1. Introduction
Depression has become one of the most prevalent and widespread psychiatric disorders worldwide. Patients typically exhibit symptoms such as a depressed mood, slowed thinking, and significant cognitive impairment, and severe cases may involve self-harm behaviors or life-threatening conditions [
1,
2]. Neuroimaging studies have confirmed that depression is associated with functional abnormalities in the prefrontal-limbic neural circuitry, particularly involving disrupted functional connectivity in regions such as the anterior cingulate cortex and dorsolateral prefrontal cortex. These neurophysiological alterations result in persistent deficits in emotional regulation and cognitive processing, and are also closely linked to the high recurrence rate of depression. According to the World Health Organization, depression currently affects more than 350 million people globally and is projected to become the leading cause of disease burden by 2030 [
3,
4,
5]. With its rising prevalence, early diagnosis and intervention for depression are becoming increasingly critical.
Currently, depression diagnosis primarily relies on clinicians’ subjective assessments, often based on standardized symptom rating scales [
6]. However, this approach has inherent limitations and may result in misdiagnosis or missed diagnoses [
7,
8]. Consequently, researchers have been actively exploring objective diagnostic methods. In recent years, advancements in neuroimaging technologies—such as EEG, functional magnetic resonance imaging (fMRI), and positron emission tomography (PET)—have provided powerful tools for studying and diagnosing depression [
9,
10]. These techniques allow for the monitoring and analysis of brain activity from different perspectives, offering more objective diagnostic criteria [
11]. Nevertheless, practical challenges such as high costs and complicated equipment requirements have restricted their widespread accessibility and application.
In contrast, EEG, as a non-invasive, low-cost modality with exceptionally high temporal resolution, has emerged as a promising direction in research aimed at diagnosing brain disorders, particularly depression [
12,
13]. EEG-based depression classification methods generally fall into two categories: the first relies on handcrafted feature extraction combined with traditional machine learning algorithms [
14,
15,
16,
17]. Although this approach has achieved progress in certain scenarios, its effectiveness remains limited due to the inherently low spatial resolution and inter-individual variability of EEG signals. In recent years, deep learning techniques have demonstrated increasing advantages in EEG processing, significantly enhancing accuracy and robustness in depression classification through automated feature learning and high-dimensional data representation [
18,
19,
20,
21]. Deep learning methods overcome the limitations of traditional approaches through end-to-end training, promoting EEG application in depression classification tasks.
Despite valuable contributions from existing EEG-based depression classification methods, two key challenges remain inadequately addressed, which are outlined below.
First, the low spatial resolution of EEG signals restricts the capture of spatiotemporal features. Many studies extract features directly from scalp EEG signals. For instance, Acharya et al. [
22] proposed a method for constructing a Depression Diagnosis Index based on multiple nonlinear features, which, combined with a Support Vector Machine (SVM), achieved high-accuracy automatic classification of depression. Zhang et al. [
23] developed a brain functional network based on resting-state EEG and employed a Random Forest classifier, reaching a maximum classification accuracy of 93.31%. Yang et al. [
24] introduced a fusion approach that combined Lempel-Ziv complexity features under both eyes-open and eyes-closed paradigms, and applied multiple classifiers for cross-subject depression recognition, achieving an accuracy of 94.03% using an SVM. Liu et al. [
25] proposed a depression classification method combining spatiotemporal features, utilizing a GCN and an adjacency matrix based on channel correlations for resting-state EEG processing. Ying et al. [
26] introduced the EEG-based Depression Transformer, extracting temporal, spatial, and frequency domain features to distinguish depressed patients from healthy controls. Seal et al. [
27] developed a deep learning model based on convolutional neural networks (CNN), which achieved an accuracy of 99.37% on their private dataset. Other studies, such as those by Liu et al. [
28] and Lu et al. [
29], have also employed deep learning architectures to automatically extract spatiotemporal features, achieving impressive classification performance. However, the volume conduction effect of EEG often leads to pseudo-connectivity and inaccurate regional information [
30,
31,
32], making it difficult for existing feature extraction methods to fully capture the spatiotemporal characteristics of EEG signals.
Second, class imbalance negatively impacts classification effectiveness. Due to the significant disparity in sample sizes between depressed patients and healthy controls, many studies have focused on improving model generalizability. For example, Ye et al. [
33] integrated deep similarity learning and adversarial learning, proposing a cross-subject emotional recognition method. Song et al. [
34] combined CNN with LSTM, employing a domain discriminator to reduce differences between training and testing datasets. Jia et al. [
35] proposed the MSTGCN model with domain generalization capability to extract subject-invariant sleep features. Additionally, Mohammed et al. [
36] applied domain adaptation techniques to mitigate inter-subject feature distribution discrepancies, enhancing depression classification performance. He et al. [
37] designed three alignment mechanisms—domain alignment, semantic alignment, and structural alignment—within a deep neural network framework to minimize domain gaps. Jin et al. [
38] proposed a method that combined unsupervised semantic segmentation with multi-level feature space adversarial transfer learning, significantly improving localization accuracy and segmentation quality in real-world scenarios. Ayodele et al. [
39] employed a domain generalization strategy to integrate multi-source EEG data and utilized a recurrent convolutional network for epilepsy detection, achieving 72.5% sensitivity and a low false positive rate on an independent dataset. Ganin et al. [
40] proposed a deep domain adaptation method incorporating a GRL, which enabled joint training on labeled source-domain data and unlabeled target-domain data, thereby promoting the learning of discriminative and domain-invariant features. Li et al. [
41] employed a convolutional neural network integrated with transfer learning to recognize mild depression, leveraging spectral, spatial, and temporal information from EEG signals. However, existing studies generally overlook the class imbalance problem, potentially a critical factor contributing to suboptimal classification performance.
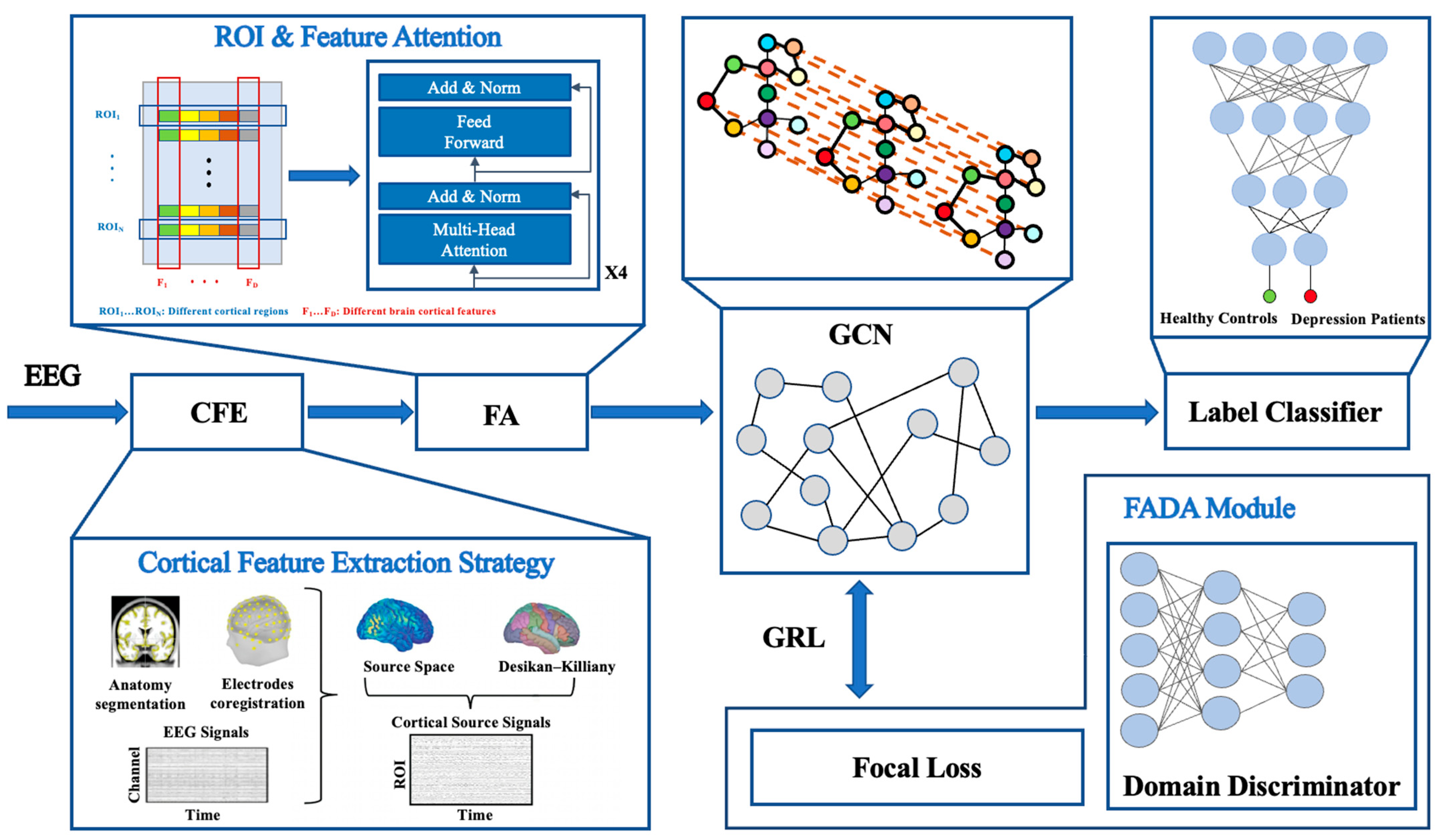
Therefore, this paper proposes a multi-stage deep learning model for EEG-based depression classification, which integrates a CFE strategy, FA module, GCN, and FADA module to improve classification accuracy and generalizability. Specifically, the CFE strategy reconstructs brain cortical source signals using the sLORETA algorithm and extracts multi-dimensional features to characterize cortical activity. The FA module leverages a multi-head self-attention mechanism to enhance the representation of spatiotemporal dependencies among brain regions. The GCN models functional connectivity between brain regions to capture higher-order structural information, while the FADA module alleviates class imbalance and domain shift issues through the use of Focal Loss and GRL mechanisms. Experimental validation on the publicly available PRED+CT dataset demonstrates that the proposed model achieves an accuracy of 85.33%, representing a 2.16% improvement over current mainstream methods, thereby highlighting its effectiveness and potential in cross-subject EEG analysis for depression classification.
2. Methods
This section first provides an overview of the proposed model framework, followed by detailed descriptions of the CFE strategy, FA module, and GCN. Finally, this paper discusses the FADA module to enhance the model’s robustness and generalization capability.
2.1. Overall Framework of Our Model
Figure 1 illustrates the overall framework of the proposed depression classification model. First, raw multi-channel EEG signals are processed through the CFE strategy, mapping EEG signals to high-resolution cortical source signals using the sLORETA algorithm. Subsequently, multi-dimensional features including frequency-domain, time-domain, spatial, and nonlinear characteristics are extracted, forming feature matrices that reflect cortical activation patterns. Next, brain cortical features enter the FA module, which is based on a Transformer architecture, employing a multi-head attention mechanism to enhance key regional and temporal features. A GCN, integrated with the cortical connectivity graph, further models structured high-dimensional features to extract discriminative representations. Finally, classification is performed using a classifier. To enhance the model’s generalization ability, the FADA module employs a GRL to mask domain-specific information and incorporates Focal Loss to balance the samples.
2.2. Cortical Feature Extraction Strategy
The CFE strategy aims to extract biologically meaningful brain cortical features from EEG signals, reflecting cortical activities relevant for depression classification. A two-step procedure is adopted: firstly, cortical source signals are reconstructed using sLORETA [
42] to derive current-source distributions; secondly, multi-dimensional features are extracted from cortical signals across different frequency bands, encompassing temporal, spectral, spatial, and nonlinear characteristics.
2.2.1. Cortical Source Reconstruction
Cortical source signals are reconstructed via sLORETA using the Brainstorm toolbox (v3.4) in MATLAB R2021a [
43]. The reconstruction process begins by building a four-layer boundary element head model, comprising the scalp, outer skull, inner skull, and cortex [
44]. This model is based on the ICBM152 MNI standard template, defining a cortical source space with 15,002 vertices (~5 mm spacing). Each vertex is represented by a current dipole oriented perpendicular to the cortical surface.
Source reconstruction solves the EEG inverse problem, inferring cortical source distributions
(source points
, time points
) from scalp potentials
(electrodes
). The process is described by the lead field matrix
:
where
denotes measurement noise. Due to the severe ill-posedness of the inverse problem, sLORETA introduces Tikhonov regularization:
where
denotes the Frobenius norm and the closed-form solution of the formula is
where
denotes the identity matrix. The regularization parameter
is estimated using generalized cross-validation to balance the data-fitting term and the norm constraint of the solution. To eliminate the dependence of source localization on current amplitude, sLORETA standardizes the current density:
where
is the estimated dipole moment at source
,
denotes the noise covariance (usually
), and
is the transfer submatrix for source
.
The sLORETA implementation is performed using the Brainstorm toolbox (v3.4) within MATLAB R2021a. The electrode-level EEG data
, recorded via the standard 10–20 electrode system, were spatially co-registered with the head model to construct the forward model. A four-layer boundary element method (BEM) was utilized for the head model, comprising scalp, outer skull, inner skull, and cortical surfaces, with the cortical surface serving as the defined source space
. The cortical source space, constructed based on the ICBM152 template, contains 15,002 vertices, each assigned a dipole source oriented perpendicular to the cortical surface. The lead-field matrix
was generated using the aforementioned BEM model, with conductivity values assigned as 1 S/m for cerebrospinal fluid, 0.0125 S/m for the skull, and 1 S/m for the scalp. The regularization parameter
was set to 0.05 to balance data-fitting and source-norm constraints. After source estimation, the vertex-level current densities
were mapped onto 68 regions of interest (ROIs) defined by the Desikan–Killiany atlas [
45], and the averaged current density across all vertices within each region was used as the final feature representation for that region. Details of the ROIs are provided in
Table 1.
Figure 2a illustrates the distribution of original EEG signals in the scalp space, while
Figure 2b depicts the corresponding cortical source signals after source reconstruction.
2.2.2. Feature Extraction
Based on evidence from EEG studies on depression, multi-dimensional features across frequency bands of ROI signals (theta 4–8 Hz, alpha 8–13 Hz, beta 13–30 Hz, gamma 31–80 Hz) are extracted, which contain linear features (spatial, temporal, and spectral domains) and nonlinear features (dynamic complexity metric), aiming to comprehensively characterize the EEG signal and provide effective input information for depression classification. The complete feature list is shown in
Table 2.
Spatial features are extracted by constructing a brain network, where each ROI acts as a node and the connection strength between nodes is quantified using the Phase Locking Value (PLV) [
46]. The PLV measures the degree of phase synchronization between a pair of signals in the frequency band
, which is computed as
where
denotes the instantaneous phase of the signal in the frequency band
and time point
(obtained by Hilbert transform), and
is the total number of time points. The PLV ranges from 0 to 1, quantifying the degree of phase synchronization—where values closer to 1 indicate stronger phase synchronization.
Based on the PLV adjacency matrix, the Clustering Coefficient (Cp) and Local Efficiency (Eloc) are further calculated. The Cp measures the degree to which the neighbors of a node are interconnected, and is defined as
where
denotes the actual number of edges among the neighbors of node
, and
is the degree (number of neighboring nodes) of node
. Eloc quantifies the efficiency of information transfer within the local subgraph composed of a node’s immediate neighbors, and is defined as
where
and
are the elements of the adjacency matrix (1 if the connection exists, 0 otherwise), and
is the shortest path length through the neighbors of node
.
Finally, all brain cortical features extracted from different frequency bands and dimensions are organized into a feature matrix
, where
is the number of ROIs, and
is the sum of the feature dimensions extracted from each ROI. In this paper,
includes spectral features [
47], temporal features, spatial features [
48], and nonlinear features [
49].
2.3. Feature Attention Module
To further enhance the feature representation of EEG data, we introduce the FA module based on the Transformer architecture. This module enhances the input features through a multi-head self-attention mechanism, aiming to effectively capture the spatiotemporal relationships between different brain regions and thus improve the accuracy of depression classification. The FA module employs a 4-layer multi-head self-attention mechanism, which is able to enrich feature representations while preserving the original feature dimensionality.
Module Architecture
The core of the FA module is a Transformer architecture based on the multi-head self-attention mechanism. In this architecture, each layer contains multiple parallel self-attentive heads, which independently capture relationships among input features within different subspaces. Specifically, the FA module takes as input the feature matrix of ROI signals, processes the features using the multiple self-attention mechanism, and outputs an enhanced feature matrix .
First, the input feature matrix
is mapped to the query, key, and value matrices by different linear transformations, which are expressed as
where
are learnable weight matrices, and
denote the query, key, and value matrices, respectively. Next, the attention scores are computed and the value matrix is weighted and summed to generate the output for each header. The attention mechanism is computed as
where
is the scaling factor and
is the dimension of the key vectors. The outputs from all attention heads are concatenated and passed through a linear transformation to produce the final output matrix
. The computation is formulated as
where
denotes the number of attention heads, and
is the linear transformation matrix of the output. By adding the input from the previous layer and applying a ReLU activation function, nonlinear transformations of the features are ensured, completing the processing of each layer.
The process is repeated for four layers, resulting in a deep feature enhancement network. The output of each layer is used as input to the next layer, thus capturing more complex feature relationships at multiple levels.
2.4. Graph Convolution Neural Network
GCN is a deep learning model specifically designed for processing graph-structured data, which can directly model the topological relationships among nodes in the data compared to traditional CNN. In this paper, GCN is used to process multi-dimensional features extracted from ROI signals, learning the functional connectivity relationships between brain regions, and to effectively capture spatiotemporal characteristics of neural activity through graph convolution operations.
2.4.1. Feature Input and Adjacency Matrix Construction
In the GCN model, the input feature matrix
consists of the multi-dimensional features obtained from the CFE strategy described in the previous section. These features effectively reflect the spatiotemporal patterns in the ROI signals. To further capture the spatial structural relationships between brain regions, we construct an adjacency matrix
. Previous studies have shown that the correlation coefficient between different brain regions is an effective indicator for depression classification [
50]. This adjacency matrix is constructed as follows:
where
denotes the pearson correlation coefficient between the
th and
th ROI signals, and
is the predefined correlation threshold. If
exceeds this threshold, then it is considered that there is a connection between the two ROIs, and the corresponding element
in the adjacency matrix is set to 1; otherwise,
is set to 0.
2.4.2. Graph Convolution Operation
The graph convolution operation is the core mechanism of the GCN, integrating both the topological structure of the graph and the feature information of nodes, and is able to efficiently learn both local and global structural patterns among nodes. In this paper, the graph convolution operation is performed using the normalized adjacency matrix
and the feature matrix
. The formula for graph convolution is as follows:
where
denotes the input feature matrix of the graph convolution layer
,
represents the output feature matrix of the graph convolution layer
,
is the normalized adjacency matrix,
is the convolution kernel weights of the
th layer, and
is the activation function, and ReLU is used in this paper.
To avoid the adverse effects of large variations in node degrees in graph convolution on model training, the adjacency matrix
is normalized:
where
is the original adjacency matrix,
is the identity matrix, and
is the degree matrix,
, which represents the degree of node
. The normalized adjacency matrix
ensures that the contribution of each node in the convolution operation is more balanced, which helps to improve the learning ability of GCN on graph-structured data.
2.5. Focal Adversarial Domain Adaptation Module
This paper proposes a module called FADA, which integrates Focal Loss, a domain-adversarial training mechanism based on GRL, and a class-center constraint (Center Loss) to construct a robust classification strategy for cross-subject EEG analysis. The FADA is designed to simultaneously address the class imbalance problem, achieve inter-domain feature alignment, and enhance the intra-class compactness in the feature space, thus adapting to the inherent distributional heterogeneity of EEG signals across different individuals.
2.5.1. Focal Loss for Addressing Class Imbalance
In depression-related EEG data, significant class imbalance often arises due to limitations in clinical sample collection. During training, models tend to bias toward the majority class, which affects the accuracy of depression classification. The FADA introduces Focal Loss as the main classification loss function, which effectively improves the model’s ability to focus on minority class and boundary samples. It is defined as follows:
where
denotes the total number of training samples,
represents the
th sample,
denotes its ground-truth label (with 0 indicating a depressed subject and 1 indicating a healthy subject), and
denotes the predicted probability for the true class. The parameter
serves as a class-balancing factor, and
is the focal parameter, which suppresses the model’s excessive attention to easily classified samples. Focal Loss significantly enhances the model’s ability to distinguish minority class instances, providing a stable foundation for subsequent domain alignment and intra-class compactness constraints.
2.5.2. Adversarial Domain Adaptation Mechanism Based on GRL
EEG signals often exhibit substantial distributional shifts across individuals and environments, making it difficult for traditional supervised training to generalize to new domains. To address this issue, the FADA module integrates an adversarial domain adaptation mechanism based on GRL, aiming to encourage the model to learn domain-invariant discriminative features.
Let the feature extractor be , the classifier be , and the domain discriminator be . The model is trained jointly through two optimization objectives: first, minimizing the classification loss to improve predictive performance; second, maximizing the domain discriminator loss so that the extracted features are difficult to distinguish from the source domain, thereby achieving cross-domain alignment.
The adversarial loss is defined as
where
denotes the domain label of sample
,
represents the total number of source domains, and
is an indicator function used to determine whether a sample belongs to the
th source domain. The GRL connects the feature extractor and domain discriminator by reversing the gradient during backpropagation, resulting in the following update rule:
where
is the learning rate, and
is a hyperparameter that controls the strength of adversarial training. Through this mechanism, the FADA enables domain-adversarial optimization, enabling the feature extractor to learn more stable and transferable feature representations.
2.5.3. Class-Center Constraint for Enhancing Intra-Class Compactness
After domain alignment, samples of the same class in different domains may still be loosely distributed, affecting the stability of classification boundaries. To address this issue, the FADA module further introduces class-centered loss, which constrains samples of the same class to cluster around the same class center to enhance intra-class consistency. The loss is defined as
where
denotes the feature representation of sample
, and
is the center vector of class
. The norm term measures the distance between the feature and its corresponding class center. This loss term jointly optimizes the class center positions during training, guiding the model to learn discriminative and compact intra-class representations, which is particularly significant for pathological states with blurred boundaries, such as depression.
2.5.4. Joint Optimization Objective
The FADA module comprehensively considers classification accuracy, domain alignment, and intra-class consistency to construct a unified joint loss function. The overall optimization objective is as follows:
where
and
are the weight hyperparameters of class Center Loss and adversarial loss, respectively, which are used to balance the three objectives. This joint optimization strategy ensures that the model has the ability to cope with challenges such as class imbalance, cross-domain differences, and feature discreteness in EEG data for depression, thereby achieving stable and efficient classification performance.