Identification of Homogeneous Subgroups from Resting-State fMRI Data
Abstract
1. Introduction
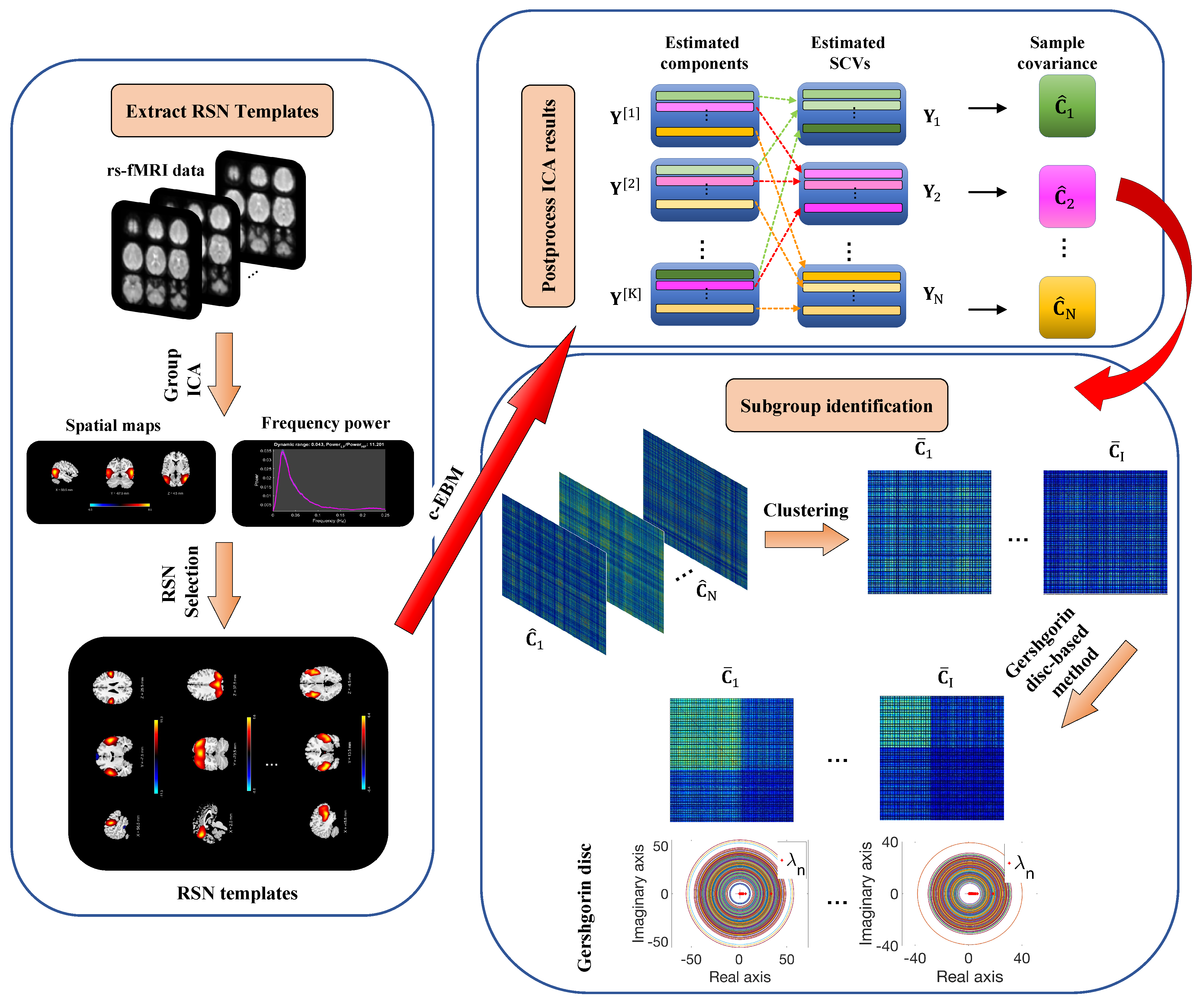
2. Materials and Methods
2.1. ICA-EBM
2.2. Constrained EBM
2.3. Subgroup Identification
2.4. Subgroup Validation
2.5. Data Preprocessing
2.5.1. Resting-State fMRI
2.5.2. Cognitive Test Scores
2.6. Reference Generation
2.6.1. Data Acquisition and Preprocessing
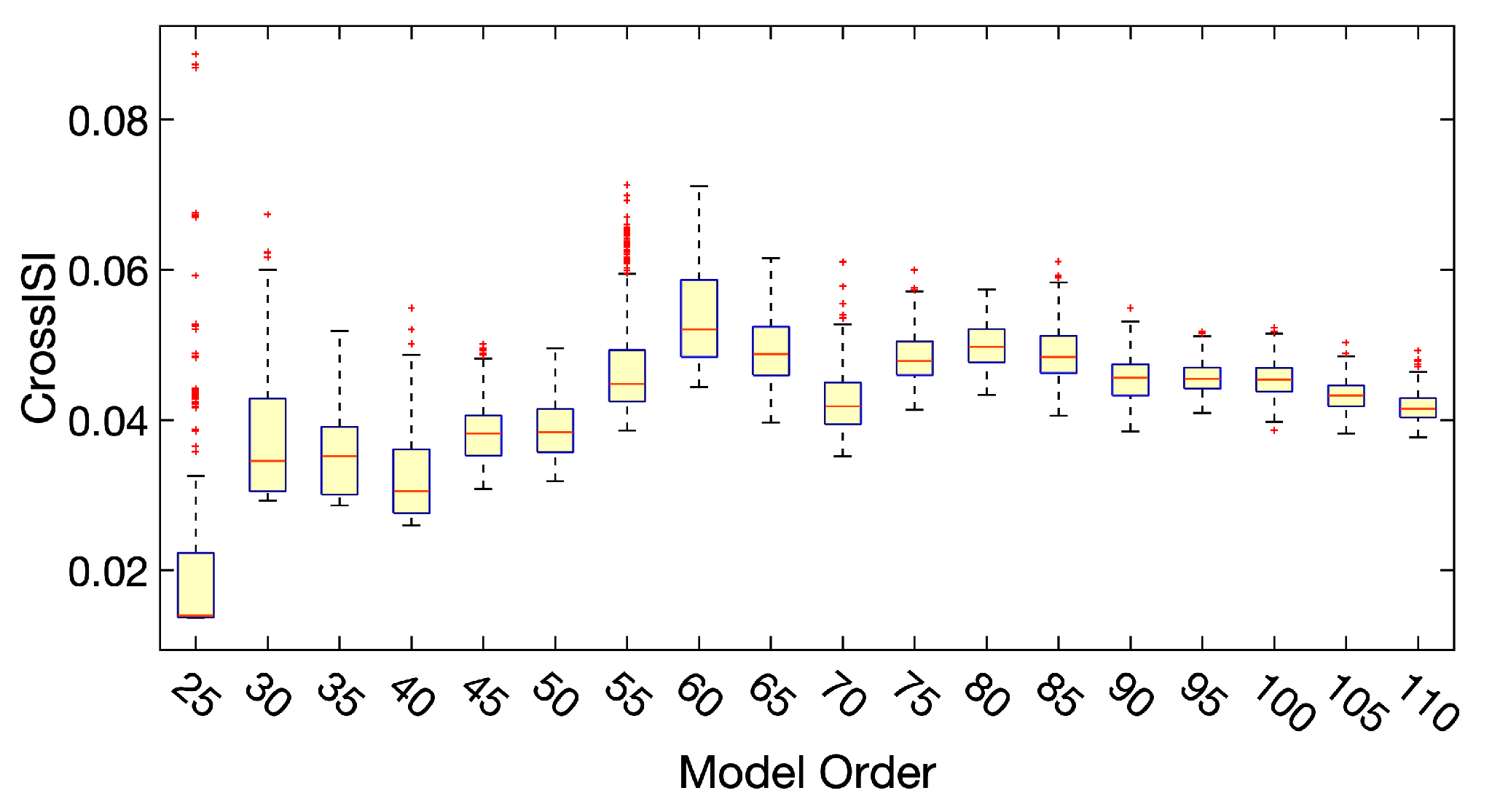
2.6.2. Model Order and RSNs Selection
- Estimate the model order by entropy-rate based order selection using the finite memory length model (ER-FM) and autoregressive model (ER-AR) [73], as and , respectively;
- Test the estimated model order ranging from 25 to 110 with step size 5;
- Perform 300 runs of ICA-EBM with random initialization on each one of the estimated model orders;
- Calculate Cross-ISI for all the runs. The distribution of Cross-ISI for all the orders is displayed in Figure 2;
- Select model orders that have relatively small values and small variance of Cross-ISI;
- Select the best run that has the smallest Cross-ISI from the selected model orders;
- Inspect the results based on the visualization of spatial activation of functional networks and the corresponding spectral summary.
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ICA | Independent component analysis |
| GICA | Group independent component analysis |
| c-ICA | Constrained independent component analysis |
| EBM | Entropy bound minimization |
| IVA | Independent vector analysis |
| SCV | Source component vector |
| RSN | Resting-state network |
| GDM | Global difference map |
| Cross-ISI | Cross inter-symbol interference |
References
- National Research Council. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease; The National Academies Press: Washington, DC, USA, 2011; pp. 23–36.
- Tonelli, M.R.; Shirts, B.H. Knowledge for precision medicine: Mechanistic reasoning and methodological pluralism. JAMA 2017, 318, 1649–1650. [Google Scholar] [CrossRef]
- Shao, Y.; Cuccaro, M.; Hauser, E.; Raiford, K.; Menold, M.; Wolpert, C.; Ravan, S.; Elston, L.; Decena, K.; Donnelly, S.; et al. Fine mapping of autistic disorder to chromosome 15q11-q13 by use of phenotypic subtypes. Am. J. Hum. Genet. 2003, 72, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Dekker, M.; Bonifati, V.; Van Duijn, C. Parkinson’s disease: Piecing together a genetic jigsaw. Brain 2003, 126, 1722–1733. [Google Scholar] [CrossRef]
- Scott, W.K.; Hauser, E.R.; Schmechel, D.E.; Welsh-Bohmer, K.A.; Small, G.W.; Roses, A.D.; Saunders, A.M.; Gilbert, J.R.; Vance, J.M.; Haines, J.L.; et al. Ordered-subsets linkage analysis detects novel Alzheimer disease loci on chromosomes 2q34 and 15q22. Am. J. Hum. Genet. 2003, 73, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, D.B.; Cabral, C.; Kambeitz-Ilankovic, L.; Sanfelici, R.; Kambeitz, J.; Calhoun, V.; Falkai, P.; Pantelis, C.; Meisenzahl, E.; Koutsouleris, N. Brain subtyping enhances the neuroanatomical discrimination of schizophrenia. Schizophr. Bull. 2018, 44, 1060–1069. [Google Scholar] [CrossRef]
- Tsuang, M.T.; Lyons, M.J.; Faraone, S.V. Heterogeneity of schizophrenia. Br. J. Psychiatry 1990, 156, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Biederman, J.; Mick, E.; Faraone, S.V.; Spencer, T.; Wilens, T.E.; Wozniak, J. Pediatric mania: A developmental subtype of bipolar disorder? Biol. Psychiatry 2000, 48, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Fried, E.I.; Nesse, R.M. Depression is not a consistent syndrome: An investigation of unique symptom patterns in the STAR* D study. J. Affect. Disord. 2015, 172, 96–102. [Google Scholar] [CrossRef]
- Payne, J.L.; Palmer, J.T.; Joffe, H. A reproductive subtype of depression: Conceptualizing models and moving toward etiology. Harv. Rev. Psychiatry 2009, 17, 72–86. [Google Scholar] [CrossRef]
- Voineagu, I.; Wang, X.; Johnston, P.; Lowe, J.K.; Tian, Y.; Horvath, S.; Mill, J.; Cantor, R.M.; Blencowe, B.J.; Geschwind, D.H. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 2011, 474, 380–384. [Google Scholar] [CrossRef]
- Veatch, O.; Veenstra-VanderWeele, J.; Potter, M.; Pericak-Vance, M.; Haines, J. Genetically meaningful phenotypic subgroups in autism spectrum disorders. Genes Brain Behav. 2014, 13, 276–285. [Google Scholar] [CrossRef]
- Bitsika, V.; Sharpley, C.; Orapeleng, S. An exploratory analysis of the use of cognitive, adaptive and behavioural indices for cluster analysis of ASD subgroups. J. Intellect. Disabil. Res. 2008, 52, 973–985. [Google Scholar] [CrossRef] [PubMed]
- Mestre, T.A.; Fereshtehnejad, S.M.; Berg, D.; Bohnen, N.I.; Dujardin, K.; Erro, R.; Espay, A.J.; Halliday, G.; Van Hilten, J.J.; Hu, M.T.; et al. Parkinson’s disease subtypes: Critical appraisal and recommendations. J. Parkinsons Dis. 2021, 11, 395–404. [Google Scholar] [CrossRef] [PubMed]
- McGuire, P.K.; Matsumoto, K. Functional neuroimaging in mental disorders. World Psychiatry 2004, 3, 6. [Google Scholar] [PubMed]
- Finn, E.S.; Shen, X.; Scheinost, D.; Rosenberg, M.D.; Huang, J.; Chun, M.M.; Papademetris, X.; Constable, R.T. Functional connectome fingerprinting: Identifying individuals using patterns of brain connectivity. Nat. Neurosci. 2015, 18, 1664–1671. [Google Scholar] [CrossRef]
- Bell, A.J.; Sejnowski, T.J. An information-maximization approach to blind separation and blind deconvolution. Neural Comput. 1995, 7, 1129–1159. [Google Scholar] [CrossRef]
- McKeown, M.J.; Makeig, S.; Brown, G.G.; Jung, T.P.; Kindermann, S.S.; Bell, A.J.; Sejnowski, T.J. Analysis of fMRI data by blind separation into independent spatial components. Hum. Brain Mapp. 1998, 6, 160–188. [Google Scholar] [CrossRef]
- Calhoun, V.D.; Adali, T. Multisubject independent component analysis of fMRI: A decade of intrinsic networks, default mode, and neurodiagnostic discovery. IEEE Rev. Biomed. Eng. 2012, 5, 60–73. [Google Scholar] [CrossRef]
- Bhinge, S.; Long, Q.; Calhoun, V.D.; Adali, T. Spatial dynamic functional connectivity analysis identifies distinctive biomarkers in schizophrenia. Front. Neurosci. 2019, 13, 1006. [Google Scholar] [CrossRef]
- Du, Y.; Fu, Z.; Sui, J.; Gao, S.; Xing, Y.; Lin, D.; Salman, M.; Abrol, A.; Rahaman, M.A.; Chen, J.; et al. NeuroMark: An automated and adaptive ICA based pipeline to identify reproducible fMRI markers of brain disorders. NeuroImage Clin. 2020, 28, 102375. [Google Scholar] [CrossRef]
- Calhoun, V.D.; Maciejewski, P.K.; Pearlson, G.D.; Kiehl, K.A. Temporal lobe and “default” hemodynamic brain modes discriminate between schizophrenia and bipolar disorder. Hum. Brain Mapp. 2008, 29, 1265–1275. [Google Scholar] [CrossRef] [PubMed]
- Sorg, C.; Riedl, V.; Mühlau, M.; Calhoun, V.D.; Eichele, T.; Läer, L.; Drzezga, A.; Förstl, H.; Kurz, A.; Zimmer, C.; et al. Selective changes of resting-state networks in individuals at risk for Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2007, 104, 18760–18765. [Google Scholar] [CrossRef] [PubMed]
- Greicius, M.D.; Srivastava, G.; Reiss, A.L.; Menon, V. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: Evidence from functional MRI. Proc. Natl. Acad. Sci. USA 2004, 101, 4637–4642. [Google Scholar] [CrossRef] [PubMed]
- Adali, T.; Anderson, M.; Fu, G.S. Diversity in independent component and vector analyses: Identifiability, algorithms, and applications in medical imaging. IEEE Signal Process. Mag. 2014, 31, 18–33. [Google Scholar] [CrossRef]
- Calhoun, V.D.; Adali, T.; Pearlson, G.D.; Pekar, J.J. A method for making group inferences from functional MRI data using independent component analysis. Hum. Brain Mapp. 2001, 14, 140–151. [Google Scholar] [CrossRef]
- Allen, E.A.; Erhardt, E.B.; Wei, Y.; Eichele, T.; Calhoun, V.D. Capturing inter-subject variability with group independent component analysis of fMRI data: A simulation study. Neuroimage 2012, 59, 4141–4159. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.M.; Anderson, M.; Miller, R.L.; Adalı, T.; Calhoun, V.D. Preserving subject variability in group fMRI analysis: Performance evaluation of GICA vs. IVA. Front. Syst. Neurosci. 2014, 8, 106. [Google Scholar] [CrossRef]
- Laney, J.; Westlake, K.P.; Ma, S.; Woytowicz, E.; Calhoun, V.D.; Adalı, T. Capturing subject variability in fMRI data: A graph-theoretical analysis of GICA vs. IVA. J. Neurosci. Methods 2015, 247, 32–40. [Google Scholar] [CrossRef]
- Dea, J.T.; Anderson, M.; Allen, E.; Calhoun, V.D.; Adalı, T. IVA for multi-subject fMRI analysis: A comparative study using a new simulation toolbox. In Proceedings of the IEEE International Workshop on Machine Learning for Signal Processing, Beijing, China, 18–21 September 2011; pp. 1–6. [Google Scholar]
- Ma, S.; Phlypo, R.; Calhoun, V.D.; Adalı, T. Capturing group variability using IVA: A simulation study and graph-theoretical analysis. In Proceedings of the IEEE International Conference Acoustics, Speech and Signal Processing, Vancouver, BC, Canada, 26–31 May 2013; pp. 3128–3132. [Google Scholar]
- Kim, T.; Eltoft, T.; Lee, T.W. Independent vector analysis: An extension of ICA to multivariate components. In Proceedings of the Independent Component Analysis and Blind Signal Separation: 6th International Conference, ICA 2006, Charleston, SC, USA, 5–8 March 2006; Springer: Berlin/Heidelberg, Germany, 2006; pp. 165–172. [Google Scholar]
- Anderson, M.; Adali, T.; Li, X.L. Joint blind source separation with multivariate Gaussian model: Algorithms and performance analysis. IEEE Trans. Signal Process 2011, 60, 1672–1683. [Google Scholar] [CrossRef]
- Long, Q.; Bhinge, S.; Calhoun, V.D.; Adali, T. Independent vector analysis for common subspace analysis: Application to multi-subject fMRI data yields meaningful subgroups of schizophrenia. NeuroImage 2020, 216, 116872. [Google Scholar] [CrossRef]
- Yang, H.; Akhonda, M.; Ghayem, F.; Long, Q.; Calhoun, V.; Adali, T. Independent Vector Analysis Based Subgroup Identification from Multisubject fMRI Data. In Proceedings of the International Conference on Acoustics, Speech and Signal Processing, Virtual, 7–13 May 2022; pp. 1471–1475. [Google Scholar]
- De Vos, M.; De Lathauwer, L.; Van Huffel, S. Spatially constrained ICA algorithm with an application in EEG processing. Signal Process. 2011, 91, 1963–1972. [Google Scholar] [CrossRef]
- Lu, W.; Rajapakse, J.C. ICA with reference. Neurocomputing 2006, 69, 2244–2257. [Google Scholar] [CrossRef]
- Lu, W.; Rajapakse, J. Constrained independent component analysis. Adv. Neural Inf. Process. Syst. 2000, 13, 570–576. [Google Scholar]
- Lin, Q.H.; Liu, J.; Zheng, Y.R.; Liang, H.; Calhoun, V.D. Semiblind spatial ICA of fMRI using spatial constraints. Hum. Brain Mapp. 2010, 31, 1076–1088. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.A.; Anderson, M.; Li, X.L.; Adalı, T. General non-orthogonal constrained ICA. IEEE Trans. Signal Process 2014, 62, 2778–2786. [Google Scholar]
- Du, Y.; Fan, Y. Group information guided ICA for fMRI data analysis. NeuroImage 2013, 69, 157–197. [Google Scholar] [CrossRef]
- Li, X.L.; Adali, T. Independent component analysis by entropy bound minimization. IEEE Trans. Signal Process. 2010, 58, 5151–5164. [Google Scholar] [CrossRef]
- Devinsky, O.; Morrell, M.J.; Vogt, B.A. Contributions of anterior cingulate cortex to behaviour. Brain 1995, 118, 279–306. [Google Scholar] [CrossRef]
- Benes, F.M. Relationship of cingulate cortex to schizophrenia and other psychiatric disorders. In Neurobiology of Cingulate Cortex and Limbic Thalamus; Springer: Berlin/Heidelberg, Germany, 1993; pp. 581–605. [Google Scholar]
- Matsumoto, H.; Simmons, A.; Williams, S.; Hadjulis, M.; Pipe, R.; Murray, R.; Frangou, S. Superior temporal gyrus abnormalities in early-onset schizophrenia: Similarities and differences with adult-onset schizophrenia. Am. J. Psychiatry 2001, 158, 1299–1304. [Google Scholar] [CrossRef]
- Birchwood, M.; Smith, J.; Cochrane, R.; Wetton, S.; Copestake, S. The social functioning scale the development and validation of a new scale of social adjustment for use in family intervention programmes with schizophrenic patients. Br. J. Psychiatry 1990, 157, 853–859. [Google Scholar] [CrossRef]
- Keefe, R.S.; Goldberg, T.E.; Harvey, P.D.; Gold, J.M.; Poe, M.P.; Coughenour, L. The Brief Assessment of Cognition in Schizophrenia: Reliability, sensitivity, and comparison with a standard neurocognitive battery. Schizophr. Res. 2004, 68, 283–297. [Google Scholar] [CrossRef]
- Kay, S.R.; Fiszbein, A.; Opler, L.A. The positive and negative syndrome scale (PANSS) for schizophrenia. Schizophr. Bull. 1987, 13, 261–276. [Google Scholar] [CrossRef] [PubMed]
- Oja, E.; Hyvarinen, A. Independent component analysis: Algorithms and applications. Neural Netw. 2000, 13, 411–430. [Google Scholar]
- Long, Q.; Bhinge, S.; Levin-Schwartz, Y.; Boukouvalas, Z.; Calhoun, V.D.; Adalı, T. The role of diversity in data-driven analysis of multi-subject fMRI data: Comparison of approaches based on independence and sparsity using global performance metrics. Hum. Brain Mapp. 2019, 40, 489–504. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.; Li, X.L.; Rodriguez, P.; Adali, T. An effective decoupling method for matrix optimization and its application to the ICA problem. In Proceedings of the International Conference on Acoustics, Speech and Signal Processing, Kyoto, Japan, 25–30 March 2012; pp. 1885–1888. [Google Scholar]
- Damaraju, E.; Allen, E.A.; Belger, A.; Ford, J.M.; McEwen, S.; Mathalon, D.; Mueller, B.; Pearlson, G.; Potkin, S.; Preda, A.; et al. Dynamic functional connectivity analysis reveals transient states of dysconnectivity in schizophrenia. NeuroImage Clin. 2014, 5, 298–308. [Google Scholar] [CrossRef]
- Benjamini, Y.; Yekutieli, D. False discovery rate–adjusted multiple confidence intervals for selected parameters. J. Acoust. Soc. Am. 2005, 100, 71–81. [Google Scholar] [CrossRef]
- Levin-Schwartz, Y.; Calhoun, V.D.; Adalı, T. A method to compare the discriminatory power of data-driven methods: Application to ICA and IVA. J. Neurosci. Methods 2019, 311, 267–276. [Google Scholar] [CrossRef]
- Tamminga, C.A.; Ivleva, E.I.; Keshavan, M.S.; Pearlson, G.D.; Clementz, B.A.; Witte, B.; Morris, D.W.; Bishop, J.; Thaker, G.K.; Sweeney, J.A. Clinical phenotypes of psychosis in the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP). Am. J. Psychiatry 2013, 170, 1263–1274. [Google Scholar] [CrossRef]
- Tamminga, C.A.; Pearlson, G.; Keshavan, M.; Sweeney, J.; Clementz, B.; Thaker, G. Bipolar and schizophrenia network for intermediate phenotypes: Outcomes across the psychosis continuum. Schizophr. Bull. 2014, 40, S131–S137. [Google Scholar] [CrossRef]
- Du, Y.; Fu, Z.; Xing, Y.; Lin, D.; Pearlson, G.; Kochunov, P.; Hong, L.E.; Qi, S.; Salman, M.; Abrol, A.; et al. Evidence of shared and distinct functional and structural brain signatures in schizophrenia and autism spectrum disorder. Commun. Biol. 2021, 4, 1–16. [Google Scholar] [CrossRef]
- Karlsgodt, K.H.; Niendam, T.A.; Bearden, C.E.; Cannon, T.D. White matter integrity and prediction of social and role functioning in subjects at ultra-high risk for psychosis. Biol. Psychiatry 2009, 66, 562–569. [Google Scholar] [CrossRef]
- Koutsouleris, N.; Kambeitz-Ilankovic, L.; Ruhrmann, S.; Rosen, M.; Ruef, A.; Dwyer, D.B.; Paolini, M.; Chisholm, K.; Kambeitz, J.; Haidl, T.; et al. Prediction models of functional outcomes for individuals in the clinical high-risk state for psychosis or with recent-onset depression: A multimodal, multisite machine learning analysis. JAMA Psychiatry 2018, 75, 1156–1172. [Google Scholar] [CrossRef]
- Krakauer, K.; Ebdrup, B.; Glenthøj, B.; Raghava, J.; Nordholm, D.; Randers, L.; Rostrup, E.; Nordentoft, M. Patterns of white matter microstructure in individuals at ultra-high-risk for psychosis: Associations to level of functioning and clinical symptoms. Psychol. Med. 2017, 47, 2689–2707. [Google Scholar] [CrossRef]
- Reniers, R.L.; Lin, A.; Yung, A.R.; Koutsouleris, N.; Nelson, B.; Cropley, V.L.; Velakoulis, D.; McGorry, P.D.; Pantelis, C.; Wood, S.J. Neuroanatomical predictors of functional outcome in individuals at ultra-high risk for psychosis. Schizophr. Bull. 2017, 43, 449–458. [Google Scholar] [CrossRef]
- Ohta, M.; Nakataki, M.; Takeda, T.; Numata, S.; Tominaga, T.; Kameoka, N.; Kubo, H.; Kinoshita, M.; Matsuura, K.; Otomo, M.; et al. Structural equation modeling approach between salience network dysfunction, depressed mood, and subjective quality of life in schizophrenia: An ICA resting-state fMRI study. Neuropsychiatr. Dis. Treat. 2018, 14, 1585. [Google Scholar] [CrossRef] [PubMed]
- Li, R.R.; Lyu, H.L.; Liu, F.; Lian, N.; Wu, R.R.; Zhao, J.P.; Guo, W.B. Altered functional connectivity strength and its correlations with cognitive function in subjects with ultra-high risk for psychosis at rest. CNS Neurosci. Ther. 2018, 24, 1140–1148. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, M.; Yoshihara, Y.; Hashimoto, R.; Yahata, N.; Ichikawa, N.; Sakai, Y.; Yamada, T.; Matsukawa, N.; Okada, G.; Tanaka, S.C.; et al. A prediction model of working memory across health and psychiatric disease using whole-brain functional connectivity. Elife 2018, 7, e38844. [Google Scholar] [CrossRef] [PubMed]
- Lancon, C.; Auquier, P.; Nayt, G.; Reine, G. Stability of the five-factor structure of the Positive and Negative Syndrome Scale (PANSS). Schizophr. Res. 2000, 42, 231–239. [Google Scholar] [CrossRef]
- Yan, W.; Zhao, M.; Fu, Z.; Pearlson, G.D.; Sui, J.; Calhoun, V.D. Mapping relationships among schizophrenia, bipolar and schizoaffective disorders: A deep classification and clustering framework using fMRI time series. Schizophr. Res. 2022, 245, 141–150. [Google Scholar] [CrossRef]
- Meda, S.A.; Gill, A.; Stevens, M.C.; Lorenzoni, R.P.; Glahn, D.C.; Calhoun, V.D.; Sweeney, J.A.; Tamminga, C.A.; Keshavan, M.S.; Thaker, G.; et al. Differences in resting-state fMRI functional network connectivity between schizophrenia and psychotic bipolar probands and their unaffected first-degree relatives. Biol. Psychiatry 2012, 71, 881. [Google Scholar] [CrossRef]
- Scott, A.; Courtney, W.; Wood, D.; De la Garza, R.; Lane, S.; Wang, R.; King, M.; Roberts, J.; Turner, J.A.; Calhoun, V.D. COINS: An innovative informatics and neuroimaging tool suite built for large heterogeneous datasets. Front. Neuroinform. 2011, 5, 33. [Google Scholar] [CrossRef]
- Cetin, M.S.; Christensen, F.; Abbott, C.C.; Stephen, J.M.; Mayer, A.R.; Cañive, J.M.; Bustillo, J.R.; Pearlson, G.D.; Calhoun, V.D. Thalamus and posterior temporal lobe show greater inter-network connectivity at rest and across sensory paradigms in schizophrenia. NeuroImage 2014, 97, 117–126. [Google Scholar] [CrossRef]
- Aine, C.; Bockholt, H.J.; Bustillo, J.R.; Cañive, J.M.; Caprihan, A.; Gasparovic, C.; Hanlon, F.M.; Houck, J.M.; Jung, R.E.; Lauriello, J.; et al. Multimodal neuroimaging in schizophrenia: Description and dissemination. Neuroinformatics 2017, 15, 343–364. [Google Scholar] [CrossRef]
- Long, Q.; Jia, C.; Boukouvalas, Z.; Gabrielson, B.; Emge, D.; Adali, T. Consistent run selection for independent component analysis: Application to fMRI analysis. In Proceedings of the International Conference on Acoustics, Speech and Signal Processing, Calgary, AB, Canada, 15–20 April 2018; pp. 2581–2585. [Google Scholar]
- Amari, S.I. Estimating functions of independent component analysis for temporally correlated signals. Neural Comput. 2000, 12, 2083–2107. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.S.; Anderson, M.; Adalı, T. Likelihood estimators for dependent samples and their application to order detection. IEEE Trans. Signal Process. 2014, 62, 4237–4244. [Google Scholar] [CrossRef]
- Allen, E.A.; Erhardt, E.B.; Damaraju, E.; Gruner, W.; Segall, J.M.; Silva, R.F.; Havlicek, M.; Rachakonda, S.; Fries, J.; Kalyanam, R.; et al. A baseline for the multivariate comparison of resting-state networks. Front. Syst. Neurosci. 2011, 5, 2. [Google Scholar] [CrossRef]
- Allen, E.A.; Damaraju, E.; Plis, S.M.; Erhardt, E.B.; Eichele, T.; Calhoun, V.D. Tracking whole-brain connectivity dynamics in the resting state. Cereb. Cortex 2014, 24, 663–676. [Google Scholar] [CrossRef]
- Lancaster, J.L.; Woldorff, M.G.; Parsons, L.M.; Liotti, M.; Freitas, C.S.; Rainey, L.; Kochunov, P.V.; Nickerson, D.; Mikiten, S.A.; Fox, P.T. Automated Talairach atlas labels for functional brain mapping. Hum. Brain Mapp. 2000, 10, 120–131. [Google Scholar] [CrossRef]
- Lancaster, J.; Rainey, L.; Summerlin, J.; Freitas, C.; Fox, P.; Evans, A.; Toga, A.; Mazziotta, J. Automated labeling of the human brain: A preliminary report on the development and evaluation of a forward-transform method. Hum. Brain Mapp. 1997, 5, 238–242. [Google Scholar] [CrossRef]
- Zhang, J.X.; Leung, H.C.; Johnson, M.K. Frontal activations associated with accessing and evaluating information in working memory: An fMRI study. Neuroimage 2003, 20, 1531–1539. [Google Scholar] [CrossRef]
- Pochon, J.; Levy, R.; Fossati, P.; Lehericy, S.; Poline, J.; Pillon, B.; Le Bihan, D.; Dubois, B. The neural system that bridges reward and cognition in humans: An fMRI study. Proc. Natl. Acad. Sci. USA 2002, 99, 5669–5674. [Google Scholar] [CrossRef]
- Abrahams, S.; Goldstein, L.H.; Simmons, A.; Brammer, M.J.; Williams, S.C.; Giampietro, V.P.; Andrew, C.M.; Leigh, P.N. Functional magnetic resonance imaging of verbal fluency and confrontation naming using compressed image acquisition to permit overt responses. Hum. Brain Mapp. 2003, 20, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Holzman, P.S. Schizophrenics show spatial working memory deficits. Arch. Gen. Psychiatry 1992, 49, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Azuma, T. Working memory and perseveration in verbal fluency. Neuropsychology 2004, 18, 69. [Google Scholar] [CrossRef] [PubMed]
- Menon, V. Salience Network. Brain Mapping: An Encyclopedic Reference; Academic Press: Cambridge, MA, USA; Elsevier: Amsterdam, The Netherlands, 2015; Volume 2, pp. 597–611. [Google Scholar]
- Das, T.K.; Kumar, J.; Francis, S.; Liddle, P.F.; Palaniyappan, L. Parietal lobe and disorganisation syndrome in schizophrenia and psychotic bipolar disorder: A bimodal connectivity study. Psychiatry Res. Neuroimaging 2020, 303, 111139. [Google Scholar] [CrossRef]
- Nekovarova, T.; Fajnerova, I.; Horacek, J.; Spaniel, F. Bridging disparate symptoms of schizophrenia: A triple network dysfunction theory. Front. Behav. Neurosci. 2014, 8, 171. [Google Scholar] [CrossRef]
- Sridharan, D.; Levitin, D.J.; Menon, V. A critical role for the right fronto-insular cortex in switching between central-executive and default-mode networks. Proc. Natl. Acad. Sci. USA 2008, 105, 12569–12574. [Google Scholar] [CrossRef]



| Name | BA | MNI |
|---|---|---|
| dlPFC | 9 | , 33, 33 |
| Broca-Operc | 44 | , 6, 18 |
| Primary somatosensory cortex | 1 | 42, , 63 |
| Primary auditory | 41 | 63, , 9 |
| Middle temporal gyrus | 21 | , , 6 |
| Superior temporal gyrus | 21 | 53, , 8 |
| Supramarginal gyrus | 40 | 48, , 57 |
| ACC | 24 | 0, 30, 18 |
| BACS | p-Value (Method in [34]) | p-Value (fSIG) |
|---|---|---|
| List Learning | ||
| Digit Sequencing | ||
| Token Motor | ||
| Verbal Fluency | ||
| Symbol Coding | ||
| Tower of London |
| SFS | p-Value (Method in [34]) | p-Value (fSIG) |
|---|---|---|
| Social Engagement | 8.48 × 10 | 9.46×10 |
| Interpersonal Communication | 8.00 × 10 | 5.71×10 |
| Independence /Competence | 1.41 × 10 | 2.57×10 |
| Recreation Performance | 5.51 × 10 | 4.41×10 |
| Independence/Performance | 1.94 × 10 | 4.88×10 |
| Prosocial Performance | 3.17 × 10 | 7.57×10 |
| Occupation/Employment | 2.49 × 10 | 1.71×10 |
| PANSS | p-Value (Method in [34]) | p-Value (fSIG) |
|---|---|---|
| Positive Total | 3.81 × 10 | 9.56×10 |
| Negative Total | 1.79 × 10 | 6.83×10 |
| General Total | 8.45 × 10 | 6.86×10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Vu, T.; Long, Q.; Calhoun, V.; Adali, T. Identification of Homogeneous Subgroups from Resting-State fMRI Data. Sensors 2023, 23, 3264. https://doi.org/10.3390/s23063264
Yang H, Vu T, Long Q, Calhoun V, Adali T. Identification of Homogeneous Subgroups from Resting-State fMRI Data. Sensors. 2023; 23(6):3264. https://doi.org/10.3390/s23063264
Chicago/Turabian StyleYang, Hanlu, Trung Vu, Qunfang Long, Vince Calhoun, and Tülay Adali. 2023. "Identification of Homogeneous Subgroups from Resting-State fMRI Data" Sensors 23, no. 6: 3264. https://doi.org/10.3390/s23063264
APA StyleYang, H., Vu, T., Long, Q., Calhoun, V., & Adali, T. (2023). Identification of Homogeneous Subgroups from Resting-State fMRI Data. Sensors, 23(6), 3264. https://doi.org/10.3390/s23063264






