU | SCS |
|---|---|---|---|---|---|---|
| FCN [4] | 0.965 ± 0.012 | 0.925 ± 0.018 | 0.945 ± 0.011 | 0.944 ± 0.013 | 0.885 ± 0.015 | 0.832 ± 0.014 |
| DeepLab [6] | 0.901 ± 0.019 | 0.932 ± 0.016 | 0.895 ± 0.017 | 0.888 ± 0.018 | 0.856 ± 0.021 | 0.857 ± 0.019 |
| HRformer [5] | 0.785 ± 0.023 | 0.928 ± 0.020 | 0.862 ± 0.022 | 0.851 ± 0.021 | 0.822 ± 0.025 | 0.848 ± 0.023 |
| UTnet [42] | 0.923 ± 0.014 | 0.985 ± 0.008 | 0.951 ± 0.010 | 0.953 ± 0.011 | 0.899 ± 0.013 | 0.908 ± 0.012 |
| SegFormer [43] | 0.922 ± 0.009 | 0.938 ± 0.012 | 0.921 ± 0.008 | 0.935 ± 0.010 | 0.877 ± 0.014 | 0.886 ± 0.011 |
| Swin-UNet [44] | 0.933 ± 0.011 | 0.958 ± 0.010 | 0.945 ± 0.009 | 0.946 ± 0.010 | 0.878 ± 0.012 | 0.894 ± 0.011 |
| MobileViT [45] | 0.906 ± 0.016 | 0.905 ± 0.015 | 0.913 ± 0.014 | 0.906 ± 0.015 | 0.851 ± 0.017 | 0.872 ± 0.016 |
| MS-UNet | 0.987 ± 0.012 | 0.959 ± 0.014 | 0.963 ± 0.012 | 0.967 ± 0.009 | 0.913 ± 0.011 | 0.935 ± 0.013 |
| Configuration | Attention | Multi- Branch | U-Net Fusion | Pre | Re | MIoU | F1 | Boundary IoU | SCS |
|---|---|---|---|---|---|---|---|---|---|
| U-Net [46] | × | × | × | 0.917 | 0.903 | 0.902 | 0.897 | 0.821 | 0.865 |
| Ours-Attention | ✓ | × | × | 0.938 | 0.921 | 0.913 | 0.921 | 0.835 | 0.878 |
| Ours-Multibranch | × | ✓ | × | 0.951 | 0.925 | 0.936 | 0.941 | 0.851 | 0.892 |
| Ours-Attention-Multibranch | ✓ | ✓ | × | 0.967 | 0.927 | 0.945 | 0.947 | 0.870 | 0.907 |
| Ours | ✓ | ✓ | ✓ | 0.987 | 0.959 | 0.963 | 0.967 | 0.913 | 0.935 |
| Configuration | Attention | Multi- Branch | U-Net Fusion | Parameters (M) | FLOPs (G) | Maximum Memory Usage (MB) | Inference Time (ms) |
|---|---|---|---|---|---|---|---|
| U-Net [46] | × | × | × | 7.76 | 6.2 | 642 | 12.4 |
| Ours-Attention | ✓ | × | × | 10.23 | 14.8 | 798 | 16.7 |
| Ours-Multibranch | × | ✓ | × | 15.67 | 18.6 | 1245 | 20.8 |
| Ours-Attention-Multibranch | ✓ | ✓ | × | 18.12 | 28.4 | 1487 | 25.3 |
| Ours | ✓ | ✓ | ✓ | 19.21 | 31.2 | 1576 | 27.6 |
| Threshold | Pre | Re | MIoU | F1 | Boundary IoU | SCS |
|---|---|---|---|---|---|---|
| 0.15 | 0.891 | 0.973 | 0.896 | 0.935 | 0.831 | 0.862 |
| 0.30 | 0.915 | 0.965 | 0.912 | 0.943 | 0.849 | 0.878 |
| 0.45 | 0.938 | 0.961 | 0.927 | 0.951 | 0.867 | 0.894 |
| 0.60 | 0.956 | 0.954 | 0.939 | 0.955 | 0.882 | 0.909 |
| 0.75 | 0.987 | 0.959 | 0.963 | 0.967 | 0.913 | 0.935 |
| 0.90 | 0.989 | 0.928 | 0.948 | 0.96 | 0.894 | 0.917 |
| Methods | Parameters (M) | FLOPs (G) | Maximum Memory Usage (MB) | Inference Time (ms) |
|---|---|---|---|---|
| SegFormer [43] | 3.7 | 8.4 | 648 | 11.2 |
| MobileViT [45] | 6 | 4.9 | 512 | 9.6 |
| UTnet [42] | 9.53 | 15.8 | 782 | 16.4 |
| Swin-UNet [44] | 27 | 67.3 | 2184 | 39.8 |
| HRformer [5] | 37.9 | 89.2 | 3048 | 52.7 |
| DeepLab [6] | 41.2 | 52.7 | 1724 | 26.5 |
| FCN [4] | 134.3 | 148.6 | 2856 | 18.3 |
| MS-UNet | 19.21 | 31.2 | 1576 | 27.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.; Sun, L.; Wu, Q.; Zou, Q.; Su, P.; Xie, P. MS-UNet: A Hybrid Network with a Multi-Scale Vision Transformer and Attention Learning Confusion Regions for Soybean Rust Fungus. Sensors 2025, 25, 5582. https://doi.org/10.3390/s25175582
Liu T, Sun L, Wu Q, Zou Q, Su P, Xie P. MS-UNet: A Hybrid Network with a Multi-Scale Vision Transformer and Attention Learning Confusion Regions for Soybean Rust Fungus. Sensors. 2025; 25(17):5582. https://doi.org/10.3390/s25175582
Chicago/Turabian StyleLiu, Tian, Liangzheng Sun, Qiulong Wu, Qingquan Zou, Peng Su, and Pengwei Xie. 2025. "MS-UNet: A Hybrid Network with a Multi-Scale Vision Transformer and Attention Learning Confusion Regions for Soybean Rust Fungus" Sensors 25, no. 17: 5582. https://doi.org/10.3390/s25175582
APA StyleLiu, T., Sun, L., Wu, Q., Zou, Q., Su, P., & Xie, P. (2025). MS-UNet: A Hybrid Network with a Multi-Scale Vision Transformer and Attention Learning Confusion Regions for Soybean Rust Fungus. Sensors, 25(17), 5582. https://doi.org/10.3390/s25175582







