Improved Resting-State Functional MRI Using Multi-Echo Echo-Planar Imaging on a Compact 3T MRI Scanner with High-Performance Gradients
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Data Preprocessing for fMRI Datasets
2.3. Data Analysis
2.3.1. Explained Variance Evaluation
2.3.2. ROI-Based FC Analysis
2.3.3. Seed-Based FC Map
2.3.4. ICA-Based FC Detection
3. Results
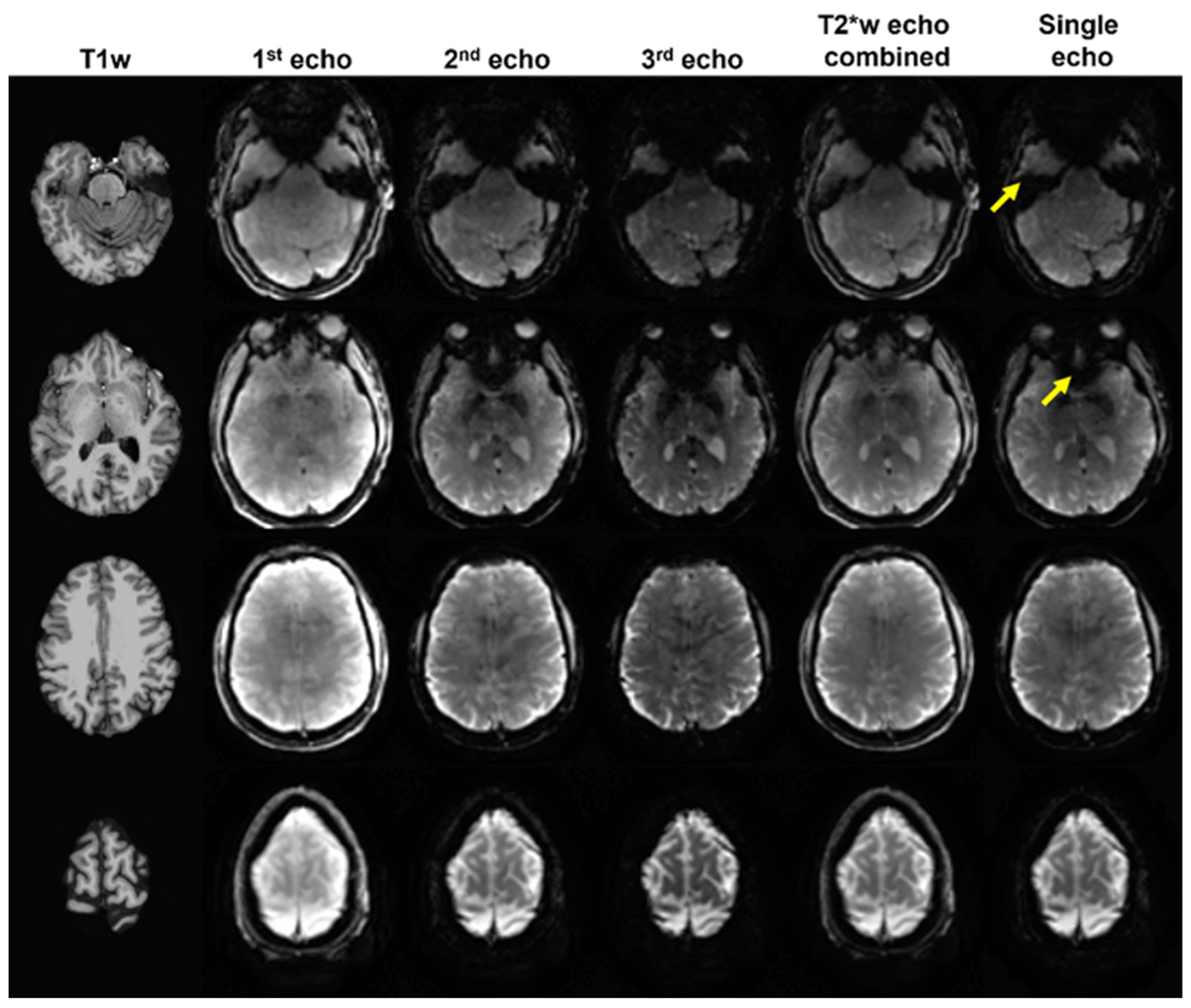
3.1. Data Selection for Analysis
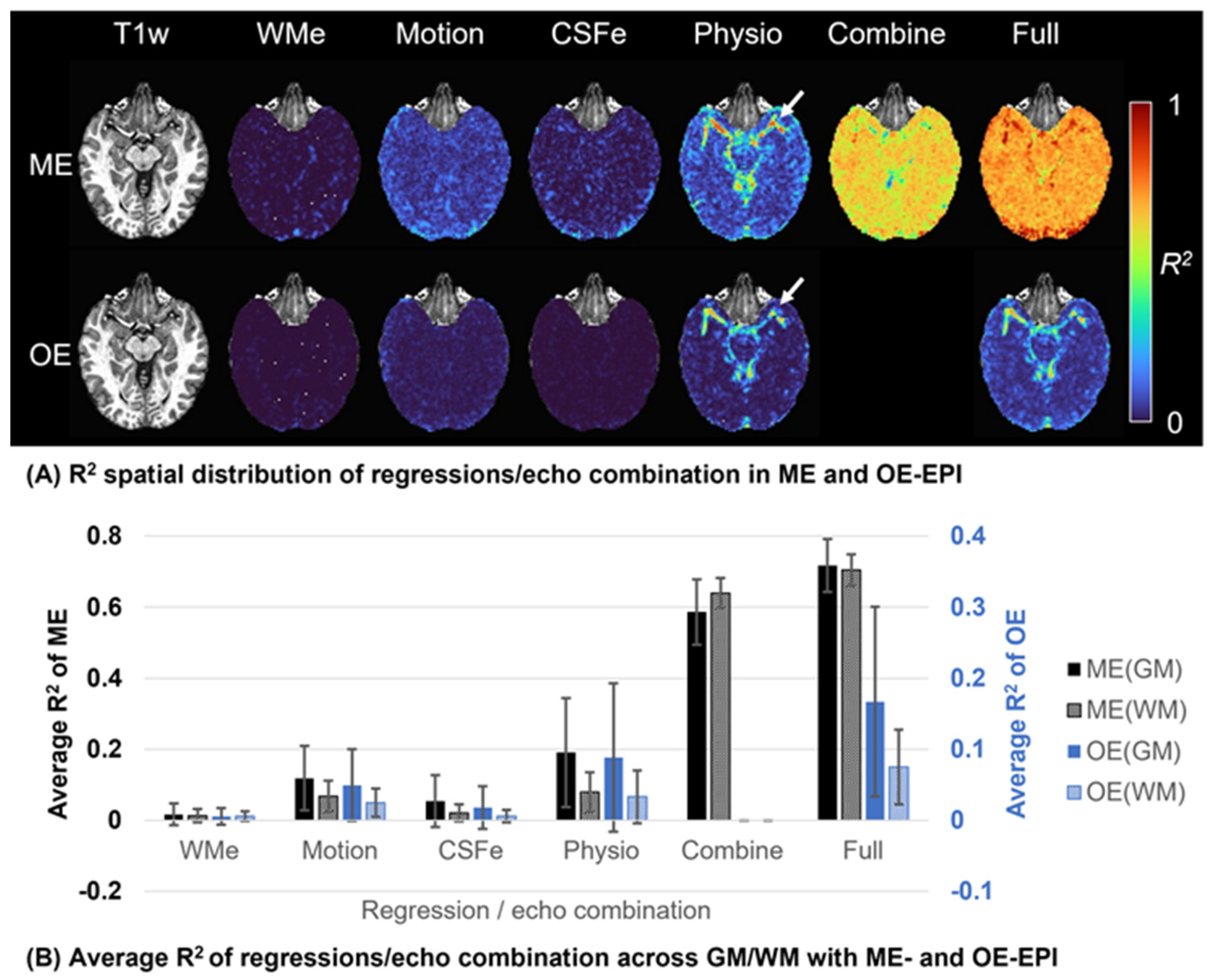
3.2. Explained Variance
3.3. ROI-Based FC Difference
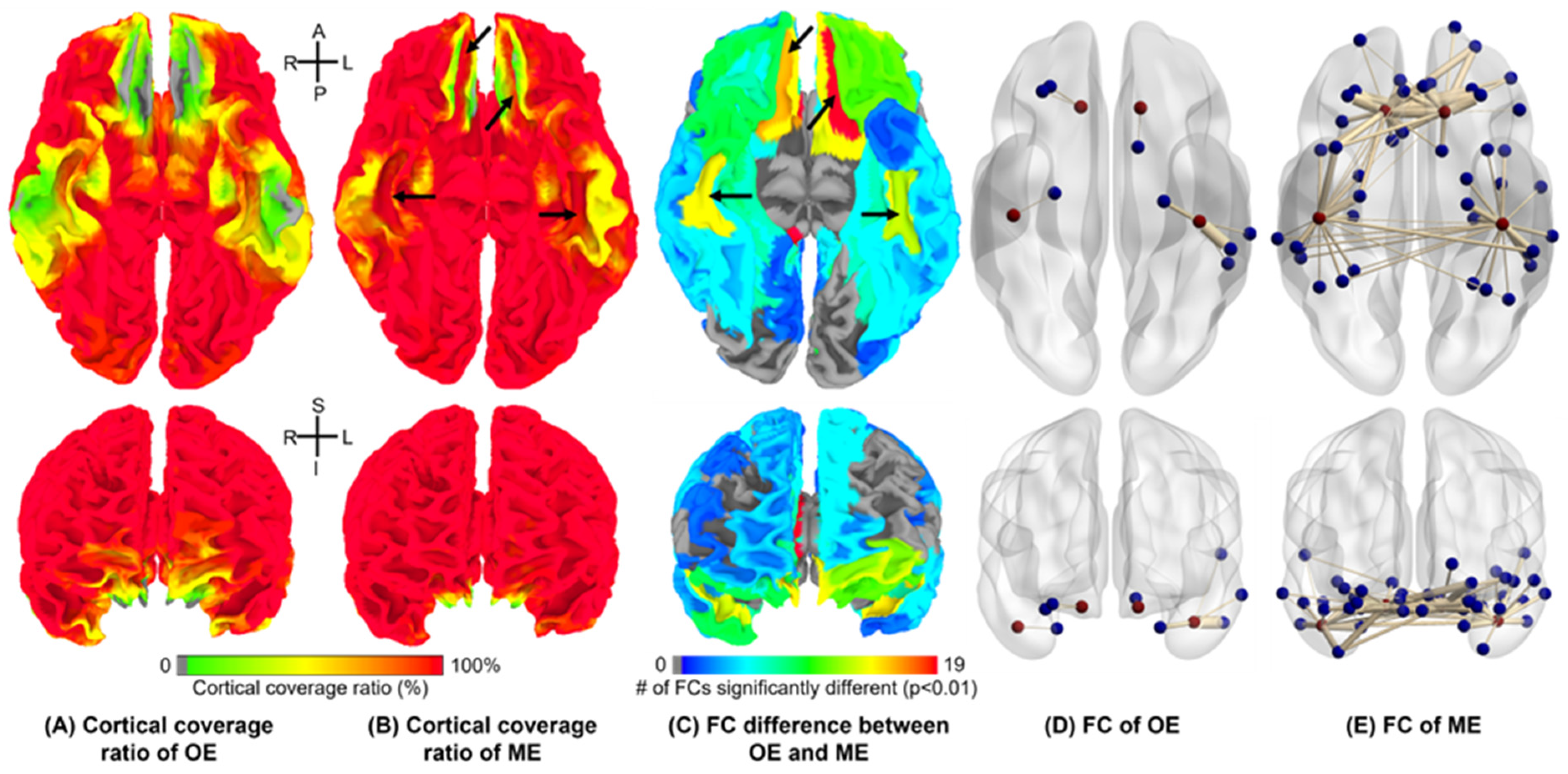
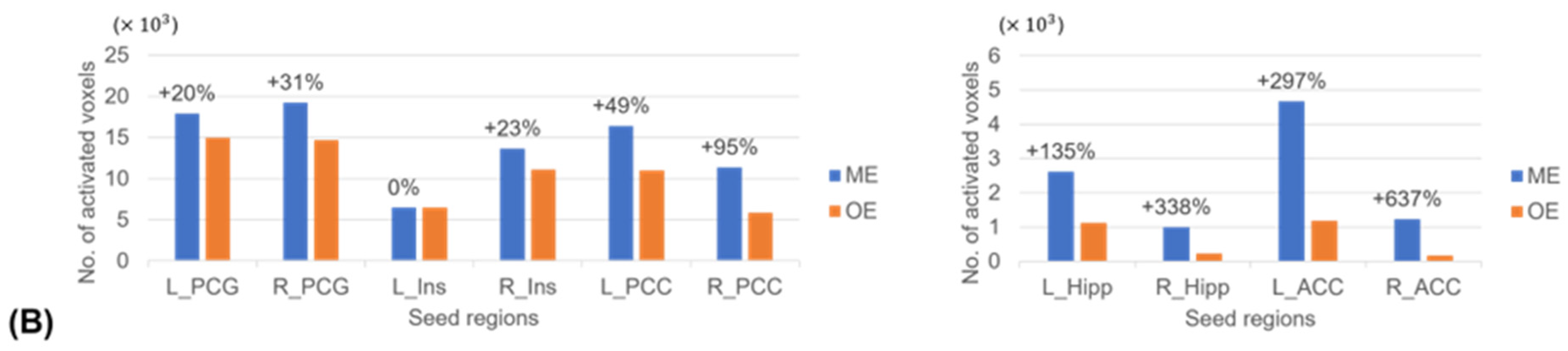
3.4. Seed-Based FC Extents
3.5. ICA-Based FC Detection
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ogawa, S.; Lee, T. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc. Natl. Acad. Sci. USA 1990, 87, 9868–9872. [Google Scholar] [CrossRef] [PubMed]
- Menon, R.S.; Ogawa, S.; Tank, D.W.; Uğurbil, K. Tesla gradient recalled echo characteristics of photic stimulation-induced signal changes in the human primary visual cortex. Magn. Reson. Med. 1993, 30, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Speck, O.; Hennig, J. Functional imaging by I0- and T2*-parameter mapping using multi-image EPI. Magn. Reson. Med. J. Soc. Magn. Reson. Med. Soc. Magn. Reson. Med. 1998, 40, 243–248. [Google Scholar] [CrossRef]
- Poser, B.a.; Versluis, M.J.; Hoogduin, J.M.; Norris, D.G. BOLD contrast sensitivity enhancement and artifact reduction with multiecho EPI: Parallel-acquired inhomogeneity-desensitized fMRI. Magn. Reson. Med. 2006, 55, 1227–1235. [Google Scholar] [CrossRef]
- Kang, D.; Sung, Y.W.; Shioiri, S. Estimation of physiological sources of nonlinearity in blood oxygenation level-dependent contrast signals. Magn. Reson. Imaging 2018, 46, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Norris, D.G. Principles of magnetic resonance assessment of brain function. J. Magn. Reson. Imaging 2006, 23, 794–807. [Google Scholar] [CrossRef]
- Casey, B.J.; Cannonier, T.; Conley, M.I.; Cohen, A.O.; Barch, D.M.; Heitzeg, M.M.; Soules, M.E.; Teslovich, T.; Dellarco, D.V.; Garavan, H.; et al. The Adolescent Brain Cognitive Development (ABCD) study: Imaging acquisition across 21 sites. Dev. Cogn. Neuros-Neth 2018, 32, 43–54. [Google Scholar] [CrossRef]
- Gunter, J.L.; Borowski, B.J.; Thostenson, K.; Arani, A.; Reid, R.I.; Cash, D.M.; Thomas, D.L.; Zhang, H.; DeCarli, C.S.; Fox, N.C.; et al. ADNI-3 MRI PROTOCOL. Alzheimer’s Dement. 2017, 13, P104–P105. [Google Scholar] [CrossRef]
- Glasser, M.F.; Sotiropoulos, S.N.; Wilson, J.A.; Coalson, T.S.; Fischl, B.; Andersson, J.L.; Xu, J.; Jbabdi, S.; Webster, M.; Polimeni, J.R.; et al. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 2013, 80, 105–124. [Google Scholar] [CrossRef]
- Setsompop, K.; Gagoski, B.A.; Polimeni, J.R.; Witzel, T.; Wedeen, V.J.; Wald, L.L. Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magn. Reson. Med. 2012, 67, 1210–1224. [Google Scholar] [CrossRef]
- Griswold, M.A.; Jakob, P.M.; Heidemann, R.M.; Nittka, M.; Jellus, V.; Wang, J.; Kiefer, B.; Haase, A. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn. Reson. Med. 2002, 47, 1202–1210. [Google Scholar] [CrossRef] [PubMed]
- Pruessmann, K.P.; Weiger, M.; Scheidegger, M.B.; Boesiger, P. SENSE: Sensitivity encoding for fast MRI. Magn. Reson. Med. 1999, 42, 952–962. [Google Scholar] [CrossRef]
- Todd, N.; Moeller, S.; Auerbach, E.J.; Yacoub, E.; Flandin, G.; Weiskopf, N. Evaluation of 2D multiband EPI imaging for high-resolution, whole-brain, task-based fMRI studies at 3T: Sensitivity and slice leakage artifacts. Neuroimage 2016, 124, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Triantafyllou, C.; Hoge, R.D.; Krueger, G.; Wiggins, C.J.; Potthast, A.; Wiggins, G.C.; Wald, L.L. Comparison of physiological noise at 1.5 T, 3 T and 7 T and optimization of fMRI acquisition parameters. NeuroImage 2005, 26, 243–250. [Google Scholar] [CrossRef]
- Murphy, K.; Bodurka, J.; Bandettini, P.A. How long to scan? The relationship between fMRI temporal signal to noise ratio and necessary scan duration. Neuroimage 2007, 34, 565–574. [Google Scholar] [CrossRef]
- Raimondo, L.; Priovoulos, N.; Passarinho, C.; Heij, J.; Knapen, T.; Dumoulin, S.O.; Siero, J.C.W.; van der Zwaag, W. Robust high spatio-temporal line-scanning fMRI in humans at 7T using multi-echo readouts, denoising and prospective motion correction. J. Neurosci. Methods 2023, 384, 109746. [Google Scholar] [CrossRef]
- Kuppers, F.; Yun, S.D.; Shah, N.J. Development of a novel 10-echo multi-contrast sequence based on EPIK to deliver simultaneous quantification of T(2) and T(2)(*) with application to oxygen extraction fraction. Magn. Reson. Med. 2022, 88, 1608–1623. [Google Scholar] [CrossRef]
- Devi, R.; Lepsien, J.; Lorenz, K.; Schlumm, T.; Mildner, T.; Moller, H.E. Multi-echo investigations of positive and negative CBF and concomitant BOLD changes. Neuroimage 2022, 263, 119661. [Google Scholar] [CrossRef]
- Yang, Q.; Ma, L.; Zhou, Z.; Bao, J.; Yang, Q.; Huang, H.; Cai, S.; He, H.; Chen, Z.; Zhong, J.; et al. Rapid high-fidelity T2* mapping using single-shot overlapping-echo acquisition and deep learning reconstruction. Magn. Reson. Med. 2023, 89, 2157–2170. [Google Scholar] [CrossRef]
- Valsamis, J.J.; Luciw, N.J.; Haq, N.; Atwi, S.; Duchesne, S.; Cameron, W.; MacIntosh, B.J. An imaging-based method of mapping multi-echo BOLD intracranial pulsatility. Magn. Reson. Med. 2023; online early view. [Google Scholar] [CrossRef]
- Alvand, A.; Kuruvilla-Mathew, A.; Kirk, I.J.; Roberts, R.P.; Pedersen, M.; Purdy, S.C. Altered brain network topology in children with auditory processing disorder: A resting-state multi-echo fMRI study. Neuroimage Clin. 2022, 35, 103139. [Google Scholar] [CrossRef] [PubMed]
- Power, J.D.; Plitt, M.; Gotts, S.J.; Kundu, P.; Voon, V.; Bandettini, P.A.; Martin, A. Ridding fMRI data of motion-related influences: Removal of signals with distinct spatial and physical bases in multiecho data. Proc. Natl. Acad. Sci. USA 2018, 115, E2105–E2114. [Google Scholar] [CrossRef] [PubMed]
- Dipasquale, O.; Sethi, A.; Lagana, M.M.; Baglio, F.; Baselli, G.; Kundu, P.; Harrison, N.A.; Cercignani, M. Comparing resting state fMRI de-noising approaches using multi- and single-echo acquisitions. PLoS ONE 2017, 12, e0173289. [Google Scholar] [CrossRef] [PubMed]
- Moia, S.; Termenon, M.; Urunuela, E.; Chen, G.; Stickland, R.C.; Bright, M.G.; Caballero-Gaudes, C. ICA-based denoising strategies in breath-hold induced cerebrovascular reactivity mapping with multi echo BOLD fMRI. Neuroimage 2021, 233, 117914. [Google Scholar] [CrossRef]
- Lynch, C.J.; Power, J.D.; Scult, M.A.; Dubin, M.; Gunning, F.M.; Liston, C. Rapid Precision Functional Mapping of Individuals Using Multi-Echo fMRI. Cell Rep. 2020, 33, 108540. [Google Scholar] [CrossRef] [PubMed]
- Foo, T.K.F.; Laskaris, E.; Vermilyea, M.; Xu, M.; Thompson, P.; Conte, G.; Van Epps, C.; Immer, C.; Lee, S.K.; Tan, E.T.; et al. Lightweight, compact, and high-performance 3T MR system for imaging the brain and extremities. Magn. Reson. Med. 2018, 80, 2232–2245. [Google Scholar] [CrossRef] [PubMed]
- Weavers, P.T.; Shu, Y.; Tao, S.; Huston, J., 3rd; Lee, S.K.; Graziani, D.; Mathieu, J.B.; Trzasko, J.D.; Foo, T.K.; Bernstein, M.A. Technical Note: Compact three-tesla magnetic resonance imager with high-performance gradients passes ACR image quality and acoustic noise tests. Med. Phys. 2016, 43, 1259–1264. [Google Scholar] [CrossRef]
- Weavers, P.T.; Tao, S.; Trzasko, J.D.; Frigo, L.M.; Shu, Y.; Frick, M.A.; Lee, S.K.; Foo, T.K.; Bernstein, M.A. B0 concomitant field compensation for MRI systems employing asymmetric transverse gradient coils. Magn. Reson. Med. 2018, 79, 1538–1544. [Google Scholar] [CrossRef]
- Tao, S.; Weavers, P.T.; Trzasko, J.D.; Shu, Y.; Huston, J., 3rd; Lee, S.K.; Frigo, L.M.; Bernstein, M.A. Gradient pre-emphasis to counteract first-order concomitant fields on asymmetric MRI gradient systems. Magn. Reson. Med. 2017, 77, 2250–2262. [Google Scholar] [CrossRef]
- Tao, S.; Trzasko, J.D.; Gunter, J.L.; Weavers, P.T.; Shu, Y.; Huston, J.; Lee, S.K.; Tan, E.T.; Bernstein, M.A. Gradient nonlinearity calibration and correction for a compact, asymmetric magnetic resonance imaging gradient system. Phys. Med. Biol. 2017, 62, N18–N31. [Google Scholar] [CrossRef]
- Lee, S.K.; Mathieu, J.B.; Graziani, D.; Piel, J.; Budesheim, E.; Fiveland, E.; Hardy, C.J.; Tan, E.T.; Amm, B.; Foo, T.K.; et al. Peripheral nerve stimulation characteristics of an asymmetric head-only gradient coil compatible with a high-channel-count receiver array. Magn. Reson. Med. 2016, 76, 1939–1950. [Google Scholar] [CrossRef] [PubMed]
- In, M.H.; Shu, Y.; Trzasko, J.D.; Yarach, U.; Kang, D.; Gray, E.M.; Huston, J.; Bernstein, M.A. Reducing PNS with minimal performance penalties via simple pulse sequence modifications on a high-performance compact 3T scanner. Phys. Med. Biol. 2020, 65, 15NT02. [Google Scholar] [CrossRef] [PubMed]
- Tan, E.T.; Lee, S.K.; Weavers, P.T.; Graziani, D.; Piel, J.E.; Shu, Y.; Huston, J., 3rd; Bernstein, M.A.; Foo, T.K. High slew-rate head-only gradient for improving distortion in echo planar imaging: Preliminary experience. J. Magn. Reson. Imaging 2016, 44, 653–664. [Google Scholar] [CrossRef]
- In, M.H.; Tan, E.T.; Trzasko, J.D.; Shu, Y.; Kang, D.; Yarach, U.; Tao, S.; Gray, E.M.; Huston, J., 3rd; Bernstein, M.A. Distortion-free imaging: A double encoding method (DIADEM) combined with multiband imaging for rapid distortion-free high-resolution diffusion imaging on a compact 3T with high-performance gradients. J. Magn. Reson. Imaging 2020, 51, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Jo, H.J.; In, M.H.; Yarach, U.; Meyer, N.K.; Bardwell Speltz, L.J.; Gray, E.M.; Trzasko, J.D.; Huston, J., III; Bernstein, M.A.; et al. The benefit of high-performance gradients on echo planar imaging for BOLD-based resting-state functional MRI. Phys. Med. Biol. 2020, 65, 235024. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.K.; Oshio, K.; Panych, L.P. Improved image reconstruction for partial Fourier gradient-echo echo-planar imaging (EPI). Magn. Reson. Med. 2008, 59, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.D.; Yang, B.; Fernandez, B.; Banerjee, S.; Wang, Y. Improved resting state functional connectivity sensitivity and reproducibility using a multiband multi-echo acquisition. Neuroimage 2021, 225, 117461. [Google Scholar] [CrossRef]
- Heunis, S.; Breeuwer, M.; Caballero-Gaudes, C.; Hellrung, L.; Huijbers, W.; Jansen, J.F.; Lamerichs, R.; Zinger, S.; Aldenkamp, A.P. The effects of multi-echo fMRI combination and rapid T2*-mapping on offline and real-time BOLD sensitivity. Neuroimage 2021, 238, 118244. [Google Scholar] [CrossRef]
- Fernandez, B.; Leuchs, L.; Samann, P.G.; Czisch, M.; Spoormaker, V.I. Multi-echo EPI of human fear conditioning reveals improved BOLD detection in ventromedial prefrontal cortex. Neuroimage 2017, 156, 65–77. [Google Scholar] [CrossRef]
- Lynch, C.J.; Elbau, I.; Liston, C. Improving precision functional mapping routines with multi-echo fMRI. Curr. Opin. Behav. Sci. 2021, 40, 113–119. [Google Scholar] [CrossRef]
- Cox, R.W. AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed. Res. 1996, 29, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Glover, G.H.; Li, T.Q.; Ress, D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn. Reson. Med. 2000, 44, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Birn, R.M.; Smith, M.A.; Jones, T.B.; Bandettini, P.A. The respiration response function: The temporal dynamics of fMRI signal fluctuations related to changes in respiration. Neuroimage 2008, 40, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Jo, H.J.; Saad, Z.S.; Simmons, W.K.; Milbury, L.A.; Cox, R.W. Mapping sources of correlation in resting state FMRI, with artifact detection and removal. Neuroimage 2010, 52, 571–582. [Google Scholar] [CrossRef]
- Jo, H.J.; Gotts, S.J.; Reynolds, R.C.; Bandettini, P.A.; Martin, A.; Cox, R.W.; Saad, Z.S. Effective Preprocessing Procedures Virtually Eliminate Distance-Dependent Motion Artifacts in Resting State FMRI. J. Appl. Math. 2013, 2013, 935154. [Google Scholar] [CrossRef]
- Jo, H.J.; Reynolds, R.C.; Gotts, S.J.; Handwerker, D.A.; Balzekas, I.; Martin, A.; Cox, R.W.; Bandettini, P.A. Fast detection and reduction of local transient artifacts in resting-state fMRI. Comput. Biol. Med. 2020, 120, 103742. [Google Scholar] [CrossRef]
- Posse, S.; Wiese, S.; Gembris, D.; Mathiak, K.; Kessler, C.; Grosse-Ruyken, M.L.; Elghahwagi, B.; Richards, T.; Dager, S.R.; Kiselev, V.G. Enhancement of BOLD-contrast sensitivity by single-shot multi-echo functional MR imaging. Magn. Reson. Med. 1999, 42, 87–97. [Google Scholar] [CrossRef]
- Cho, J.W.; Korchmaros, A.; Vogelstein, J.T.; Milham, M.P.; Xu, T. Impact of concatenating fMRI data on reliability for functional connectomics. Neuroimage 2021, 226, 117549. [Google Scholar] [CrossRef]
- Welvaert, M.; Rosseel, Y. On the definition of signal-to-noise ratio and contrast-to-noise ratio for FMRI data. PLoS ONE 2013, 8, e77089. [Google Scholar] [CrossRef]
- Saad, Z.S.; Reynolds, R.C.; Argall, B.; Japee, S.; Cox, R.W. SUMA: An interface for surface-based intra- and inter-subject analysis with AFNI. In Proceedings of the 2004 2nd IEEE International Symposium on Biomedical Imaging: Macro to Nano, Arlington, VA, USA, 18 April 2004; Volume 2, pp. 1510–1513. [Google Scholar]
- Reuter, M.; Schmansky, N.J.; Rosas, H.D.; Fischl, B. Within-subject template estimation for unbiased longitudinal image analysis. Neuroimage 2012, 61, 1402–1418. [Google Scholar] [CrossRef]
- Destrieux, C.; Fischl, B.; Dale, A.; Halgren, E. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage 2010, 53, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Rolls, E.T.; Huang, C.C.; Lin, C.P.; Feng, J.; Joliot, M. Automated anatomical labelling atlas 3. Neuroimage 2020, 206, 116189. [Google Scholar] [CrossRef] [PubMed]
- Kundu, P.; Inati, S.J.; Evans, J.W.; Luh, W.-M.M.; Bandettini, P.A. Differentiating BOLD and non-BOLD signals in fMRI time series using multi-echo EPI. NeuroImage 2012, 60, 1759–1770. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.D.; Nencka, A.S.; Lebel, R.M.; Wang, Y. Multiband multi-echo imaging of simultaneous oxygenation and flow timeseries for resting state connectivity. PLoS ONE 2017, 12, e0169253. [Google Scholar] [CrossRef] [PubMed]
- Saad, Z.S.; Reynolds, R.C.; Jo, H.J.; Gotts, S.J.; Chen, G.; Martin, A.; Cox, R.W. Correcting brain-wide correlation differences in resting-state FMRI. Brain Connect 2013, 3, 339–352. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, C.F.; DeLuca, M.; Devlin, J.T.; Smith, S.M. Investigations into resting-state connectivity using independent component analysis. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, L.D.; Smith, S.M.; Ongur, D.; Beckmann, C.F. Using Dual Regression to Investigate Network Shape and Amplitude in Functional Connectivity Analyses. Front Neurosci. 2017, 11, 115. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, M.; Beckmann, C.F.; Behrens, T.E.; Woolrich, M.W.; Smith, S.M. FSL. Neuroimage 2012, 62, 782–790. [Google Scholar] [CrossRef]
- Beckmann, C.F.; Mackay, C.E.; Filippini, N.; Smith, S.M. Group comparison of resting-state FMRI data using multi-subject ICA and dual regression. Neuroimage 2009, 47, S148. [Google Scholar] [CrossRef]
- Power, J.D.; Barnes, K.A.; Snyder, A.Z.; Schlaggar, B.L.; Petersen, S.E. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage 2012, 59, 2142–2154. [Google Scholar] [CrossRef]
- Xia, M.; Wang, J.; He, Y. BrainNet Viewer: A network visualization tool for human brain connectomics. PLoS ONE 2013, 8, e68910. [Google Scholar] [CrossRef] [PubMed]
- Foo, T.K.F.; Tan, E.T.; Vermilyea, M.E.; Hua, Y.H.; Fiveland, E.W.; Piel, J.E.; Park, K.; Ricci, J.; Thompson, P.S.; Graziani, D.; et al. Highly efficient head-only magnetic field insert gradient coil for achieving simultaneous high gradient amplitude and slew rate at 3.0T (MAGNUS) for brain microstructure imaging. Magn. Reson. Med. 2020, 83, 2356–2369. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.Y.; Witzel, T.; Keil, B.; Scholz, A.; Davids, M.; Dietz, P.; Rummert, E.; Ramb, R.; Kirsch, J.E.; Yendiki, A.; et al. Connectome 2.0: Developing the next-generation ultra-high gradient strength human MRI scanner for bridging studies of the micro-, meso- and macro-connectome. Neuroimage 2021, 243, 118530. [Google Scholar] [CrossRef] [PubMed]
- Peters, A.M.; Brookes, M.J.; Hoogenraad, F.G.; Gowland, P.a.; Francis, S.T.; Morris, P.G.; Bowtell, R. T2* measurements in human brain at 1.5, 3 and 7 T. Magn. Reson. Imaging 2007, 25, 748–753. [Google Scholar] [CrossRef]
- Li, M.; Newton, A.T.; Anderson, A.W.; Ding, Z.; Gore, J.C. Characterization of the hemodynamic response function in white matter tracts for event-related fMRI. Nat. Commun. 2019, 10, 1140. [Google Scholar] [CrossRef]
- Deichmann, R.; Josephs, O.; Hutton, C.; Corfield, D.R.; Turner, R. Compensation of susceptibility-induced BOLD sensitivity losses in echo-planar fMRI imaging. Neuroimage 2002, 15, 120–135. [Google Scholar] [CrossRef]
- Constable, R.T.; Spencer, D.D. Composite image formation in z-shimmed functional MR imaging. Magn. Reson. Med. 1999, 42, 110–117. [Google Scholar] [CrossRef]
- Pruim, R.H.R.; Mennes, M.; van Rooij, D.; Llera, A.; Buitelaar, J.K.; Beckmann, C.F. ICA-AROMA: A robust ICA-based strategy for removing motion artifacts from fMRI data. Neuroimage 2015, 112, 267–277. [Google Scholar] [CrossRef]
- Kundu, P.; Voon, V.; Balchandani, P.; Lombardo, M.V.; Poser, B.A.; Bandettini, P.A. Multi-echo fMRI: A review of applications in fMRI denoising and analysis of BOLD signals. Neuroimage 2017, 154, 59–80. [Google Scholar] [CrossRef]
- Gonzalez-Castillo, J.; Panwar, P.; Buchanan, L.C.; Caballero-Gaudes, C.; Handwerker, D.A.; Jangraw, D.C.; Zachariou, V.; Inati, S.; Roopchansingh, V.; Derbyshire, J.A.; et al. Evaluation of multi-echo ICA denoising for task based fMRI studies: Block designs, rapid event-related designs, and cardiac-gated fMRI. Neuroimage 2016, 141, 452–468. [Google Scholar] [CrossRef]
- Birn, R.M.; Molloy, E.K.; Patriat, R.; Parker, T.; Meier, T.B.; Kirk, G.R.; Nair, V.A.; Meyerand, M.E.; Prabhakaran, V. The effect of scan length on the reliability of resting-state fMRI connectivity estimates. Neuroimage 2013, 83, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Shah, L.M.; Cramer, J.A.; Ferguson, M.A.; Birn, R.M.; Anderson, J.S. Reliability and reproducibility of individual differences in functional connectivity acquired during task and resting state. Brain Behav. 2016, 6, e00456. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; In, M.-H.; Gray, E.; Foo, T.K.; Madhavan, R.; Meyer, N.K.; Bardwell-Speltz, L.J.; Ahmed, Z.; Gunter, J.; Fernandez, B.; et al. Potential Benefit of Multiband Multiecho EPI for Resting-state Functional MRI in Alzheimer’s disease on a compact 3T system: A Preliminary study. In Proceedings of the International Society for Magnetic Resonance in Medicine, Online, 15–20 May 2021; p. 1514. [Google Scholar]
- Gilmore, A.W.; Agron, A.M.; Gonzalez-Araya, E.I.; Gotts, S.J.; Martin, A. A Comparison of Single- and Multi-Echo Processing of Functional MRI Data During Overt Autobiographical Recall. Front. Neurosci. 2022, 16, 854387. [Google Scholar] [CrossRef] [PubMed]
- Bilgic, B.; Setsompop, K.; Cohen-Adad, J.; Wedeen, V.; Wald, L.L.; Adalsteinsson, E. Accelerated diffusion spectrum imaging with compressed sensing using adaptive dictionaries. Med. Image Comput Comput Assist Interv. 2012, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Polak, D.; Setsompop, K.; Cauley, S.F.; Gagoski, B.A.; Bhat, H.; Maier, F.; Bachert, P.; Wald, L.L.; Bilgic, B. Wave-CAIPI for highly accelerated MP-RAGE imaging. Magn. Reson. Med. 2018, 79, 401–406. [Google Scholar] [CrossRef]



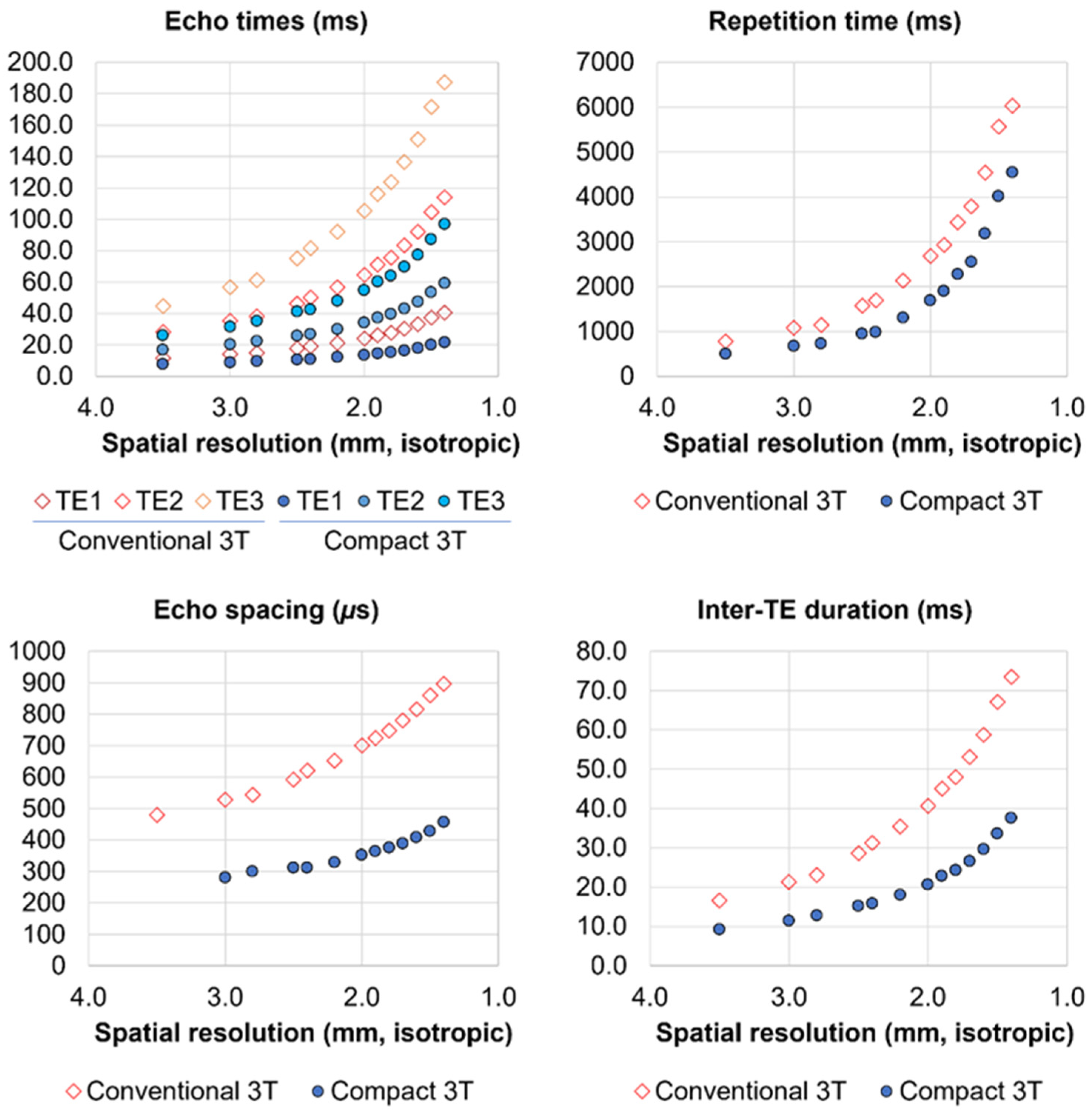
| Imaging Parameter | Single-Echo EPI | Multi-Echo EPI |
|---|---|---|
| Field of view (FOV, mm) | 224 × 224 | |
| Matrix size | 94 × 94 | |
| In-plane resolution (mm) | 2.38 × 2.38 | |
| k-space partial Fourier factor | No partial Fourier | |
| Slice thickness (mm) | 2.4 | |
| Number of slices | 52 | |
| Inter-slice gap (mm) | 0.3 | |
| Inferior-to-superior coverage (mm) | 140.4 | |
| In-plane acceleration factor | 2 | |
| Multi-band factor | 4 | |
| Echo spacing (µs) | 352 | |
| Receiver bandwidth (Hz) | ±250 k | |
| Echo time (TE, ms) | 30 | 11.9, 29.8, 47.8 |
| Repetition time (TR, ms) | 700 | 940 |
| RF flip angle (FA, °) 1 | 52 | 59 |
| Number of volumes per scan | 961 | 320 |
| Scan duration (mm:ss) | 11:12 | 5:00 |
| Note | - | Scan twice |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, D.; In, M.-H.; Jo, H.J.; Halverson, M.A.; Meyer, N.K.; Ahmed, Z.; Gray, E.M.; Madhavan, R.; Foo, T.K.; Fernandez, B.; et al. Improved Resting-State Functional MRI Using Multi-Echo Echo-Planar Imaging on a Compact 3T MRI Scanner with High-Performance Gradients. Sensors 2023, 23, 4329. https://doi.org/10.3390/s23094329
Kang D, In M-H, Jo HJ, Halverson MA, Meyer NK, Ahmed Z, Gray EM, Madhavan R, Foo TK, Fernandez B, et al. Improved Resting-State Functional MRI Using Multi-Echo Echo-Planar Imaging on a Compact 3T MRI Scanner with High-Performance Gradients. Sensors. 2023; 23(9):4329. https://doi.org/10.3390/s23094329
Chicago/Turabian StyleKang, Daehun, Myung-Ho In, Hang Joon Jo, Maria A. Halverson, Nolan K. Meyer, Zaki Ahmed, Erin M. Gray, Radhika Madhavan, Thomas K. Foo, Brice Fernandez, and et al. 2023. "Improved Resting-State Functional MRI Using Multi-Echo Echo-Planar Imaging on a Compact 3T MRI Scanner with High-Performance Gradients" Sensors 23, no. 9: 4329. https://doi.org/10.3390/s23094329
APA StyleKang, D., In, M.-H., Jo, H. J., Halverson, M. A., Meyer, N. K., Ahmed, Z., Gray, E. M., Madhavan, R., Foo, T. K., Fernandez, B., Black, D. F., Welker, K. M., Trzasko, J. D., Huston, J., III, Bernstein, M. A., & Shu, Y. (2023). Improved Resting-State Functional MRI Using Multi-Echo Echo-Planar Imaging on a Compact 3T MRI Scanner with High-Performance Gradients. Sensors, 23(9), 4329. https://doi.org/10.3390/s23094329






