Pathogenic Bacterial Detection Using Vertical-Capacitance Sensor Array Immobilized with the Antimicrobial Peptide Melittin
Abstract
1. Introduction
2. Methods
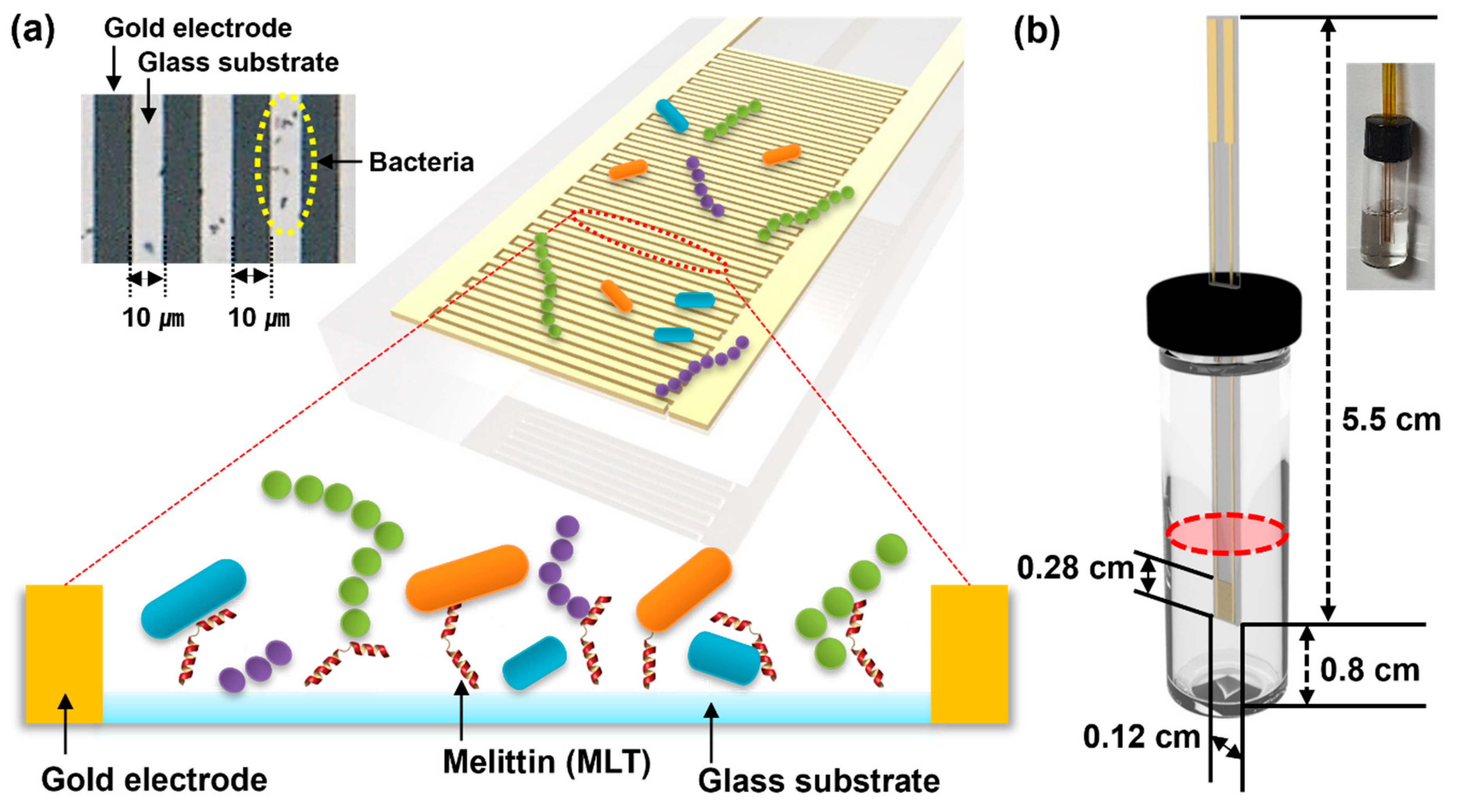
2.1. Reagents, Bacterial Cells, and Sensor Fabrication
2.2. Data Collection
3. Results and Discussion
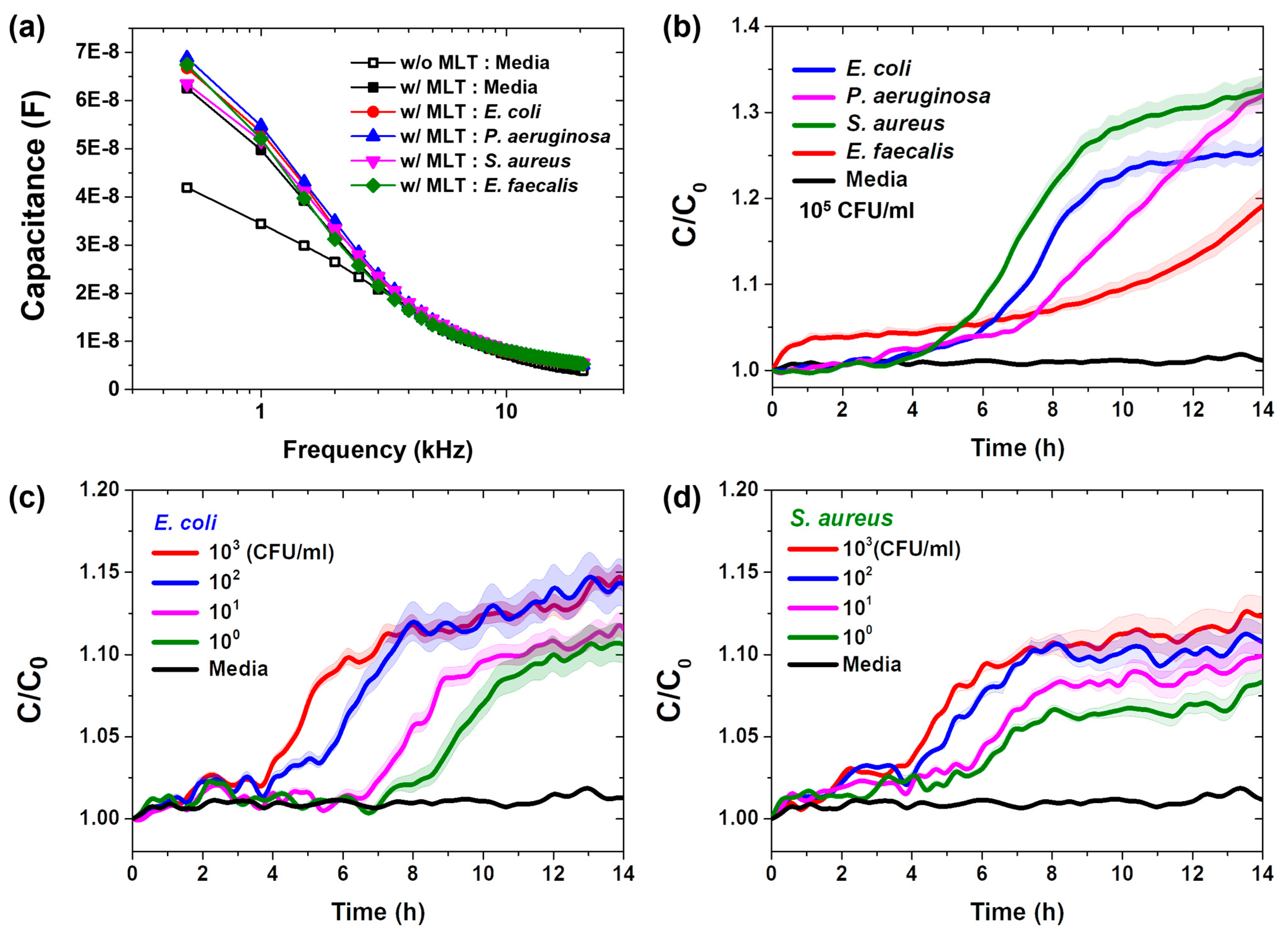
3.1. Vertical-Capacitance Sensor Characteristics
3.2. Real-Time Pathogenic Bacterial Detection in Media
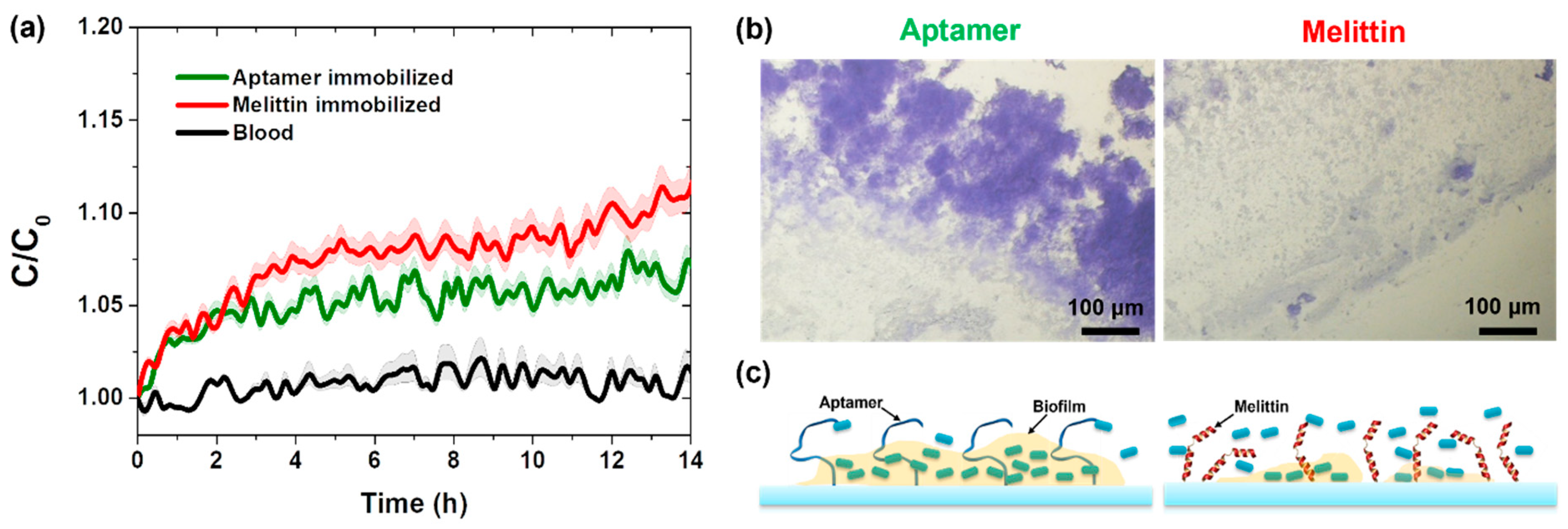
3.3. Real-Time Detection of Pathogenic Bacteria in Blood
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Deng, H.; Kong, Y.; Zhu, J.; Jiao, X.; Tong, Y.; Wan, M.; Zhao, Y.; Lin, S.; Ma, Y.; Meng, X. Proteomic analyses revealed the antibacterial mechanism of Aronia melanocarpa isolated anthocyanins against Escherichia coli O157: H7. Curr. Res. Food Sci. 2022, 5, 1559–1569. [Google Scholar] [CrossRef]
- Law, J.W.F.; Mutalib, N.S.A.; Chan, K.G.; Lee, L.H. Rapid methods for the detection of foodborne bacterial pathogens: Principles, applications, advantages and limitations. Front. Micobiol. 2015, 12, 770. [Google Scholar] [CrossRef]
- Mamun, M.A.; Wahab, Y.A.; Hossain, M.A.M.; Hashem, A.; Johan, M.R. Electrochemical biosensors with Aptamer recognition layer for the diagnosis of pathogenic bacteria: Barriers to commercialization and remediation. Trends Anal. Chem. 2021, 145, 116458. [Google Scholar] [CrossRef]
- Techakasikornpanich, M.; Jangpatarapongsa, K.; Polpanich, D.; Zine, N.; Errachid, A.; Elaissari, A. Biosensor technologies: DNA-based approaches for foodborne pathogen detection. TrAC Trends Anal. Chem. 2024, 180, 117925. [Google Scholar] [CrossRef]
- Lu, Y.; Yang, H.; Bai, J.; He, Q.; Deng, R. CRISPR-Cas based molecular diagnostics for foodborne pathogens. Crit. Rev. Food. Sci. Nutr. 2024, 64, 5269–5289. [Google Scholar] [CrossRef]
- Cuntín-Abal, C.; Jurado-Sánchez, B.; Escarpa, A. Playing with biological selectivity: Antimicrobial peptides and bacteriophages-based optical biosensors for pathogenic bacteria detection. TrAC Trends Anal. Chem. 2024, 172, 117925. [Google Scholar] [CrossRef]
- Song, J.H.; Lee, S.M.; Park, I.H.; Yong, D.; Lee, K.S.; Shin, J.S.; Yoo, K.H. Vertical capacitance aptasensors for real-time monitoring of bacterial growth and antibiotic susceptibility in blood. Biosens. Bioelectron. 2019, 143, 111623. [Google Scholar] [CrossRef]
- Lee, K.S.; Lee, S.M.; Oh, J.; Park, I.H.; Han, M.; Yong, D.; Lim, K.J.; Shin, J.S.; Yoo, K.H. Electrical antimicrobial susceptibility testing based on aptamer-functionalized capacitance sensor array for clinical isolates. Sci. Rep. 2020, 10, 13709. [Google Scholar] [CrossRef]
- Liu, S.; Lu, F.; Chen, S.; Ning, Y. Graphene oxide-based fluorescent biosensors for pathogenic bacteria detection: A review. In Analytica Chimica Acta; Elsevier: Amsterdam, The Netherlands, 2024; p. 343428. [Google Scholar]
- Zhang, B.; Asad Rahman, M.; Liu, J.; Huang, J.; Yang, Q. Real-time detection and analysis of foodborne pathogens via machine learning based fiber-optic Raman sensor. Measurement 2023, 217, 113121. [Google Scholar] [CrossRef]
- Jo, N.; Kim, B.; Lee, S.M.; Oh, J.; Park, I.H.; Lim, K.J.; Shin, J.S.; Yoo, K.H. Aptamer-functionalized capacitance sensors for real real-time monitoring of bacterial growth and antibiotic susceptibitility. Biosens. Bioelectron. 2018, 102, 164–170. [Google Scholar] [CrossRef]
- Li, Y.; Afrasiabi, R.; Fathi, F.; Wang, N.; Xiang, C.; Love, R.; She, Z.; Kraatz, H.B. Impedance based detection of pathogenic E. coli O157:H7 using a ferrocene-antimicrobial peptide modified biosensor. Biosens. Bioelectron. 2014, 58, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Eaton, B.E.; Gold, L.; Zichi, D.A. Let’s get specific: The relationship between specificity and affinity. Chem. Biol. 1995, 2, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Chung, J.; Song, M.Y.; Jurng, J.; Kim, B.C. Aptamer cocktails: Enhancement of sensing signals compared to single use of aptamers for detection of bacteria. Biosens. Bioelectron. 2014, 54, 195–198. [Google Scholar] [CrossRef] [PubMed]
- Mannoor, M.S.; Zhang, S.; Link, A.J.; McAlpine, M.C. Electrical detection of pathogenic bacteria via immobilized antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2010, 107, 19207–19212. [Google Scholar] [CrossRef]
- Lillehoj, P.B.; Kaplan, C.W.; He, K.J.; Shi, W.; Ho, C. Rapid electrical impedance detection of bacterial pathogens using immobilized antimicrobial peptides. J. Lab. Autom. 2014, 19, 42–49. [Google Scholar] [CrossRef]
- Etayash, H.; Jiang, K.; Thundat, T.; Kaur, K. Impedimetric Detection of Pathogenic Gram-Positive Bacteria Using an Antimicrobial Peptide from Class IIa Bacteriocins. Anal. Chem. 2014, 86, 1693–1700. [Google Scholar] [CrossRef]
- Wilson, D.; Materón, E.M.; Ibáñez-Redín, G.; Faria, R.C.; Correa, D.; Oliveira, O.N., Jr. Electrical detection of pathogenic bacteria in food samples using information visualization methods with a sensor based on magnetic nanoparticles functionalized with antimicrobial peptides. Talanta 2019, 194, 611–618. [Google Scholar] [CrossRef]
- Pardoux, E.; Boturyn, D.; Roupioz, Y. Antimicrobial peptides as probes in biosensors detecting whole bacteria: A review. Molecules 2020, 25, 1998. [Google Scholar] [CrossRef]
- Kulagina, N.V.; Lassman, M.E.; Ligler, F.S.; Taitt, C.R. Antimicrobial peptides for detection of bacteria in biosensor assays. Anal. Chem. 2005, 77, 6504–6508. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Rydlo, T.; Rotem, S.; Mor, A. Antibacterial properties of dermaseptin S4 derivatives under extreme incubation conditions. Antimicrob. Agents Chemother. 2006, 50, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, C.; Scott, M.G.; Karunaratne, N.; Yan, H.; Hancock, R.E.W. Salt-resistant alpha helical cationic antimicrobial peptides. Antimicrob. Agents Chemother. 1999, 43, 1542–1548. [Google Scholar] [CrossRef] [PubMed]
- Kulagina, N.V.; Shaffer, K.M.; Ligler, F.S.; Taitt, C.R. Antimicrobial peptides as new recognition molecules for screening challenging species. Sens. Actuators B Chem. 2007, 121, 150–157. [Google Scholar] [CrossRef]
- Terwilliger, T.C.; Eisenberg, D. The structure of melittin: II. Interpretation of the structure. J. Biol. Chem. 1982, 257, 6016–6022. [Google Scholar]
- Dosler, S.; Karaaslan, E.; Alev Gerceker, A. Antibacterial and anti-biofilm activities of melittin and colistin, alone and in combination with antibiotics against Gram-negative bacteria. J. Chemother. 2016, 28, 95–103. [Google Scholar] [CrossRef]
- Mirzaei, R.; Alikhani, M.Y.; Arciola, C.R.; Sedighi, I.; Yousefimashouf, R.; Bagheri, K.P. Prevention, inhibition, degradation effects of melittin alone and in combination with vancomycin and rifampin against strong biofilm producer strains of methicillin-resistant Staphylococcus epidermidis. Biomed. Pharm. 2022, 144, 112670. [Google Scholar] [CrossRef]
- Kostakioti, M.; Hadjifrangiskou, M.; Hultgren, S.J. Bacterial biofilm: Development, dispersal, and therapeutic strategies in the dawn of the post antibiotic era. Cold Spring Harb. Perspect. Med. 2013, 3, 1–23. [Google Scholar] [CrossRef]
- Galdiero, G.; Lombardi, L.; Falanga, A.; Libralato, G.; Guida, M.; Carotenuto, R. Biofilms: Novel strategies based on antibiomicorbial peptides. Pharmaceuties 2019, 11, 322. [Google Scholar] [CrossRef]
- Guidelines for the Validation of Analytical Methods for the Detection of Microbial Pathogens in Foods and Feeds; Food and Drug Administration: Silver Spring, MD, USA, 2023.
- USDA. Microbiology Laboratory Guidebook; USDA: Washington, DC, USA, 2020.
- Wellinghausen, N.; Kochem, A.J.; Disqué, C.; Mühl, H.; Gebert, S.; Winter, J.; Matten, J.; Sakka, S.G. Diagnosis of bacteremia in whole-blood samples by use of a commercial universal 16S rRNA gene-based PCR and sequence analysis. J. Clin. Microbiol. 2009, 47, 2759–2765. [Google Scholar] [CrossRef]
- Kennaugh, J.K.; Gregory, W.W.; Powell, K.R.; Hendley, J.O. The effect of dilution during culture on detection of low concentrations of bacteria in blood. Pediatr. Infect. Dis. 1984, 3, 317–318. [Google Scholar] [CrossRef]
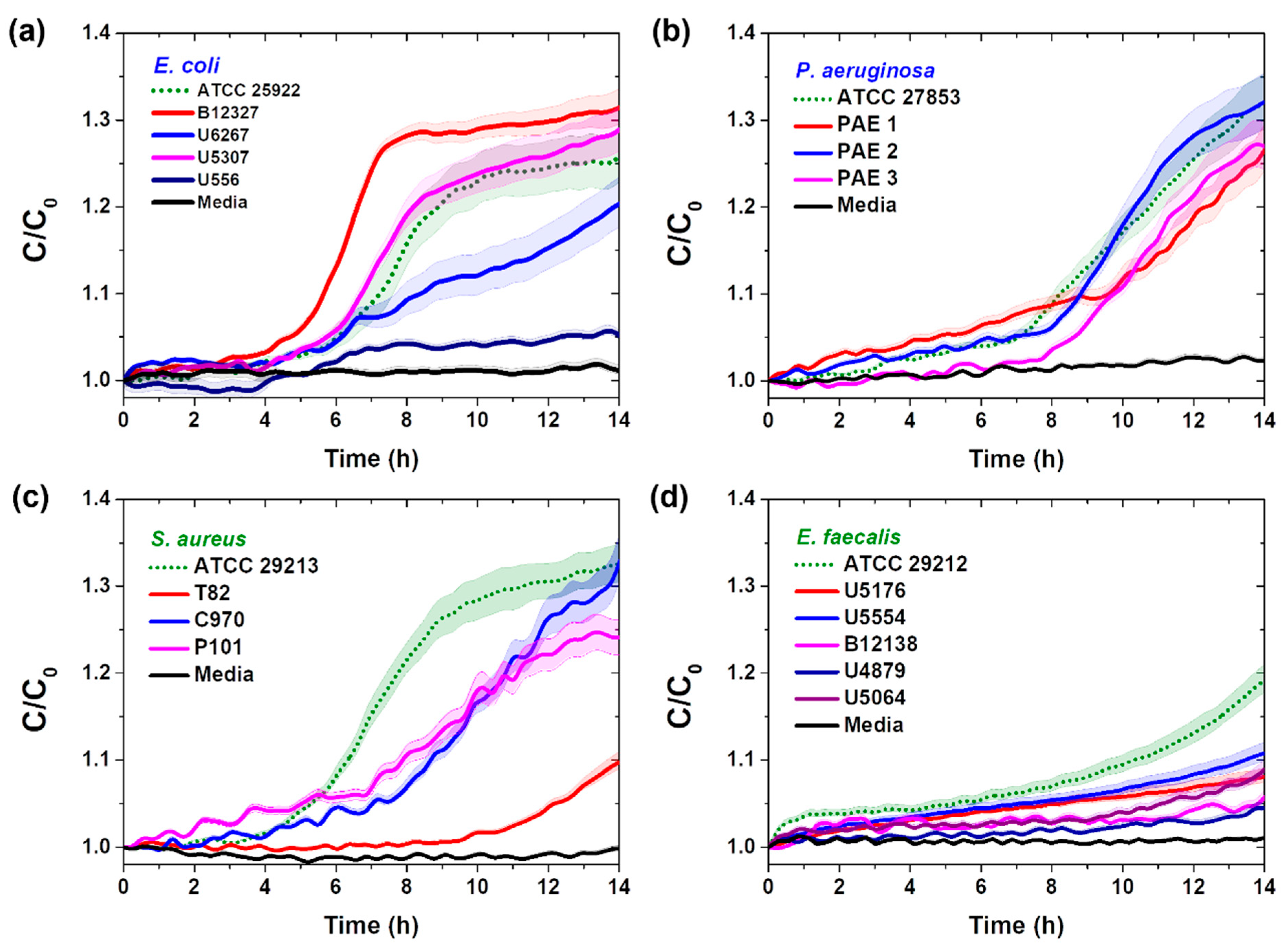
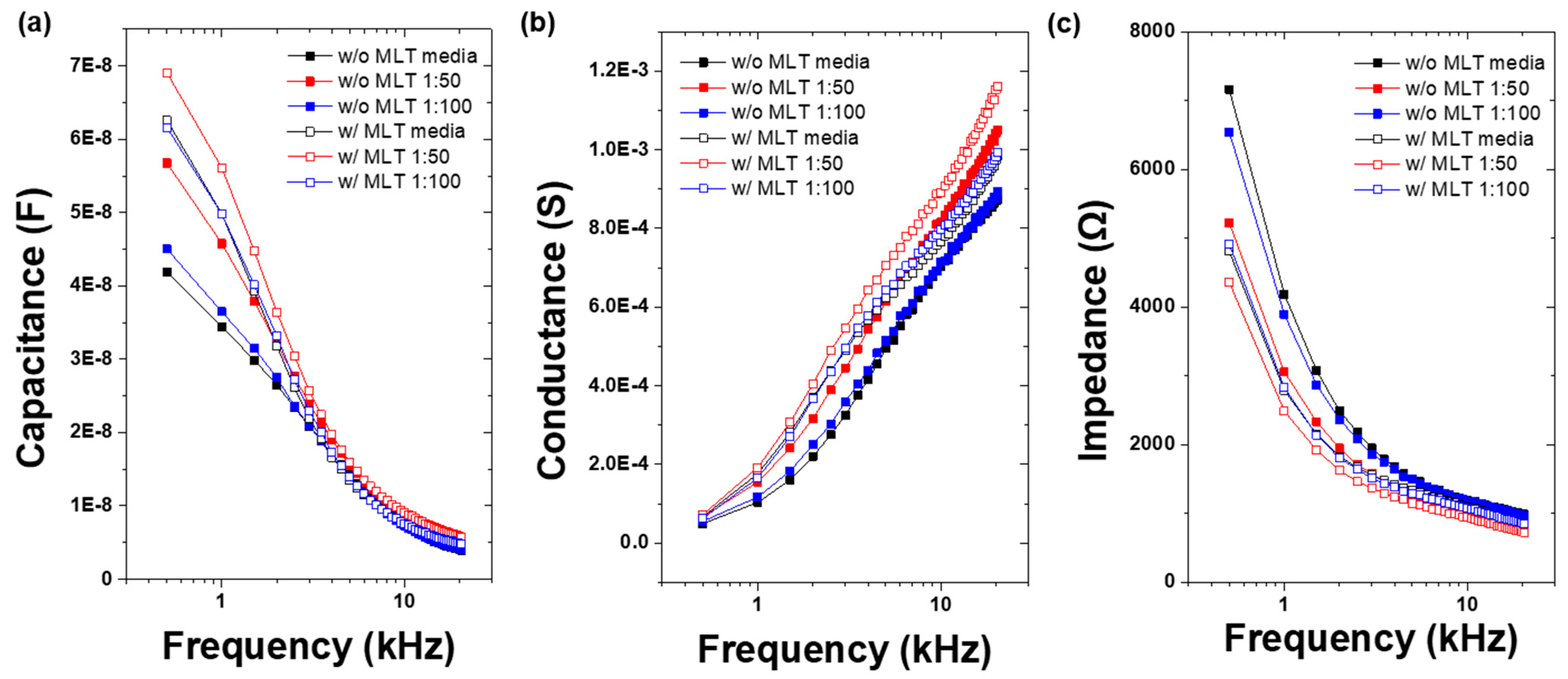

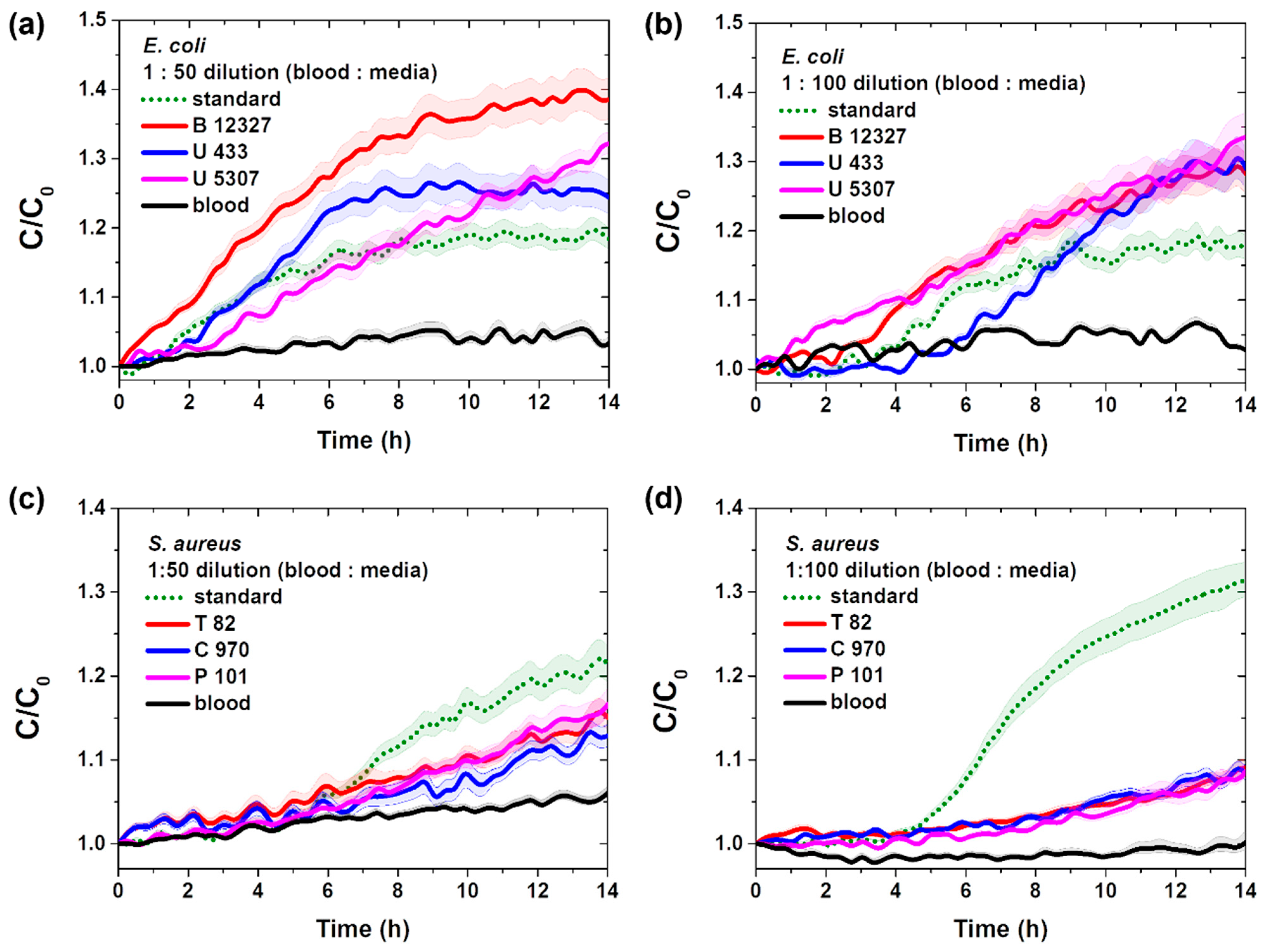
| Species of Bacteria | Standard Bacteria | Clinically Isolated Bacteria |
|---|---|---|
| Gram-negative bacteria | ||
| E. coli | ATCC 25922 | B12327, U6267, U556, U5307 |
| P. aeruginoa | ATCC 27853 | PAE1, PAE2, PAE3 |
| Gram-positive bacteria | ||
| S. aureus | ATCC 29213 | T82, C970, P101 |
| E. faecalis | ATCC 29212 | U5176, U4879, U5064, B12138, U5554 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.-M.; Song, J.-H.; Lee, K.-S.; Yoo, K.-H. Pathogenic Bacterial Detection Using Vertical-Capacitance Sensor Array Immobilized with the Antimicrobial Peptide Melittin. Sensors 2025, 25, 12. https://doi.org/10.3390/s25010012
Lee S-M, Song J-H, Lee K-S, Yoo K-H. Pathogenic Bacterial Detection Using Vertical-Capacitance Sensor Array Immobilized with the Antimicrobial Peptide Melittin. Sensors. 2025; 25(1):12. https://doi.org/10.3390/s25010012
Chicago/Turabian StyleLee, Sun-Mi, Jun-Ho Song, Kyo-Seok Lee, and Kyung-Hwa Yoo. 2025. "Pathogenic Bacterial Detection Using Vertical-Capacitance Sensor Array Immobilized with the Antimicrobial Peptide Melittin" Sensors 25, no. 1: 12. https://doi.org/10.3390/s25010012
APA StyleLee, S.-M., Song, J.-H., Lee, K.-S., & Yoo, K.-H. (2025). Pathogenic Bacterial Detection Using Vertical-Capacitance Sensor Array Immobilized with the Antimicrobial Peptide Melittin. Sensors, 25(1), 12. https://doi.org/10.3390/s25010012





