Electrocardiography Classification with Leaky Integrate-and-Fire Neurons in an Artificial Neural Network-Inspired Spiking Neural Network Framework
Abstract
1. Introduction
2. Proposed Method
2.1. Denoising ECG Signals
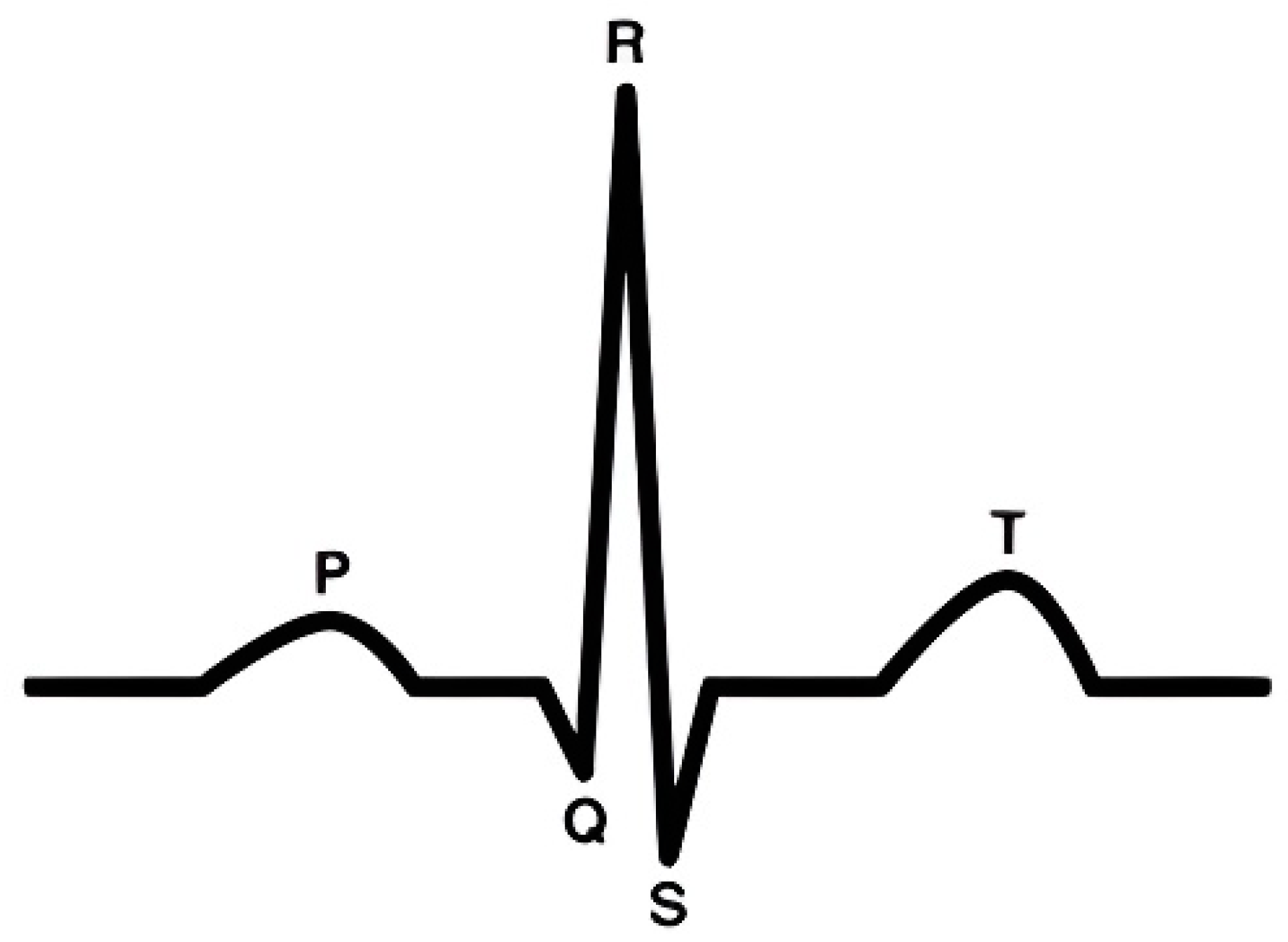
2.2. ECG Segmentation
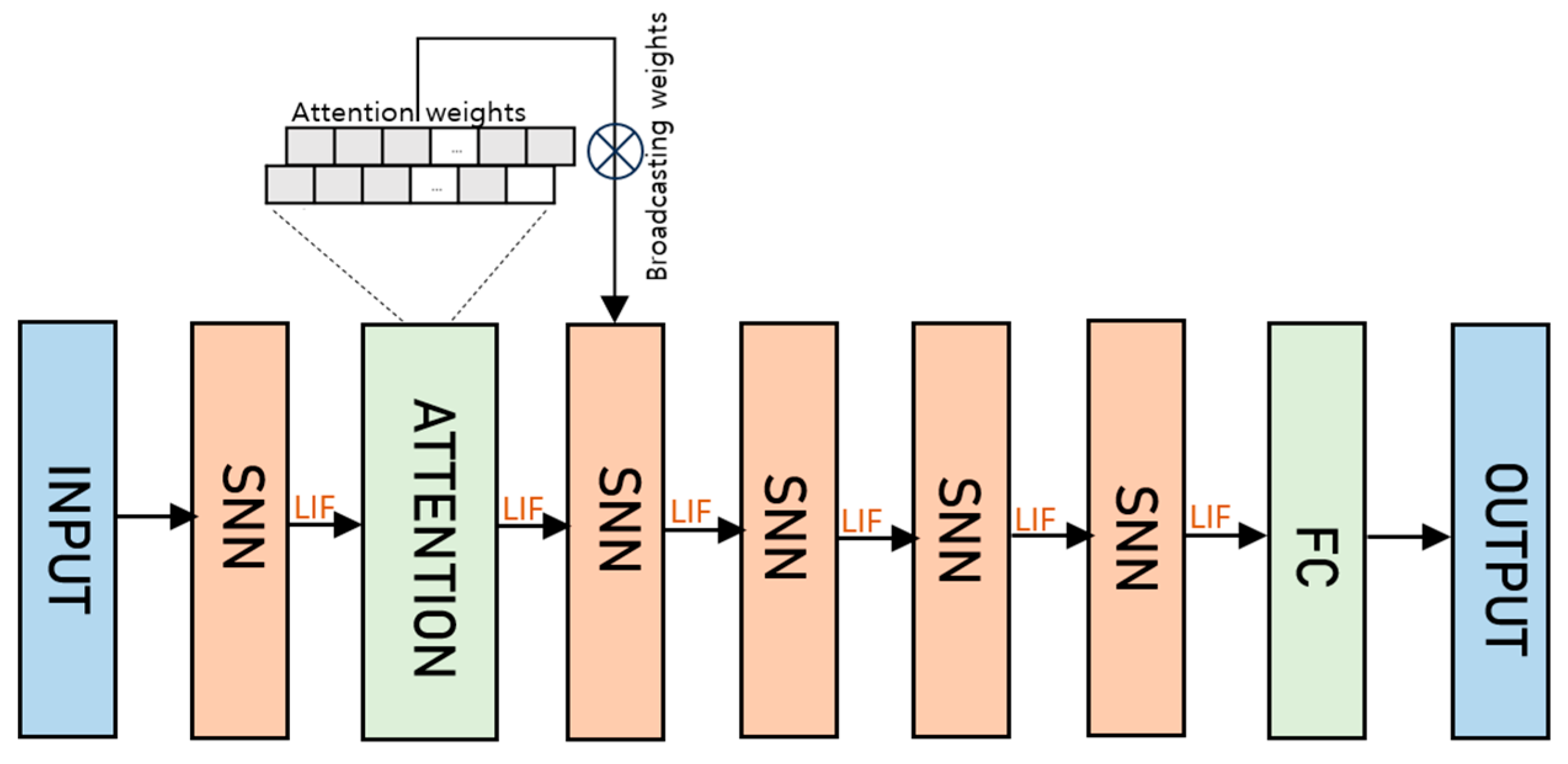
2.3. SNN
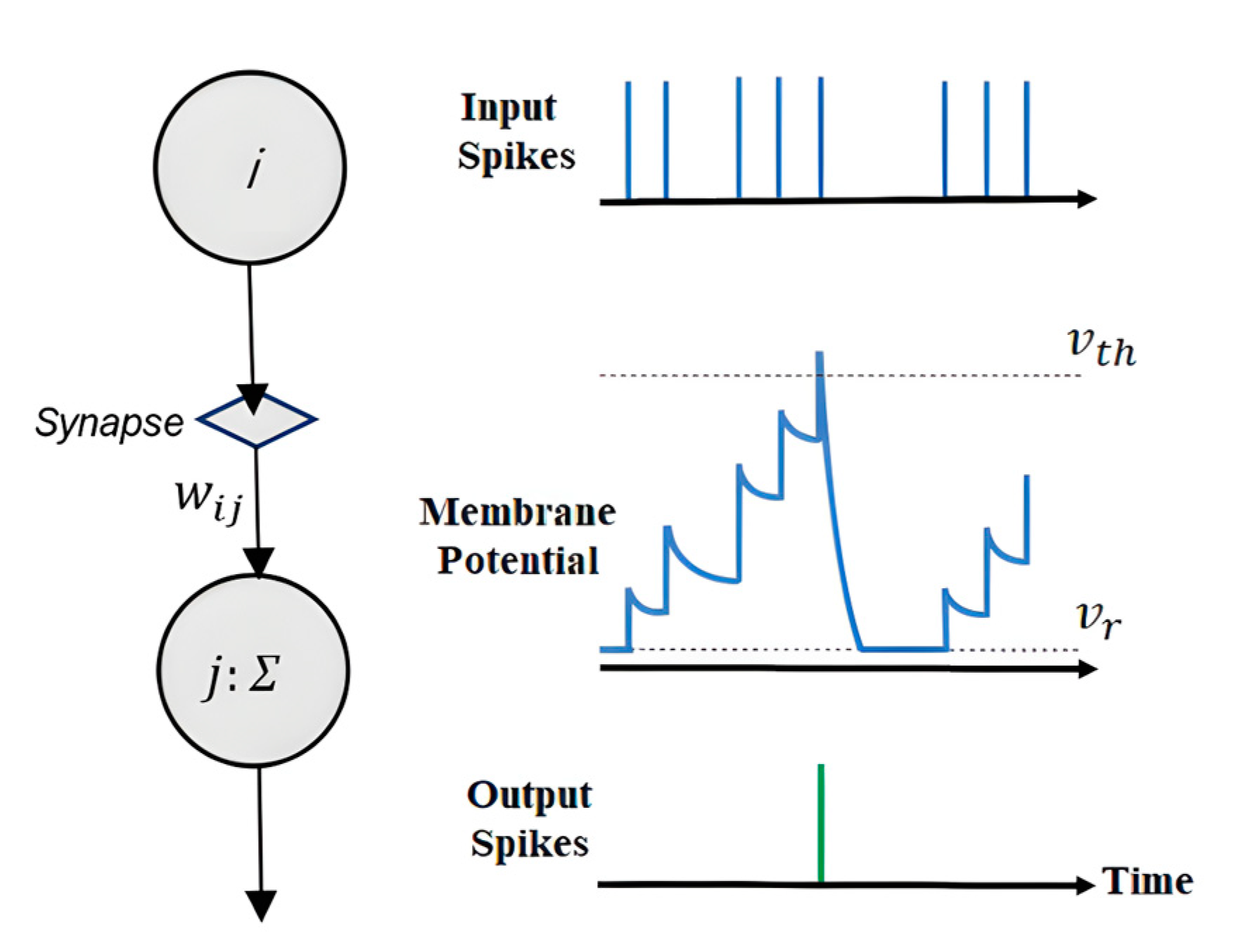
2.3.1. Leaky Integrate-and-Fire
2.3.2. Attention Module
3. Results and Discussion
3.1. Dataset Utilization and Computational Framework
3.2. Performance Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dagenais, G.R.; Leong, D.P.; Rangarajan, S.; Lanas, F.; Lopez-Jaramillo, P.; Gupta, R.; Diaz, R.; Avezum, A.; Oliveira, G.B.; Wielgosz, A.; et al. Variations in common diseases, hospital admissions, and deaths in middle-aged adults in 21 countries from five continents (PURE): A prospective cohort study. Lancet 2020, 395, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Rajni, I.K. Electrocardiogram signal analysis—An overview. Int. J. Comput. Appl. 2013, 84, 22–25. [Google Scholar] [CrossRef]
- Sopic, D.; Aminifar, A.; Aminifar, A.; Atienza, D. Real-Time Event-Driven Classification Technique for Early Detection and Prevention of Myocardial Infraction on Wearable Systems. IEEE Trans. Biomed. Circuits Syst. 2018, 12, 982–992. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Hu, Q.; Tang, W. A Real-Time QRS Detection System with PR/RT Interval and ST Segment Measurements for Wearable ECG sensors using parallel Delta Modulators. IEEE Trans. Biomed. Circuits Syst. 2018, 12, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Kiranyaz, S.; Ince, T.; Gabbouj, M. Real-Time Patient-Specific ECG Classification by 1-D Convolutional Neural Networks. IEEE Trans. Biomed. Eng. 2016, 63, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Rajpurkar, P.; Hannun, A.Y.; Haghpanahi, M.; Bourn, C.; Ng, A.Y. Cardiologist-Level Arrhythmia Detection with Convolutional Neural Networks. arXiv 2017, arXiv:1707.01836. [Google Scholar]
- Kachuee, M.; Fazeli, S.; Sarrafzadeh, M. ECG Heartbeat Classification: A Deep Transferable Representation. In Proceedings of the 2018 IEEE International Conference on Healthcare Informatics (ICHI), New York, NY, USA, 4–7 June 2018; pp. 443–444. [Google Scholar]
- Saadatnejad, S.; Oveisi, M.; Hashemi, M. LSTM-based ECG classification for continuous monitoring on personal wearable devices. IEEE J. Biomed. Health Inform. 2019, 24, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Saroj, K.P.; Rekh, R.R. Automatic Detection of Arrhythmia from Imbalanced ECG Database Using CNN Model with SMOTE. Australas. Phys. Eng. Sci. Med. 2019, 42, 1129–1139. [Google Scholar]
- Kłosowski, G.; Rymarczyk, T.; Wójcik, D.; Skowron, S.; Cieplak, T.; Adamkiewicz, P. The Use of Time-Frequency Moments as Inputs of LSTM Network for ECG Signal Classification. Electronics 2020, 9, 1452. [Google Scholar] [CrossRef]
- Tavanaei, A.; Ghodrati, M.; Kheradpisheh, S.R.; Masquelier, T.; Maida, A. Deep Learning in Spiking Neural Networks. Neural Netw. 2019, 111, 47–63. [Google Scholar] [CrossRef] [PubMed]
- Indiveri, G.; Corradi, F.; Qiao, N. Neuromorphic Architectures for Spiking Deep Neural Networks. In Proceedings of the IEEE International Electron Devices Meeting (IEDM), Washington, DC, USA, 7–9 December 2015; pp. 5–9. [Google Scholar]
- Saeed, R.K.; Timothee, M. Temporal Backpropagation for Spiking Neural Networks with One Spike per Neuron. Int. J. Neural Syst. 2020, 30, 2050027. [Google Scholar]
- Alireza, A.; Matin, H. ECG Classification Algorithm Based on STDP and R-STDP Neural Networks for Real-Time Monitoring on Ultra-Low-Power Personal Wearable Devices. IEEE Trans. Biomed. Circuits Syst. 2019, 13, 1483–1493. [Google Scholar]
- Kolagasioglu, E.; Zjajo, A. Energy Efficient Feature Extraction for Single-Lead ECG Classification Based on Spiking Neural Networks. Ph.D. Dissertation, Delft University of Technology, Delft, The Netherlands, 2018. [Google Scholar]
- Das, A.; Pradhapan, P.; Groenendaal, W.; Adiraju, P.; Rajan, R.T.; Catthoor, F.; Schaafsma, S.; Krichmar, J.L.; Dutt, N.; Van Hoof, C. Unsupervised heart-rate estimation in wearables with Liquid states and a probabilistic readout. Neural Netw. 2018, 99, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Li, Y.G.; Haider, M.R.; Massoud, Y. A low-power neuromorphic bandpass filter for biosignal processing. In Proceedings of the WAMICON 2013, Orlando, FL, USA, 7–9 April 2013. [Google Scholar]
- Cattaneo, R. Ecg Signals Classification Using Neuromorphic Hardware. Ph.D. Dissertation, ETH Zurich, Zurich, Switzerland, 2018. [Google Scholar]
- Szi-Wen, C.; Hsiao-Chen, C. A Real-Time QRS Detection Method Based on Moving-averaging Incorporating with Wavelet Denoising. Comput. Methods Programs Biomed. 2006, 82, 187–195. [Google Scholar]
- Mohammad, D. Detection of Abnormalities in Cardiac Rhythm Using Spiking Neural Networks. Thesis. 2023. Available online: https://lup.lub.lu.se/luur/download?func=downloadFile&recordOId=9113248&fileOId=9113249 (accessed on 6 April 2024).
- Bauer, F.C.; Muir, D.R.; Indiveri, G. Real-time ultra-low power ECG anomaly detection using an event-driven neuromorphic processor. IEEE Trans. Biomed. Circuits Syst. 2019, 13, 1575–1582. [Google Scholar] [CrossRef]
- Moody, G.B.; Mark, R.G. The Impact of the MIT-BIH Arrhythmia Database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Clifford, G.; Liu, C.; Moody, B.; Lehman, L.; Silva, I.; Li, Q.; Johnson, A.; Mark, R. AF classification from a short single lead ECG recording: The Physionet Computing in Cardiology Challenge 2017. In Proceedings of the 2017 Computing in Cardiology (CinC), Rennes, France, 24–27 September 2017. [Google Scholar]
- Fang, W.; Chen, Y.; Ding, J.; Yu, Z.; Masquelier, T.; Chen, D.; Huang, L.; Zhou, H.; Li, G.; Tian, Y. SpikingJelly: An Open-Source Machine Learning Infrastructure Platform for Spike-Based Intelligence. Sci. Adv. 2023, 9, eadi1480. [Google Scholar] [CrossRef]
- Yan, Z.; Zhou, J.; Wong, W.F. Energy Efficient ECG Classification with Spiking Neural Network. Biomed. Signal Process. Control. 2021, 63, 102170. [Google Scholar] [CrossRef]
- Takalo-Mattila, J.; Kiljander, J.; Soininen, J.-P. Inter-patient ECG classification using deep convolutional neural networks. In Proceedings of the 2018 21st Euromicro Conference on Digital System Design (DSD), Prague, Czech Republic, 29–31 August 2018; pp. 421–425. [Google Scholar]
- Xing, Y.; Zhang, L.; Hou, Z.; Li, X.; Shi, Y.; Yuan, Y.; Zhang, F.; Liang, S.; Li, Z.; Yan, L. Accurate ECG Classification Based on Spiking Neural Network and Attention Mechanism for Real-Time Implementation on Personal Portable Devices. Electronics 2022, 11, 1889. [Google Scholar] [CrossRef]



| Model | Overall-Acc (%) | F1 (%) | Pre (%) |
|---|---|---|---|
| 12-layer ANN (PhysioNet2017) | 84.1 | 70.1 | 73.4 |
| Proposed SNN (PhysioNet2017) | 85.8 | 70.4 | 75.7 |
| 12-layer ANN (MIT-BIH Arrhythmia) | 89.9 | 83.6 | 85.0 |
| Proposed SNN (MIT-BIH Arrhythmia) | 93.8 | 85.4 | 88.0 |
| Model | Overall-Acc (%) | F1 (%) | Pre (%) | |
|---|---|---|---|---|
| Normal | ANN | 86.8 | 90.0 | 89.6 |
| Proposed SNN | 90.8 | 92.0 | 93 | |
| Noise | ANN | 93.4 | 49.1 | 60.0 |
| Proposed SNN | 93.0 | 50.0 | 54.0 |
| Model | Acc (%) | Sen (%) | Pre (%) | F1 (%) | Power (W) |
|---|---|---|---|---|---|
| Proposed Att-SNN | 93.8 | N: 94 | N: 94.3 | N: 95.5 | 278 mW |
| V: 54 | V: 75.0 | V: 73.5 | |||
| Yan et al. [25] | 90 | N: 92 | N: 97 | N: 94.4 | 181 mW |
| V: 77 | V: 59 | V: 66.8 | |||
| Janne et al. [26] | 90 | N: 92 | N: 97 | N: 93.4 | - |
| V: 89 | V: 51 | V: 63 | |||
| Xing et al. [27] | 92.07 | N: 98.3 | N: 93.2 | N: 95.7 | 246 mW |
| V: 69.0 | V: 76.4 | V: 72.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rana, A.; Kim, K.K. Electrocardiography Classification with Leaky Integrate-and-Fire Neurons in an Artificial Neural Network-Inspired Spiking Neural Network Framework. Sensors 2024, 24, 3426. https://doi.org/10.3390/s24113426
Rana A, Kim KK. Electrocardiography Classification with Leaky Integrate-and-Fire Neurons in an Artificial Neural Network-Inspired Spiking Neural Network Framework. Sensors. 2024; 24(11):3426. https://doi.org/10.3390/s24113426
Chicago/Turabian StyleRana, Amrita, and Kyung Ki Kim. 2024. "Electrocardiography Classification with Leaky Integrate-and-Fire Neurons in an Artificial Neural Network-Inspired Spiking Neural Network Framework" Sensors 24, no. 11: 3426. https://doi.org/10.3390/s24113426
APA StyleRana, A., & Kim, K. K. (2024). Electrocardiography Classification with Leaky Integrate-and-Fire Neurons in an Artificial Neural Network-Inspired Spiking Neural Network Framework. Sensors, 24(11), 3426. https://doi.org/10.3390/s24113426







