1. Introduction
The World Health Organization (WHO) has identified cardiovascular diseases (CVDs) as a primary cause of death globally, presenting a significant health challenge worldwide [
1]. A substantial proportion of these deaths are caused by irregular heartbeats, medically known as cardiac arrhythmias [
2]. Cardiac arrhythmia happens when the electrical impulses controlling the heart’s rhythm malfunction, leading to either an elevated, diminished, or irregular heartbeat [
2]. Cardiac arrhythmias are usually detected through electrocardiogram (ECG) signals. ECG is an economical and non-invasive diagnostic tool that captures the heart’s electrical activity through electrodes positioned on the chest, upper limbs, and lower limbs [
3,
4]. The standard 12-lead ECG, obtained with the use of 10 electrodes, remains the gold standard for clinical cardiac evaluation and is routinely employed across nearly all clinical settings [
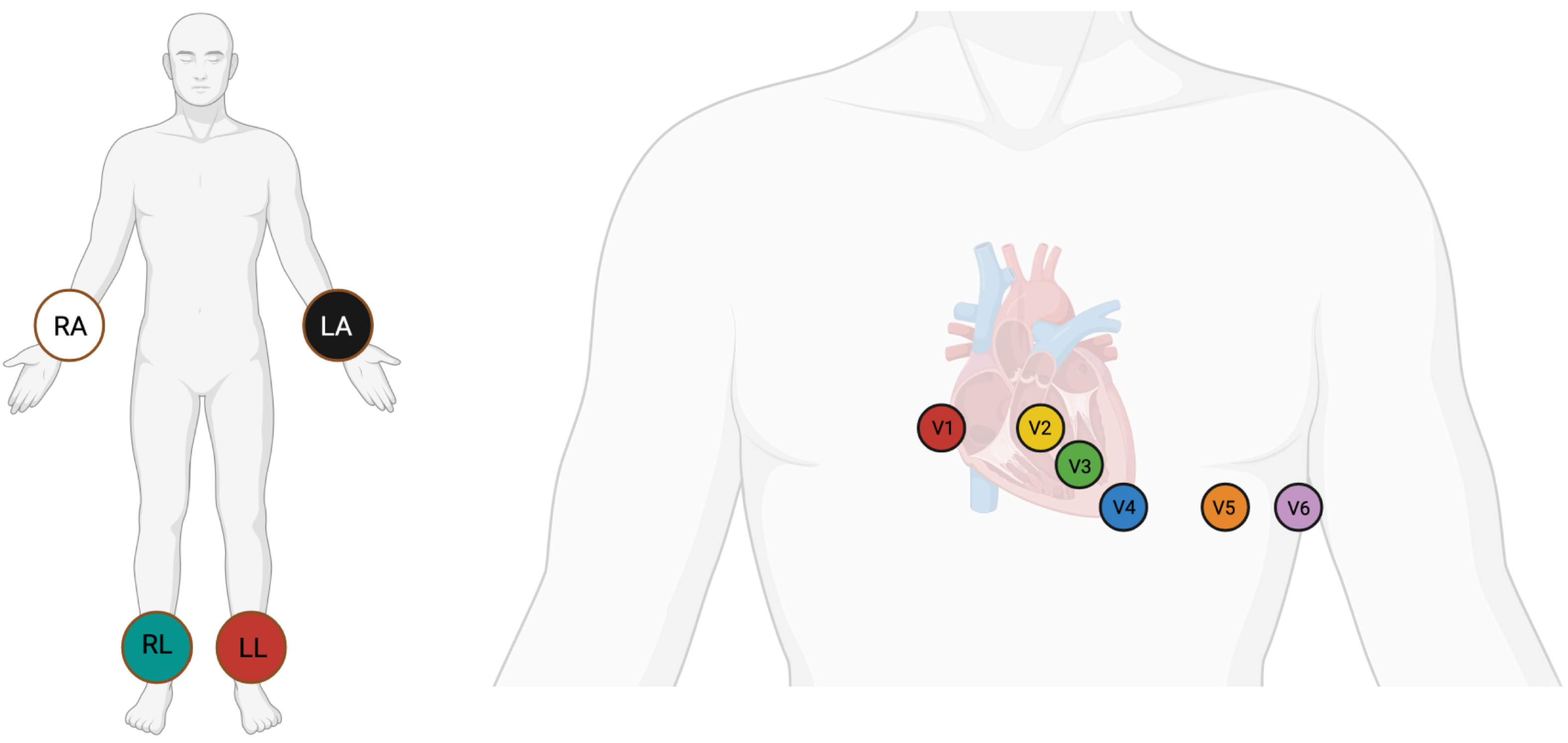
5]. The positioning of electrodes for a 12-lead ECG is shown in
Figure 1. Despite this, a sizable proportion of existing automatic ECG diagnosis models focus on single-lead analysis, which increases the risk of missed diagnoses, as many cardiac abnormalities may manifest only on specific leads and could be overlooked in single-lead setups [
6]. Additionally, while multi-lead deep learning models offer strong diagnostic performance, their computational complexity frequently restricts their practical use in real-world clinical environments [
7]. A recent review found that among studies addressing clinical ECG applications, only about 41.5% incorporated full 12-lead, 10 s ECG inputs highlighting a significant gap in comprehensive multi-lead modeling [
7]. Manual processing of ECGs requires substantial time and expertise, which typically demands years of training. This makes the process challenging and time-consuming [
8]. Therefore, accurate and reliable automated systems for ECG analysis are crucial. These challenges strongly motivate the present study to develop and validate an efficient, multi-lead deep learning model capable of supporting 12-lead ECG input, thereby bridging the clinical-technical gap between robust diagnostic accuracy and practical implementation. These systems enhance diagnostic speed, improve care quality, and expand access to care for cardiovascular disease patients.
In addition to arrhythmia detection, ECG analysis is also very important for other parts of clinical care, including postoperative monitoring and active control of blood pressure in cardiac patients. Recent studies have highlighted the incorporation of machine learning into applications such as personalized blood pressure control for remote patient monitoring and multi-modal decision support systems for predicting postoperative cardiac events [
9,
10]. These developments reflect the wider clinical impact of automated ECG analysis and encourage more study into deep learning models for accurate classification of arrhythmias. Deep learning has revolutionized cardiovascular disease identification from ECG signals through recognition of complex patterns, offering significant advantages over traditional methods. These models have demonstrated high accuracy in detecting conditions like arrhythmias.
Recent contributions from researchers in this field have resulted in the creation of advanced deep learning architectures that surpass the diagnostic performance of conventional methods by leveraging large datasets and exploring novel techniques such as hybrid models, attention mechanisms, and transfer learning. Among deep learning models, convolutional neural networks (CNNs) are the most commonly utilized for classifying cardiovascular diseases (CVDs) [
11]. A CNN with 11 layers was developed to classify myocardial infarction (MI) and normal ECG beats using the PTB database, achieving precisions of 93.53% and 95.22% with and without noise, respectively [
12]. Similarly, a CNN-based automated heart disease recognition model utilizing the MIT-BIH arrhythmia dataset attained an accuracy of 98.33% [
13]. Wang et al. used a CNN to automatically classify different types of arrhythmia, achieving 99.06% accuracy with the MIT-BIH Arrhythmia Database [
14]. Ahmed et al. utilized CNN to classify four arrhythmia classes using Lead II ECG signals from MIT-BIH arrhythmia database, achieving 99% accuracy [
15].
Hybrid models, integrating different deep learning architectures, have shown significant potentials. These models can combine CNNs, which are notable for their ability to acquire spatial features, recurrent neural networks and long short-term memory, which is popular in time-series data analysis. For instance, Oh et al. developed a CNN-LSTM model for classifying ECG signals into normal sinus rhythm, right bundle branch block, left bundle branch block, atrial premature beats, and premature ventricular contractions [
16]. Their model, evaluated with the MIT-BIH Arrhythmia Database, reached an accuracy of 98.10%. Similarly, Kusuma et al. [
17] and Tan et al. [
18] implemented CNN-LSTM models for diagnosing congestive heart failure and coronary artery disease, obtaining accuracies of 99.52% and 99.85%, respectively. Dhyani et al. proposed a ResRNN model combining ResNet and RNN for classifying nine types of arrhythmias using 12-lead ECG data from the CPSC 2018 dataset, obtaining 91% accuracy [
19].
Attention mechanisms, inspired by human biological systems that focus on key details, have become a pivotal concept in deep learning [
20]. Their success in computer vision has inspired scientists to investigate their application in ECG signal processing [
21]. Yao et al. proposed a CNN-LSTM model and subsequently improved its performance by incorporating an attention mechanism [
22]. Similarly, Li et al. suggested a deep neural network incorporating an attention mechanism to classify nine classes of arrhythmias using 12-lead ECG signals, achieving 92.8% accuracy with the PhysioNet public database [
23]. Lyu et al. introduced a new dual attention mechanism combined with hybrid network (DA-Net) for classifying five arrhythmia classes using the MIT-BIH arrhythmia dataset, obtaining 99.98% accuracy [
24].
Recent studies have further advanced multi-lead and lightweight ECG models. An et al. introduced a lightweight classification model utilizing knowledge distillation, attaining an accuracy of 96.32% while significantly minimizing the model size, making it appropriate for wearable devices [
25]. Alghieth suggested DeepECG-Net, a hybrid transformer–CNN model for arrhythmia detection with enhanced attention mechanisms for noise robustness, attaining an accuracy of 98.2% on multi-lead ECG [
26]. Baig et al. presented ArrhythmiaVision, which are lightweight models with visual interpretability that utilize 1D-CNNs for ECG classification, attaining 99% and 98% accuracies, suitable for clinical use [
27]. These recent developments highlight the trend towards computationally efficient and real-time relevant models, offering providing context and motivation for the suggested method in this work.
Publicly available datasets, including the MIT-BIH Arrhythmia Database [
28] and the St. Petersburg INCART 12-lead Arrhythmia Database [
29], have been widely utilized for training and assessing deep learning models in arrhythmia classification. However, many studies [
21,
30,
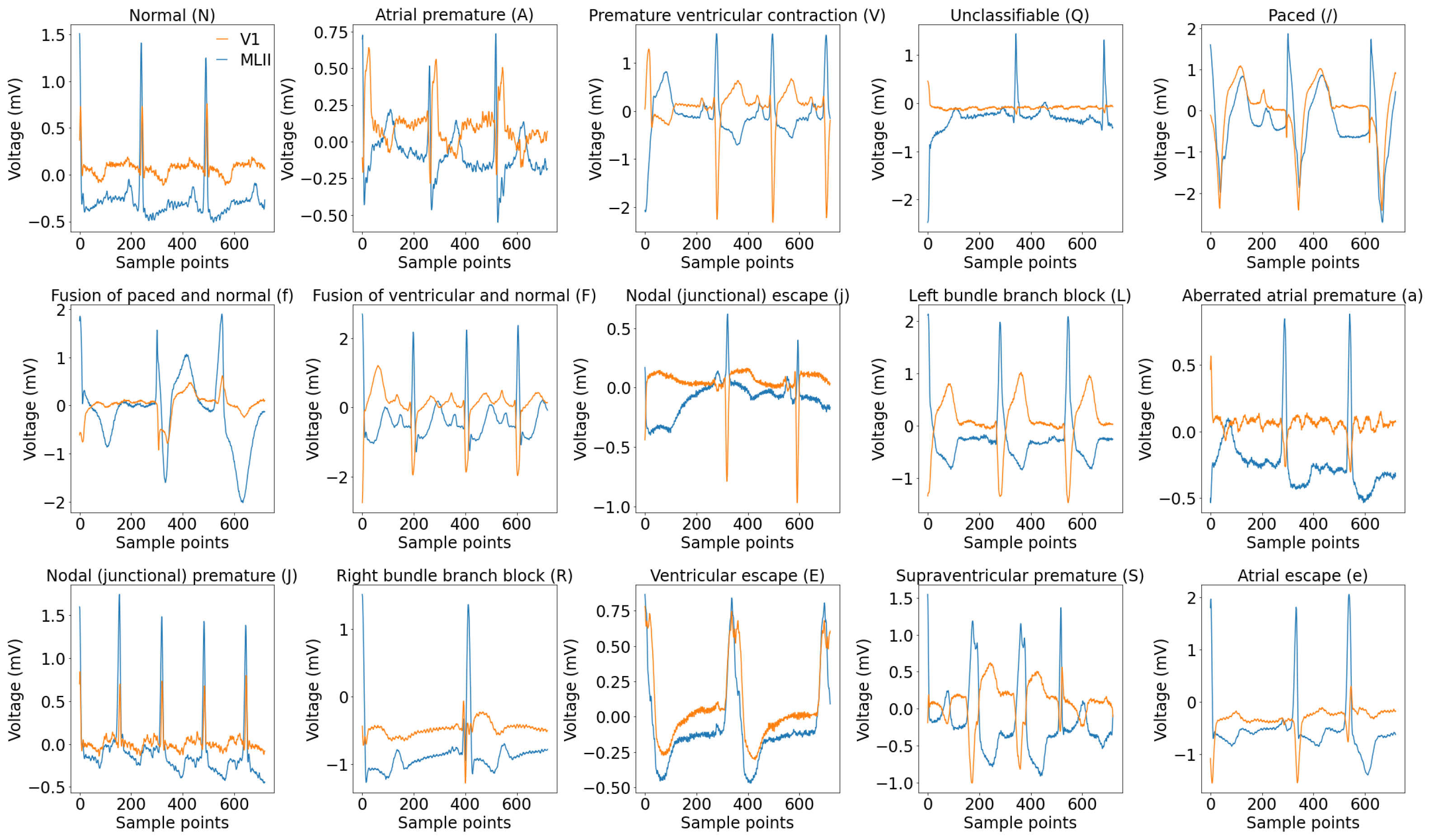
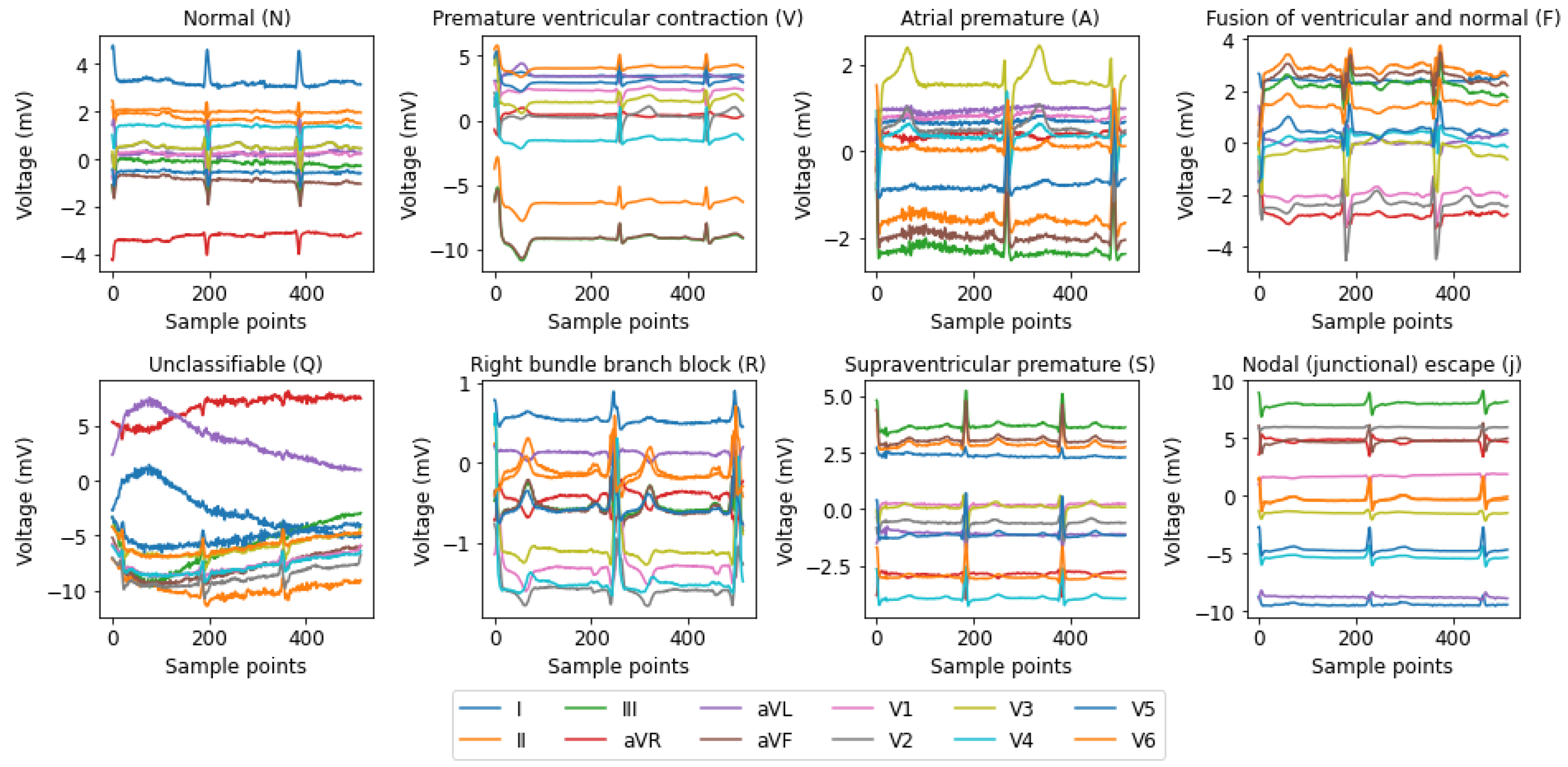
31] have focused on single-lead ECG data, such as MLII, even though multi-lead recordings are available. While this simplification aids model development, it may overlook crucial information, potentially compromising diagnostic accuracy and model generalizability. In this paper, we address these gaps by using all the available leads from the MIT-BIH Arrhythmia and INCART 12-lead Arrhythmia Databases.
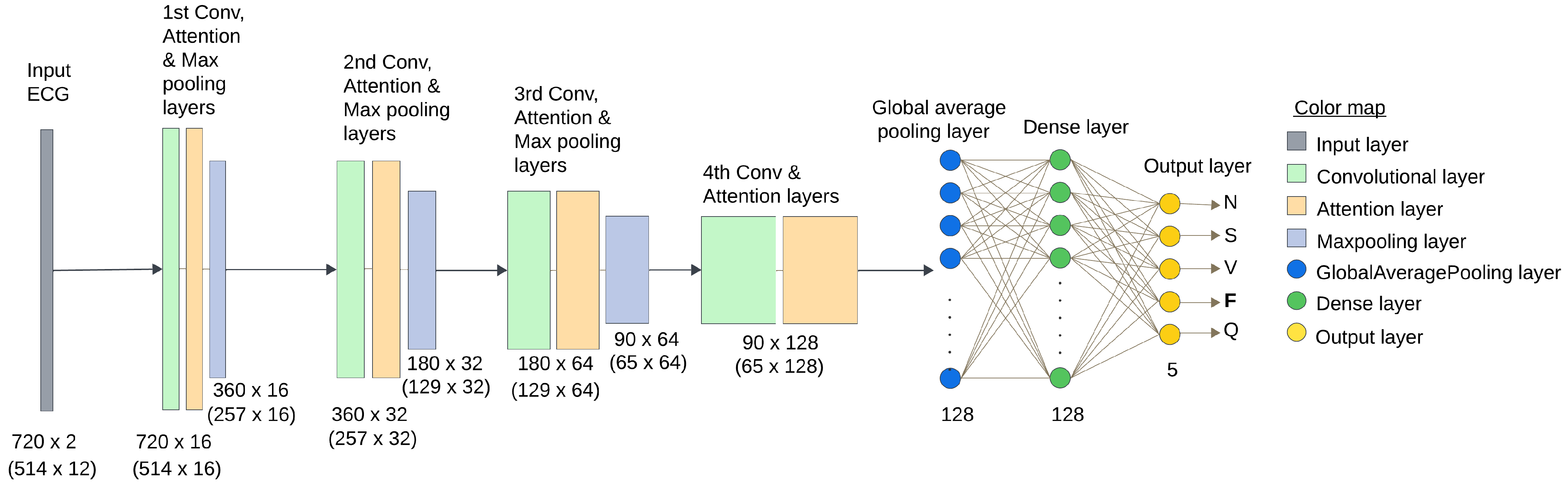
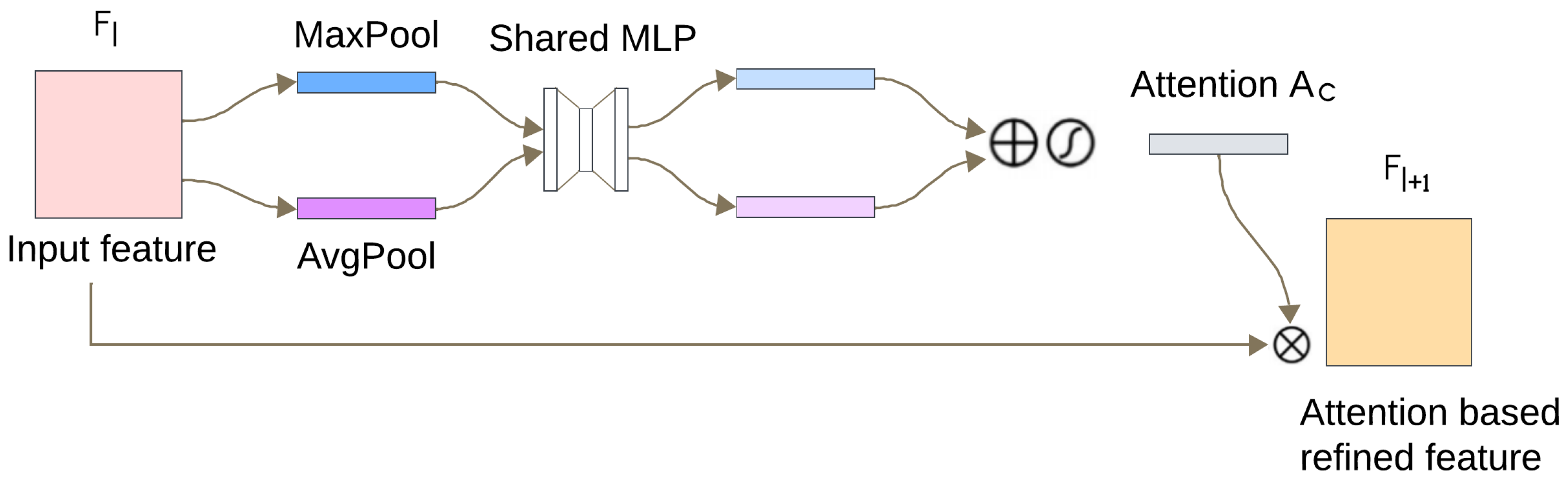
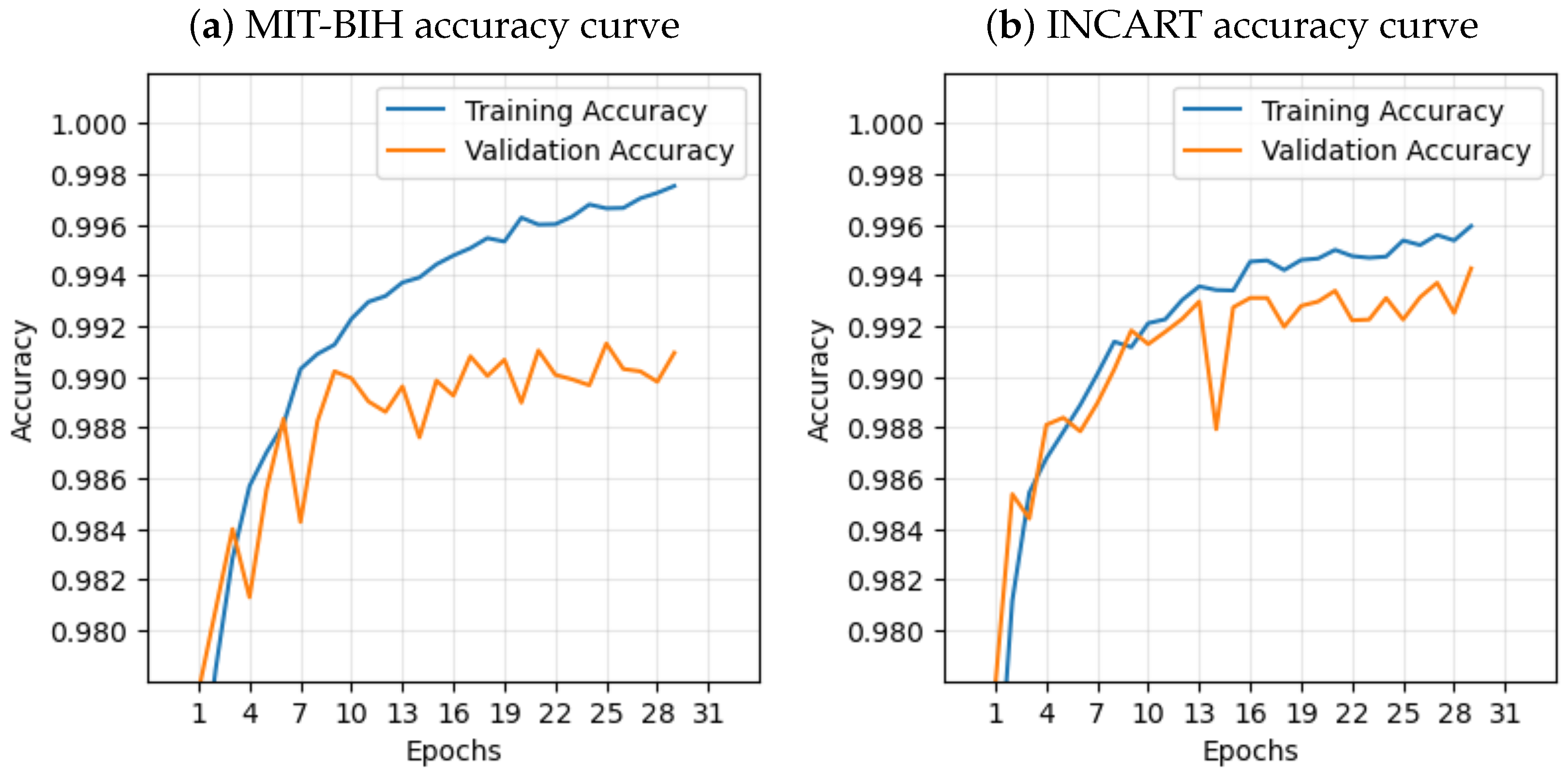
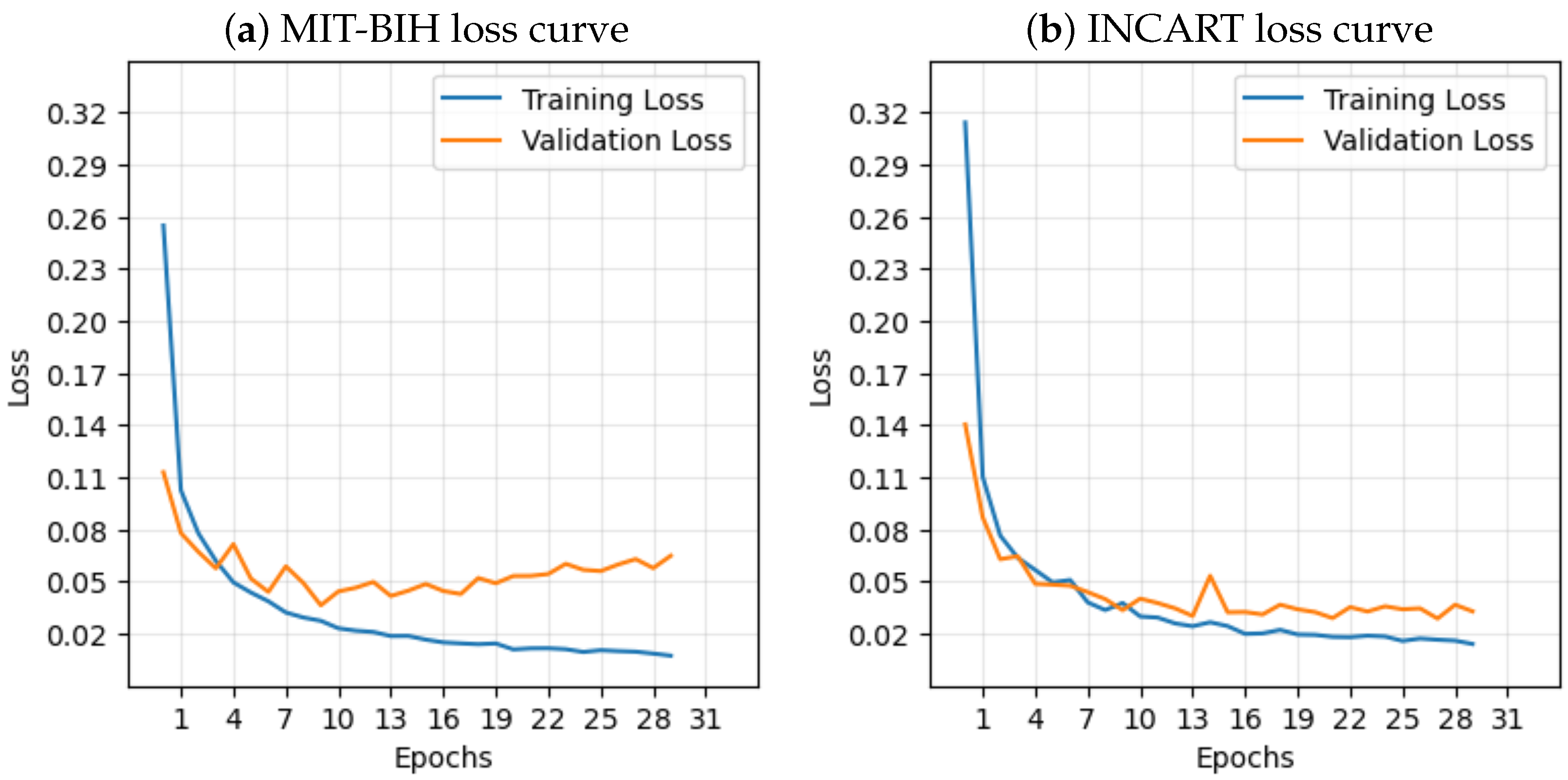
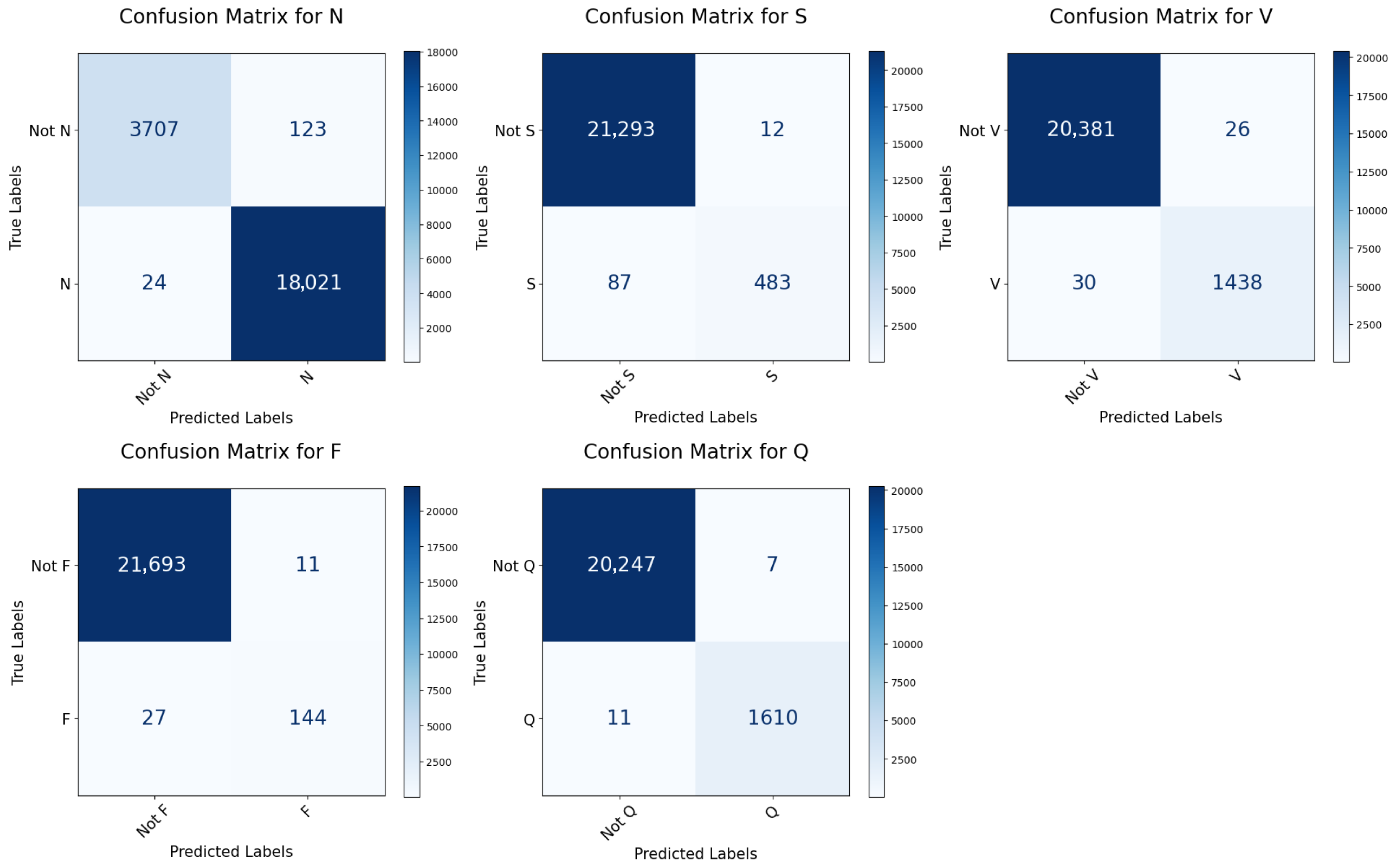
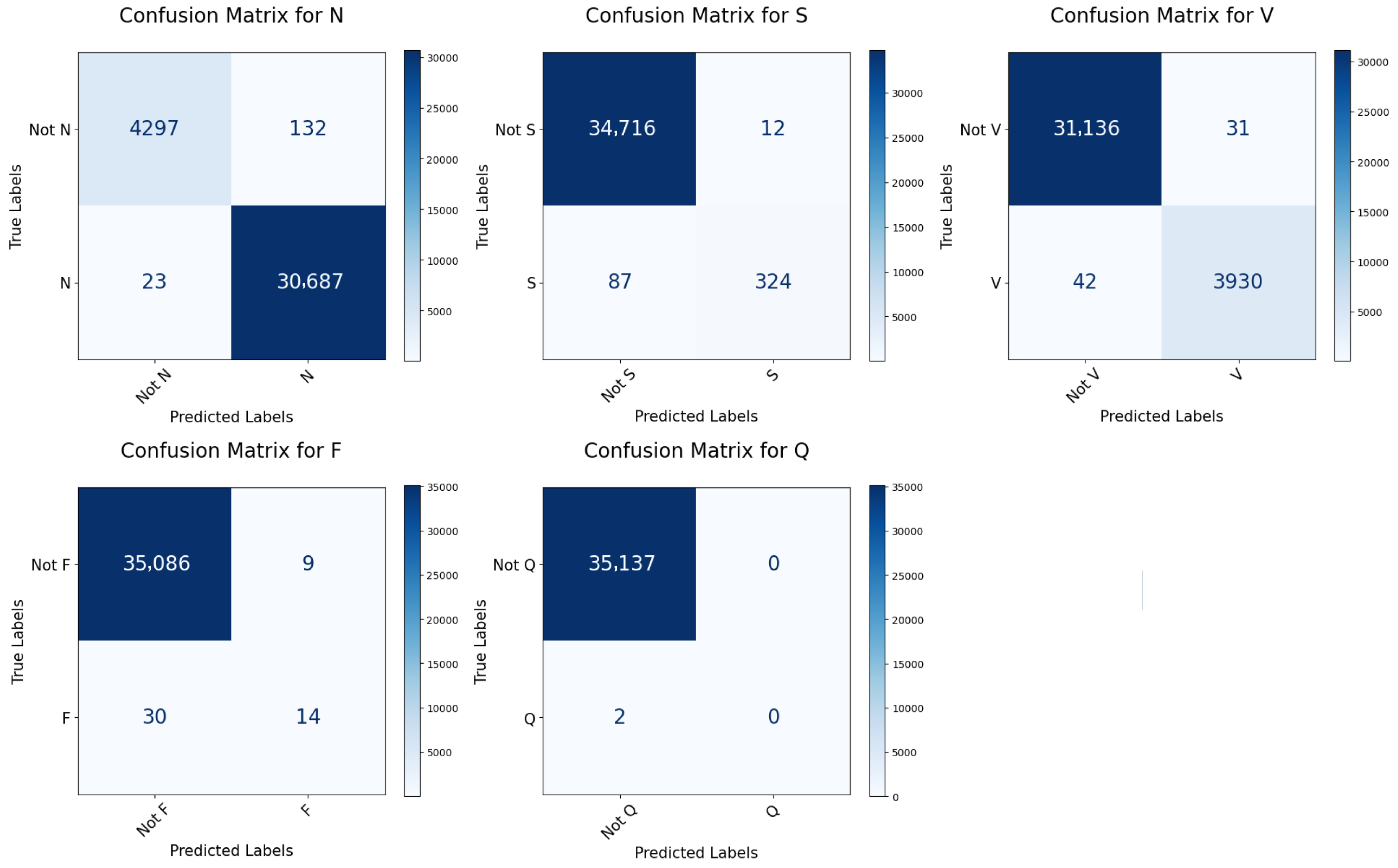
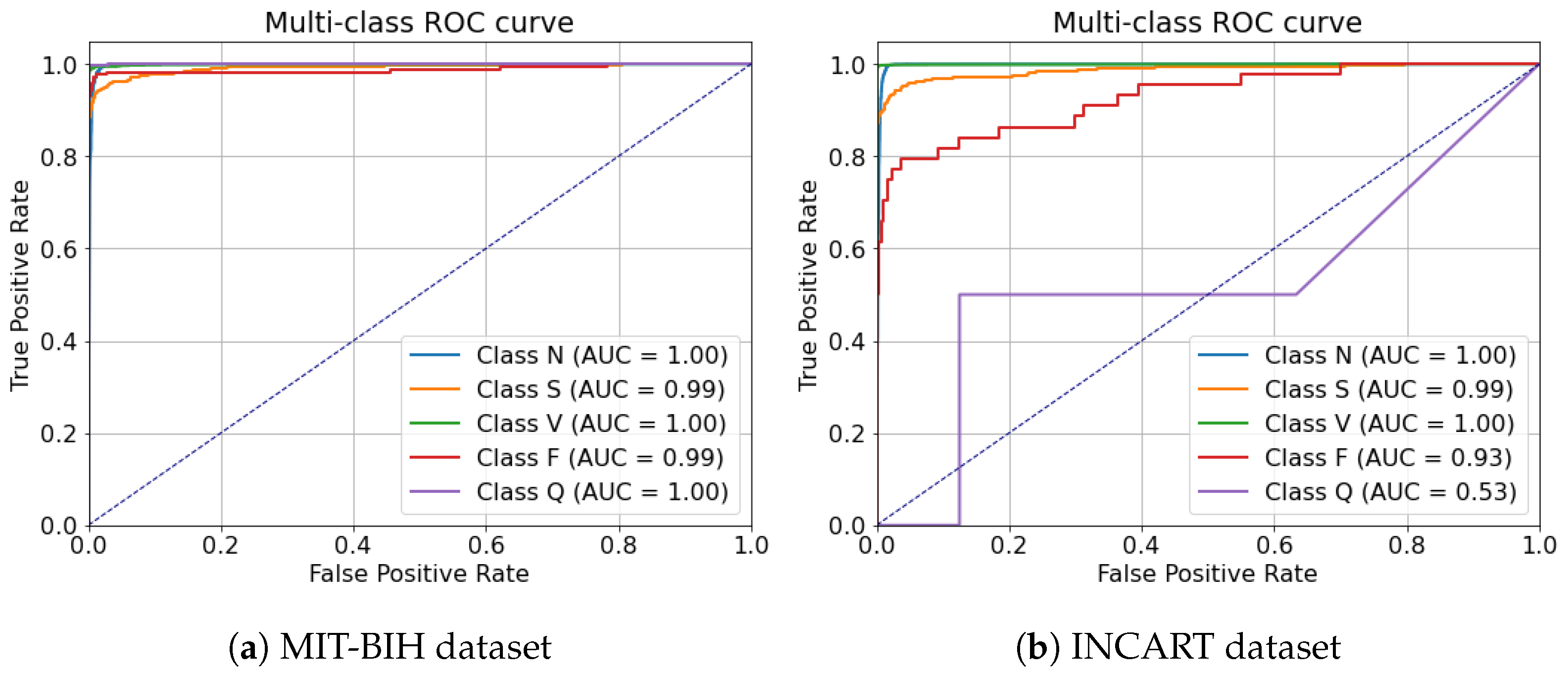
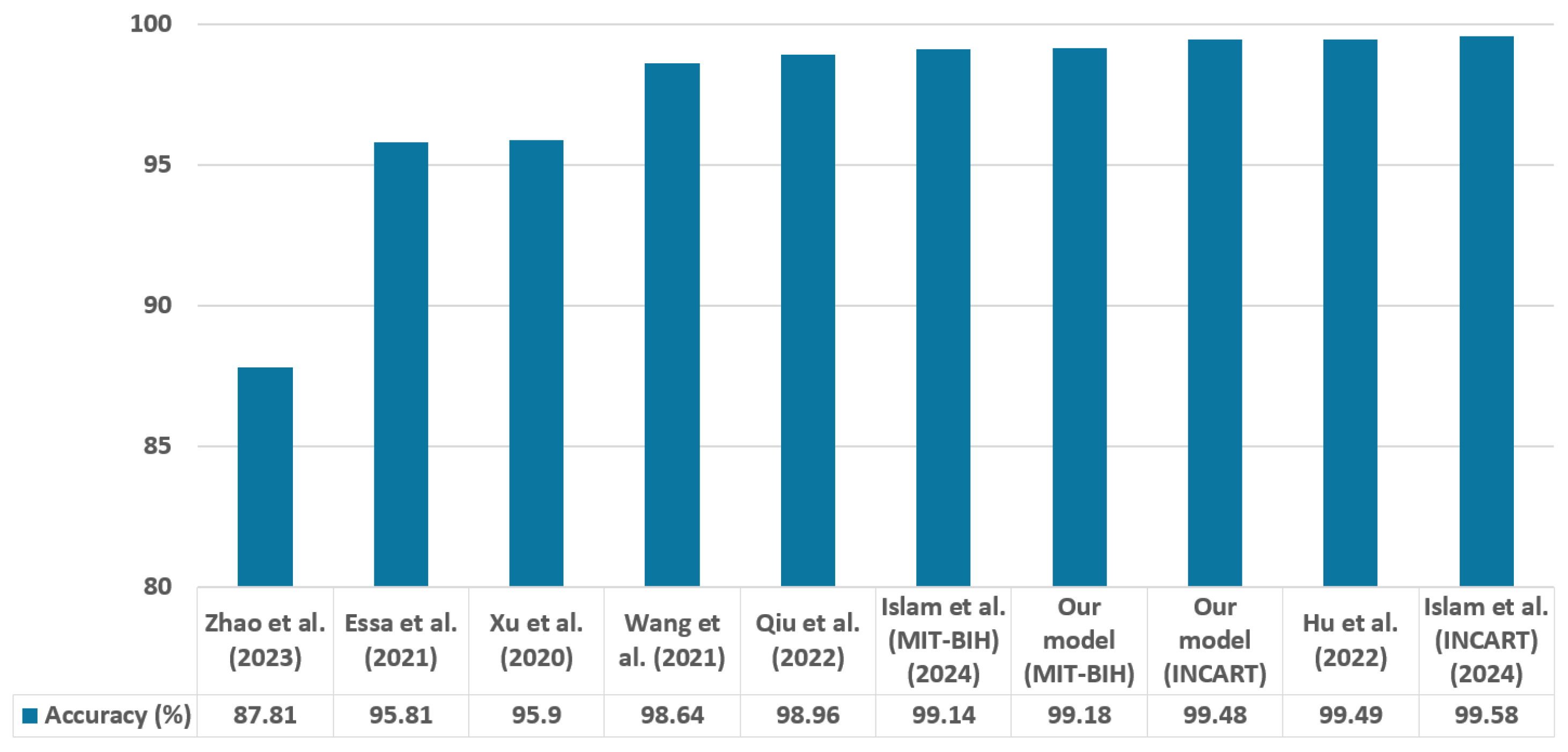
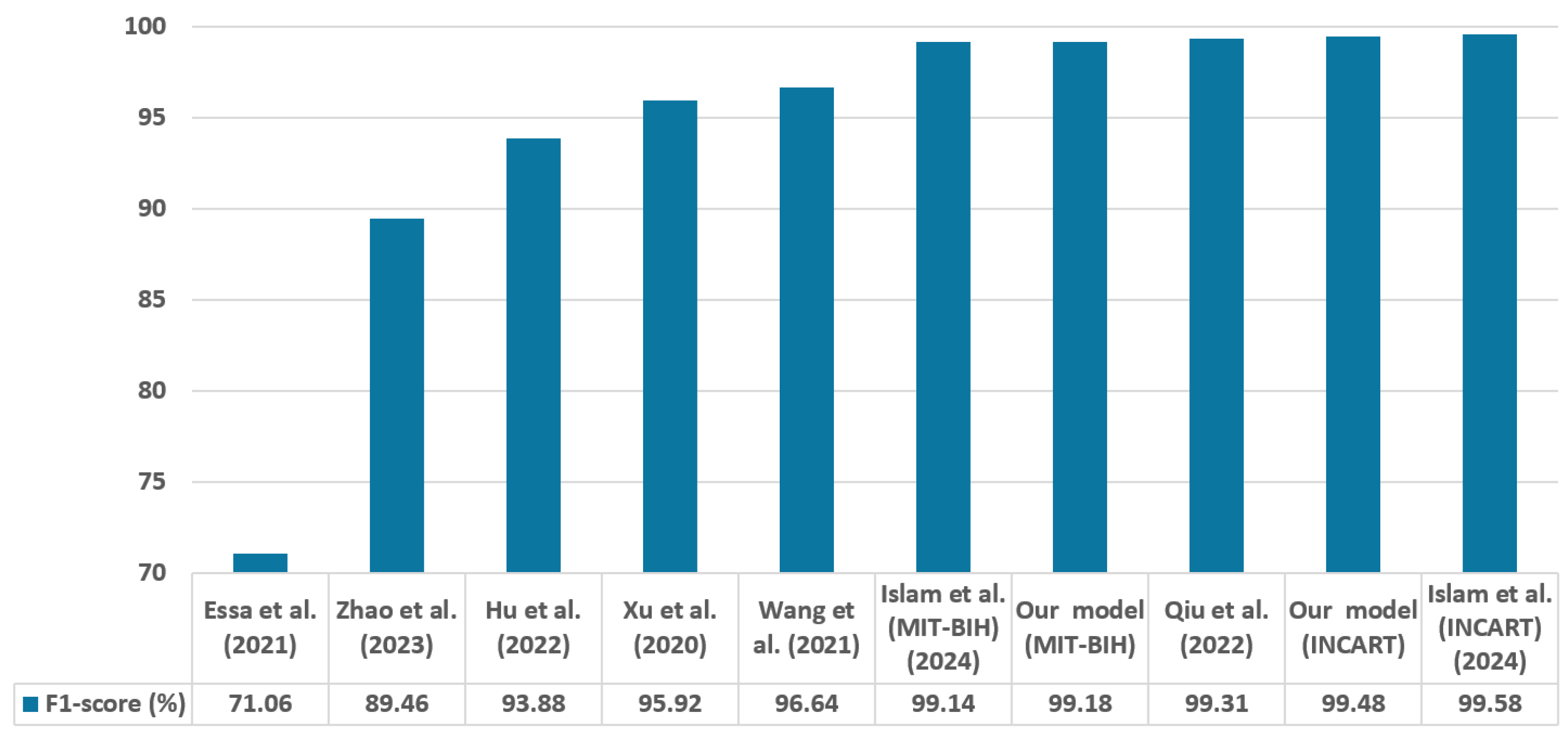
In contrast to many prior approaches that either limit their analysis to single-lead ECG signals or employ complex hybrid architectures with high computational demands, we propose a simpler and innovative deep learning model for multi-label cardiac arrhythmia classification utilizing CNN and channel attention mechanism using multiple lead ECG data. By combining the power of CNNs with the focus of attention mechanisms, our model learns to identify the most important patterns in heart rhythm data, resulting in more accurate and dependable outcomes in detecting heart problems. The proposed model can detect and classify 5 classes using MIT-BIH Arrhythmia and INCART 12-lead Arrhythmia Databases. By testing the model on the previous datasets, we obtained an accuracy of 99.18%, and 99.48%, respectively.
The novelty of this study lies in three main aspects. (1) Utilizing both 2-lead and 12-lead ECG data to better reflect diverse clinical conditions, this approach captures complementary cardiac information, in contrast to prior single-lead models [
21], and helps detect arrhythmias more comprehensively (for example, the V1 lead is particularly sensitive to atrial arrhythmias). (2) Incorporating a lightweight CNN architecture with a channel attention mechanism enhances the model’s ability to focus on diagnostically important regions in the ECG signal while maintaining low computational complexity. Quantitative indicators such as model size, number of parameters, and inference time are summarized later in the manuscript. (3) Achieving state-of-the-art performance across two benchmark datasets while remaining computationally efficient and robust to noisy signals. These contributions position our model as a robust, scalable solution for practical ECG-based diagnosis systems. The rest of the paper is structured as follows.
Section 2 presents the ECG datasets and the details of the proposed method. In
Section 3, the model’s results and performance are discussed. The paper is concluded in
Section 4.