1. Introduction
Image registration is one of the basic tasks in medical image processing. It involves the acquisition of a dense deformation field (DDF) when a moving image is matched with a fixed image so that the two to-be-aligned images and their corresponding anatomical structures are aligned accurately in space [
1]. The traditional registration method optimizes the cost function through a large number of iterations, a process that usually requires a significant amount of computation and time [
2]. With the popularization and application of deep learning in the field of medical image registration, the deep learning registration method is now faster than the traditional image registration method. Therefore, for moving and fixed images, deformation fields can be generated by training a neural network, thus achieving rapid registration for a forward pass in the testing stage. Fan et al. [
3] studied the computational costs of seven different deformable registration algorithms. The results showed that the assessed deep-learning network (BIRNet) without any iterative optimization needed the least time. Additionally, the registration accuracy improved after applying the deep learning method. For example, Cao et al. [
4] proposed a deep learning method for registering brain MRI images, and it was revealed that the method’s Dice coefficient was improved in terms of registering white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF).
The unsupervised learning image registration method has been widely applied because it is not difficult to obtain gold-standard registration [
5]. Balakrishnan et al. [
6] optimized the U-Net neural network by defining the loss function as a combination of the mean square error similarity measure and the deformation field’s smoothing constraint. de Vos et al. [
7] accomplished affine and deformable registration by superimposing several networks through unsupervised training. Kim et al. [
8,
9] used cyclic consistency to provide implicit regularization for maintaining topology and realizing 2D or 3D image registration. Moreover, a multi-scale strategy was adopted during the experiment to solve the relevant storage problem. Jiang et al. [
10] proposed an unsupervised network framework (MJ-CNN) that adopted a multi-scale joint training scheme to achieve end-to-end optimization. Kong et al. [
11] designed a cascade-connected channel attention mechanism network. During cascade registration, the attention module is incorporated to learn the features of the input image, thereby improving the expression ability of the image features. Through five iterations of the deformation field, improved bidirectional image registration was realized. Yang et al. [
12] used multiple cascaded U-Net models to form a network structure. In their structure, each U-Net is trained with smooth regularization parameters to improve the accuracy of 3D medical image registration. Zhu et al. [
13] helped a network develop high-similarity spatial correspondence by introducing a local attention model and integrated multi-scale functionality into the attention mechanism module to achieve the coarse-to-fine registration of local information. Ouyang et al. [
14] trained their designed subnetworks synergistically by training the residual recursive cascade network to realize cooperation between the subnetworks. Through the connection of the residual network, the registration speed was accelerated. Guo et al. [
15] improved the image registration accuracy and efficiency of CT-MR and used two cyclic consistency methods in a full convolution neural network to generate the spatial deformation field. Sideri-Lampretsa et al. [
16] considered that it was easy to obtain edge images, so they used the image’s edges to drive the multimodal registration training process and thus help the network learn more effective information. Qian et al. [
17] proposed a cascade framework of a registration network, and then registered images in training stages. The authors compared the performance of the cascade network framework with the traditional registration methods, subsequently, it was determined that the registration efficiency of the proposed method was significantly improved. Golkar et al. [
18] proposed a hybrid registration framework of vessel extraction and thinning for retinal image segmentation, which improved the registration accuracy of complex retinal vessels.
Inspired by the idea of two-person zero-sum game from game theory, Goodefellow et al. [
19] proposed a generation adversarial network (GAN) that used two neural networks for adversarial training and continuously improved the performance of the network in all directions during a game between the two networks. In addition to the in-depth study of the generative adversarial network (GAN), the application of an adversarial network has been integrated with techniques and aims from other fields, for instance, the combination of GAN and image processing. Therefore, GANs are also widely used in image registration. Santarossa et al. [
20] used generation adversarial networks combined with ranking loss for multimodal image registration. Fan et al. [
21,
22] implemented a GAN in the unsupervised deformable registration of 3D brain MR images. In this approach, the discrimination network identifies whether a pair of images are sufficiently similar. The resulting feedback is then used to train the registration network. Simultaneously, GANs have been applied to single- and multi-mode image registration. Zheng et al. [
23] used a GAN network to realize symmetric image registration and then transformed the symmetric registration formula of single- and multi-mode images into a conditional GAN. To align a pair of single-mode images, the registration method constitutes a cyclical process of transformation from one image to another and its inverse transformation. To align images with different modes, mode conversion should be performed before registration. In the training process, the method also adopts the semi-supervised method and trains using labeled and unlabeled images. Many registration methods have been produced based on the application of generation adversarial networks [
24,
25,
26,
27,
28]. Huang et al. [
29] fused a difficulty perception model into a cascade neural network composed of three networks. These networks are used to predict the coarse deformation field and the fine deformation field, respectively, so as to achieve accurate registration. GANs showed excellent performance in the aforementioned studies. In the previous study, a GAN based on dual attention mechanisms was proposed, which showed good registration performance in areas with relatively flat edges, but poor registration performance in narrow and long-edge areas. To this end, based on previous research, this paper proposes a method to assist GANs in realizing the registration of long and narrow regions at the peripheries of the brain, which differs from the methods of coarse registration and fine registration. Our main contributions are summarized as follows:
During training, the cascade networks are trained simultaneously to save network training time.
The second network is used as a loss function. The mean square error loss function added to the second network can constrain the deformation field output by the second network such that it tends to 0. Only the first network is used during testing, which saves testing time.
Coupled with the adversarial training of GANs, the registration performance of the first network is further improved.
The rest of this paper is organized as follows.
Section 2 introduces the networks proposed in this paper in detail.
Section 3 introduces the experimental datasets and evaluation indicators.
Section 4 introduces the experimental results obtained from the HBN and ABIDE datasets. In
Section 5, we provide a discussion. Finally, the conclusions are given in
Section 6.
2. Methodology
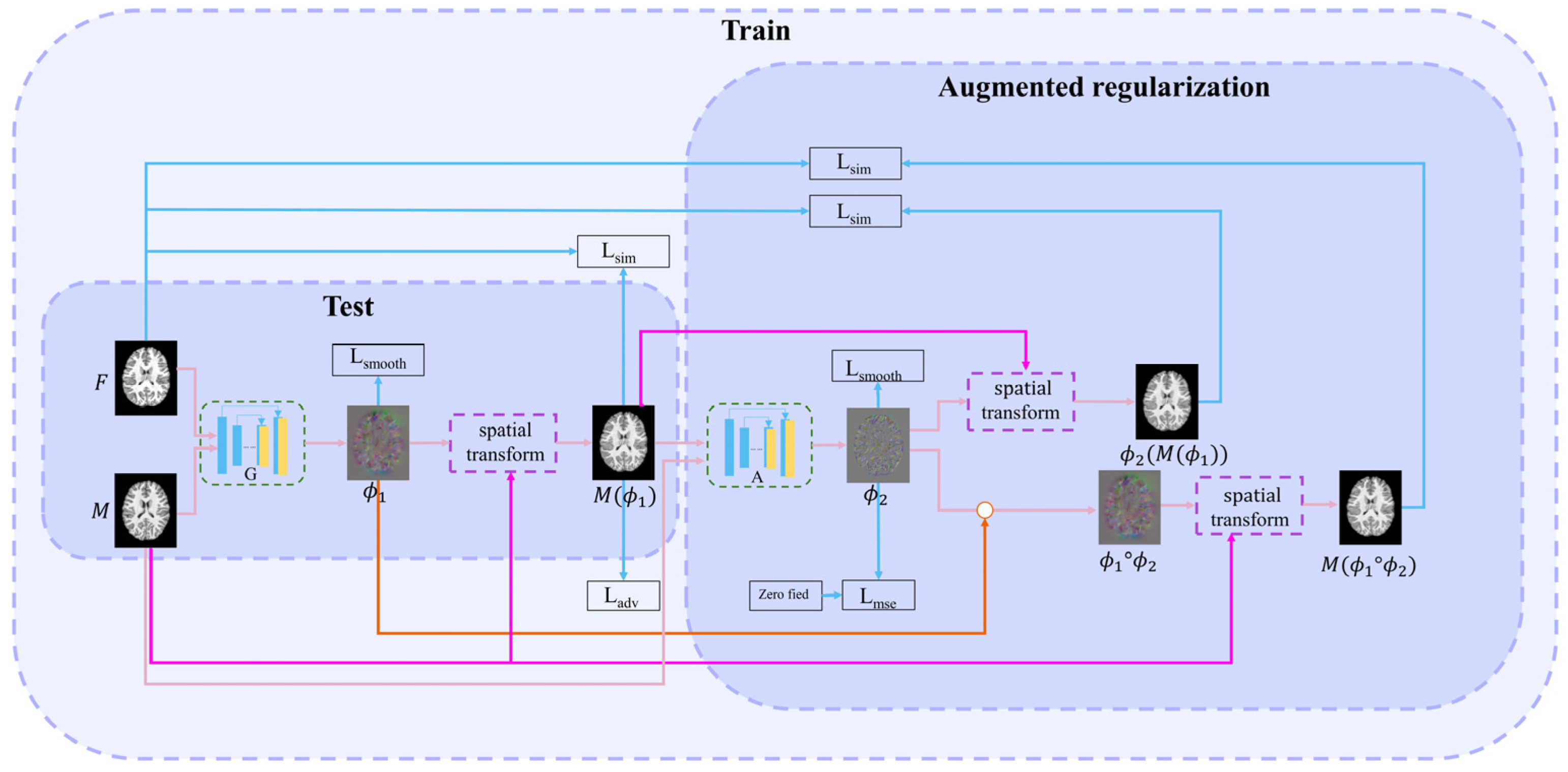
This paper proposes a method combining adversarial learning with cascade learning. Joint training of cascaded networks can allow them to predict more accurate deformation fields. The first (registration) network is used to study the deformation field
. The second (augmented) network enables the first network to learn more deformations. A discrimination network improves the first network’s performance through adversarial training. The structures of each cascading network are similar to those of VoxelMorph [
6]. The proposed overall learning framework is illustrated in
Figure 1.
2.1. First (Registration) Network
The registration network is the first network in cascading framework. Its inputs are the fixed image and the moving image . Its output is the deformation field , i.e., . This network realizes the alignment from to , i.e., = (, where () is the warped image. Subsequently, the loss function between and is calculated to drive the training process. This loss function includes three parts: intensity similarity loss Lsim, adversarial loss Ladv, and smooth regularization term Lsmooth.
The adversarial loss function of the registration network is:
where
is the output value of the discrimination network and
indicates the registration network input.
Local cross-correlation metric is used to calculate the similarity of the intensity between fixed image
and warped image
. The specific formula of the loss function is:
where
denotes the iteration of the
volume center at voxel
, and
represents a three-dimensional voxel. In this paper,
and
represents the voxel intensities of
and
at
, respectively.
and
are the local mean values of
volume. A higher CC indicates a more accurate alignment. According to the definition of CC, the intensity similarity loss L
sim is defined as follows:
Additionally, L2 regularization is implemented to smooth the deformation field
:
2.2. Successive (Augmented) Network
The inputs of the successive network are and ; the output is DDF . is used to deform to obtain . Simultaneously, to clarify the warped image, we perform a composed operation on and , i.e., . is obtained by the moving image with the composed DDF. Next, two intensity loss functions, namely, Lsim() and Lsim(), are calculated between and and between and , respectively. The DDF is also constrained as it approaches zero deformation field through the following MSE loss function, allowing the deformation field to learn more accurate deformations.
The formula of MSE loss function is defined as:
Through this function, the output effect of the first network can achieve fine registration after the two networks are connected in series.
The loss function for the registration network is as follows:
In addition, the loss function used by the second network is:
The total loss function is:
2.3. Discrimination Network
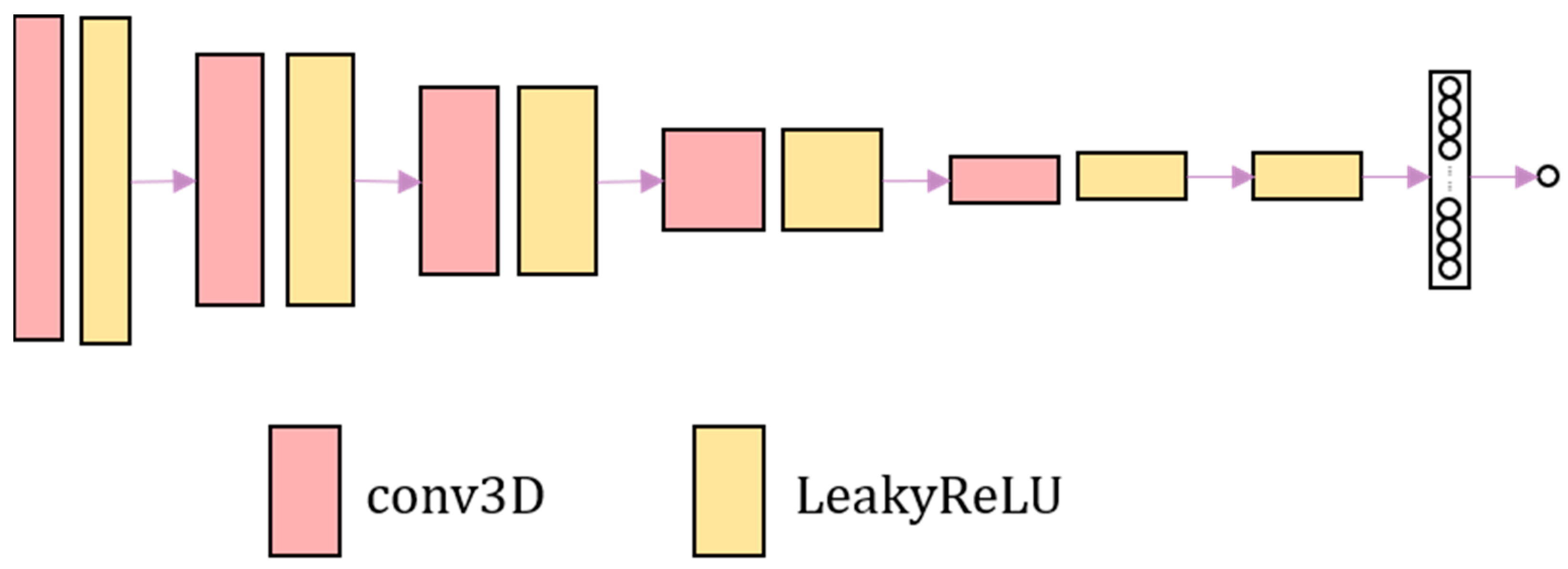
The discrimination network consists of four convolutional layers combined with leakyReLU activation layers. Finally, the sigmoid activation function is used to output the probability value. The discrimination network is shown in
Figure 2. The discrimination network distinguishes the authenticity of image. The harder it is to distinguish the warped image from the fixed image, the harder it is to judge the authenticity of the image by the discrimination network.
4. Results
The proposed methodology is compared with the following approaches: (1) Demons and SyN, two traditional registration methods; (2) Voxelmorph (VM), an unsupervised deep learning registration method; and (3) VM + A, a method consisting of a simultaneously trained registration network and augmented network.
First, the proposed GAN method (VM + A + GAN) is compared with Demons and SyN, which are two traditional methods.
Table 1 and
Table 2 summarize the test results obtained through different datasets, and all indicators show that our experimental results are the best.
Figure 3 shows the comparison of the test results of the two datasets. The first row of the experimental image represents the original image obtained from the HBN dataset, and the second row represents the segmentation image corresponding to the original image derived from the HBN dataset. Similarly, the third row represents the original image based on the ABIDE dataset, and the fourth row represents the segmentation image corresponding to the original image derived from the ABIDE dataset. Compared with Demons and SyN, the image obtained by the proposed GAN method is closer in appearance to the fixed image, and the parts with differences are shown in the enlarged image on the right.
Second, the proposed GAN method is compared with the VM and VM + A methods.
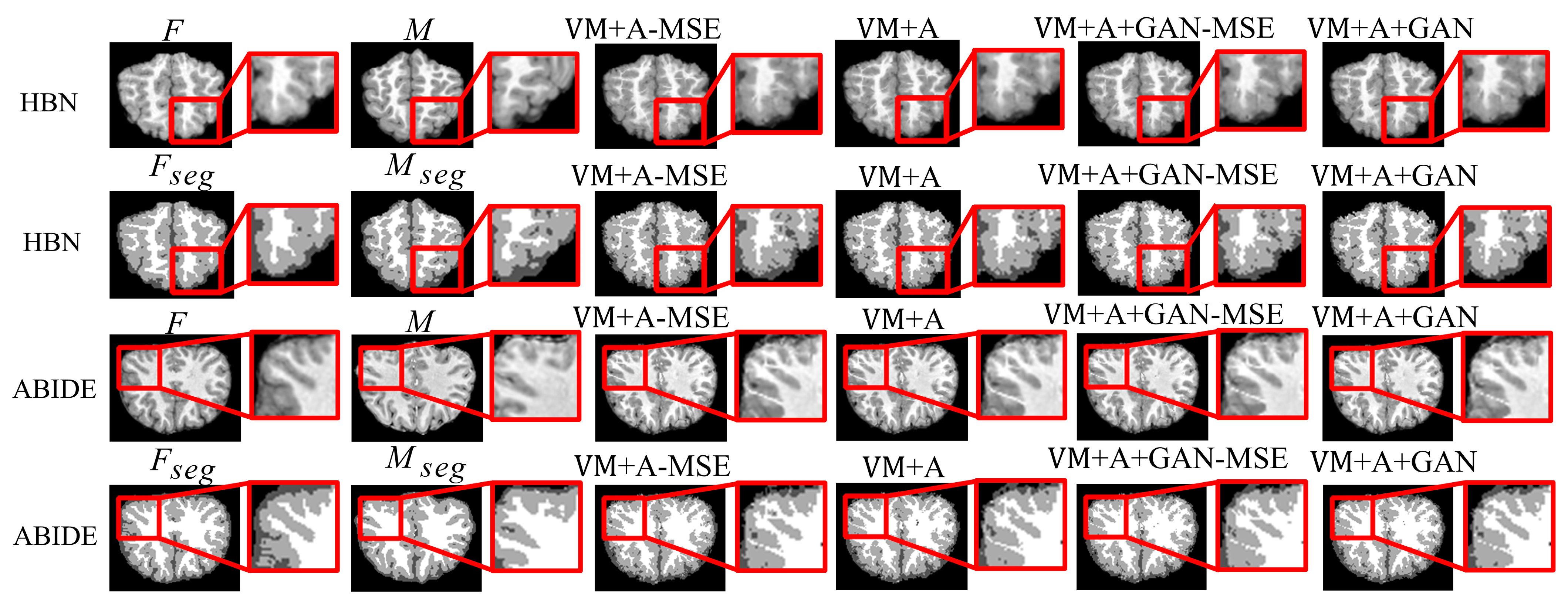
Figure 4 shows the registered moving image and the fixed image. Moreover, the first row represents the original image from the HBN dataset, and the second row represents the segmentation image corresponding to the original image from the HBN dataset. Similarly, the third row represents the original image from the ABIDE dataset, and the fourth row represents the segmentation image corresponding to the original image from the ABIDE dataset. Additionally, the enlarged figure on the right shows that the result for the proposed method regarding the training of the registration, augmented, and discrimination networks together is closer to the fixed image. Through the experimental results, the performance of the registration, augmented, and discrimination networks when trained together is verifiably better than that of the registration network trained individually and of the registration and augmented networks trained simultaneously.
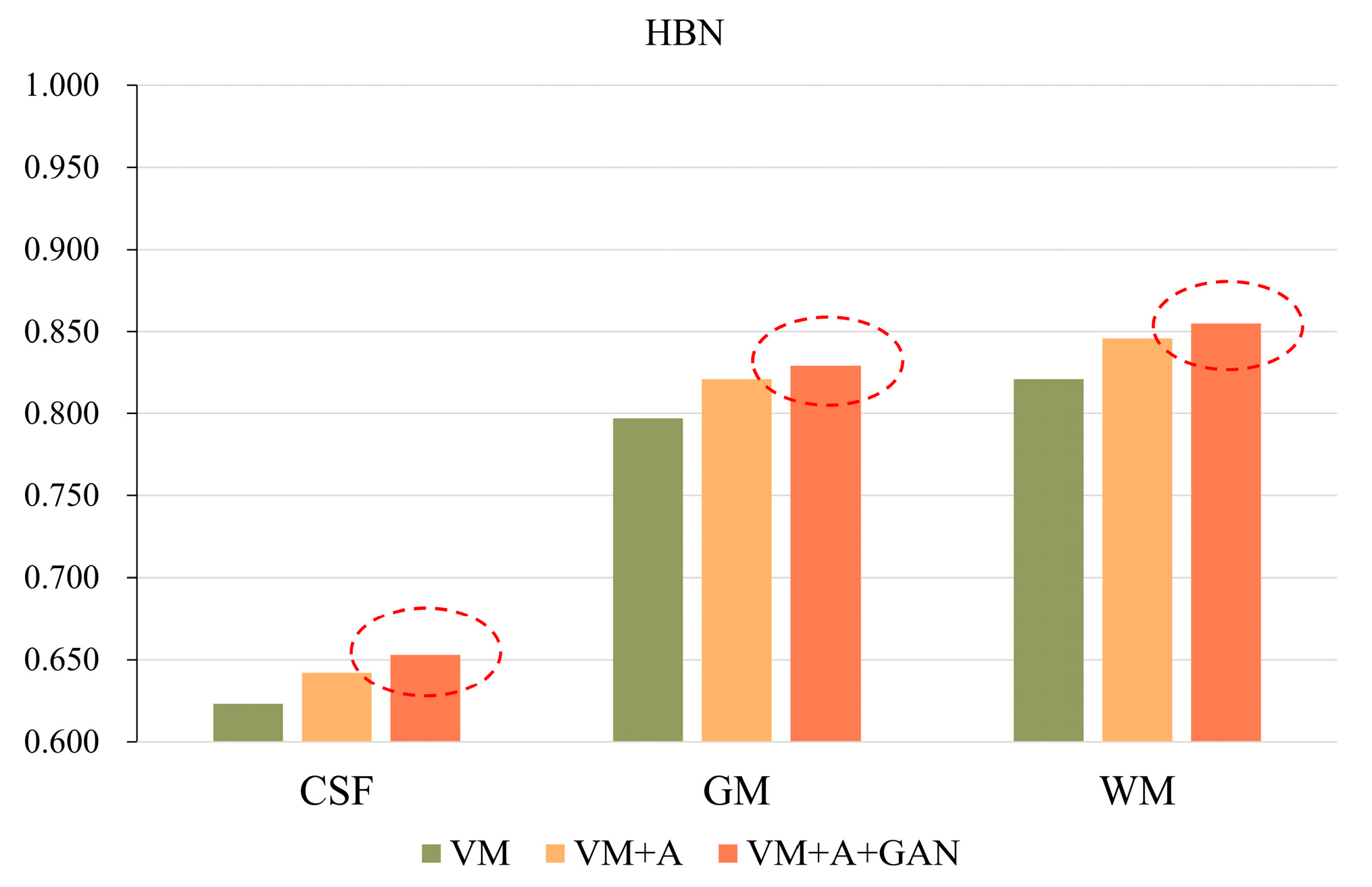
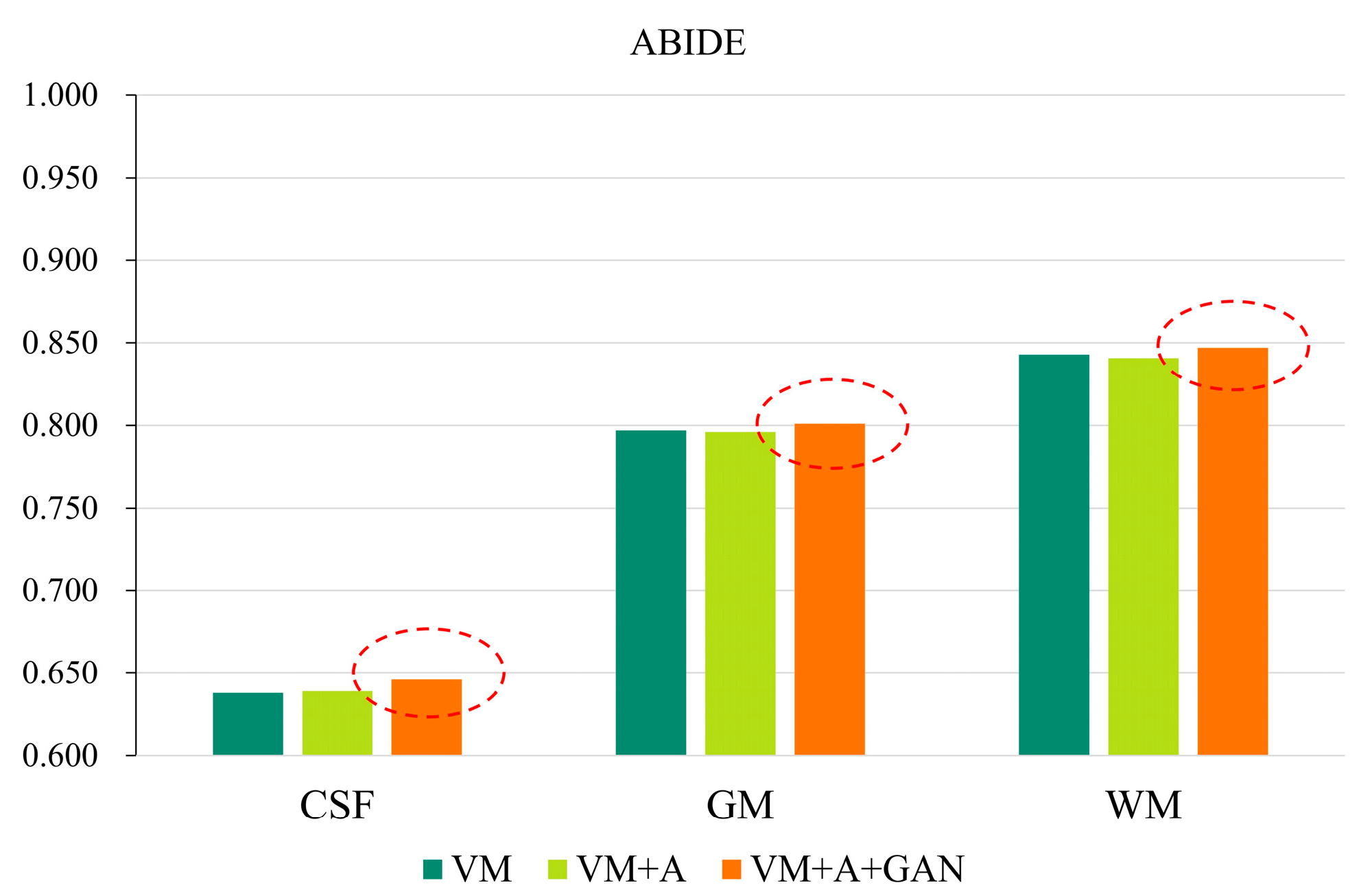
In order to more clearly highlight the effectiveness of the method proposed in this paper,
Figure 5 shows the experimental results of the three parts of the brain tissue based on the HBN dataset, and
Figure 6 shows the experimental results of the three parts of the brain tissue based on the ABIDE dataset. The dotted circle in the figure is the result obtained by the method proposed in this paper.
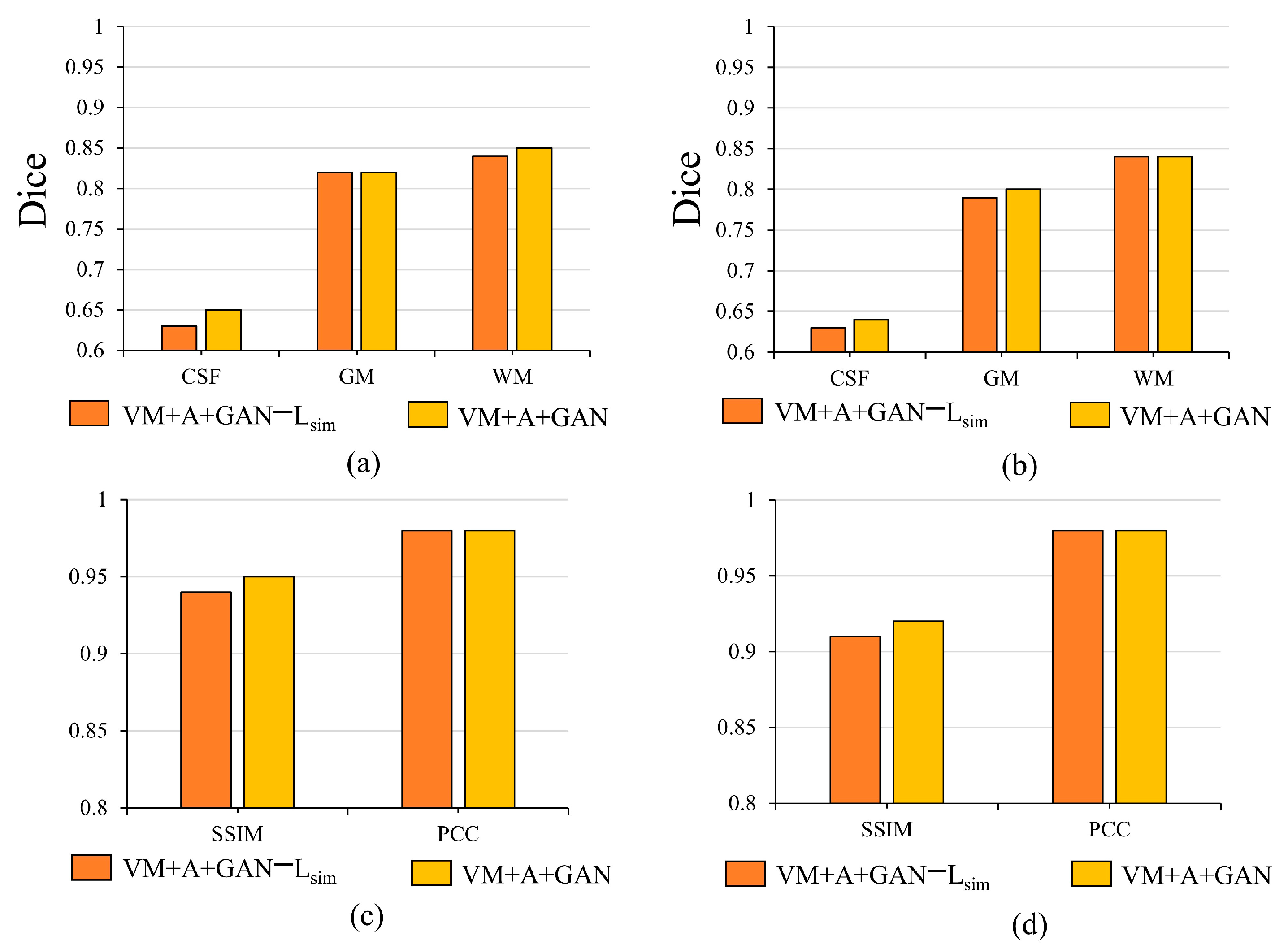
Table 3 and
Table 4 summarize the Dice, SSIM, and PCC indices corresponding to the different datasets. Considering
Table 3, for the HBN dataset, the proposed method improves the precision values by 0.030, 0.032, and 0.034 compared with the VM method. For the ABIDE dataset, the proposed method improves the accuracies by 0.008, 0.004, and 0.004 compared with the VM method. Considering
Table 4, for the HBN dataset, the proposed method increases the SSIM and PCC indices by 0.02 and 0.008, respectively, compared with the VM method. For the ABIDE dataset, the proposed method improves the SSIM and PCC indices by 0.006 and 0.003, respectively, compared with the VM method.
6. Conclusions
In this paper, a method wherein three networks (registration, augmented, and discrimination networks) are trained together is proposed, for which the MSE loss function is introduced into the augmented network to improve the registration network’s performance. It was demonstrated that the registration network’s performance was further improved when coupled with the adversarial capacity of a GAN. Then, it was proven that the proposed method offers significant advantages over the existing methods. In addition, it was clarified that the proposed training method is easy to implement, and that the implemented loss function is easy to obtain.
In the future, a more novel GAN will be used to further improve image registration performance; moreover, more indicators will be used for comparison. The developed model will then be tested on different datasets to prove its excellent generalizability.