DDM-HSA: Dual Deterministic Model-Based Heart Sound Analysis for Daily Life Monitoring
Abstract
1. Introduction
- A parallel structure algorithm based on multimodal methods improves the accuracy of heart sound detection.
- The proposed DDM-HSA outperforms existing heart sound detection methods using a single signal.
- By interpolating the S2 peak using the envelope filtering method, S2 detection accuracy can be improved.
2. Materials and Methods
2.1. Database
2.1.1. Acquisition System and Protocol
2.1.2. Subjects
2.2. Noise Reduction
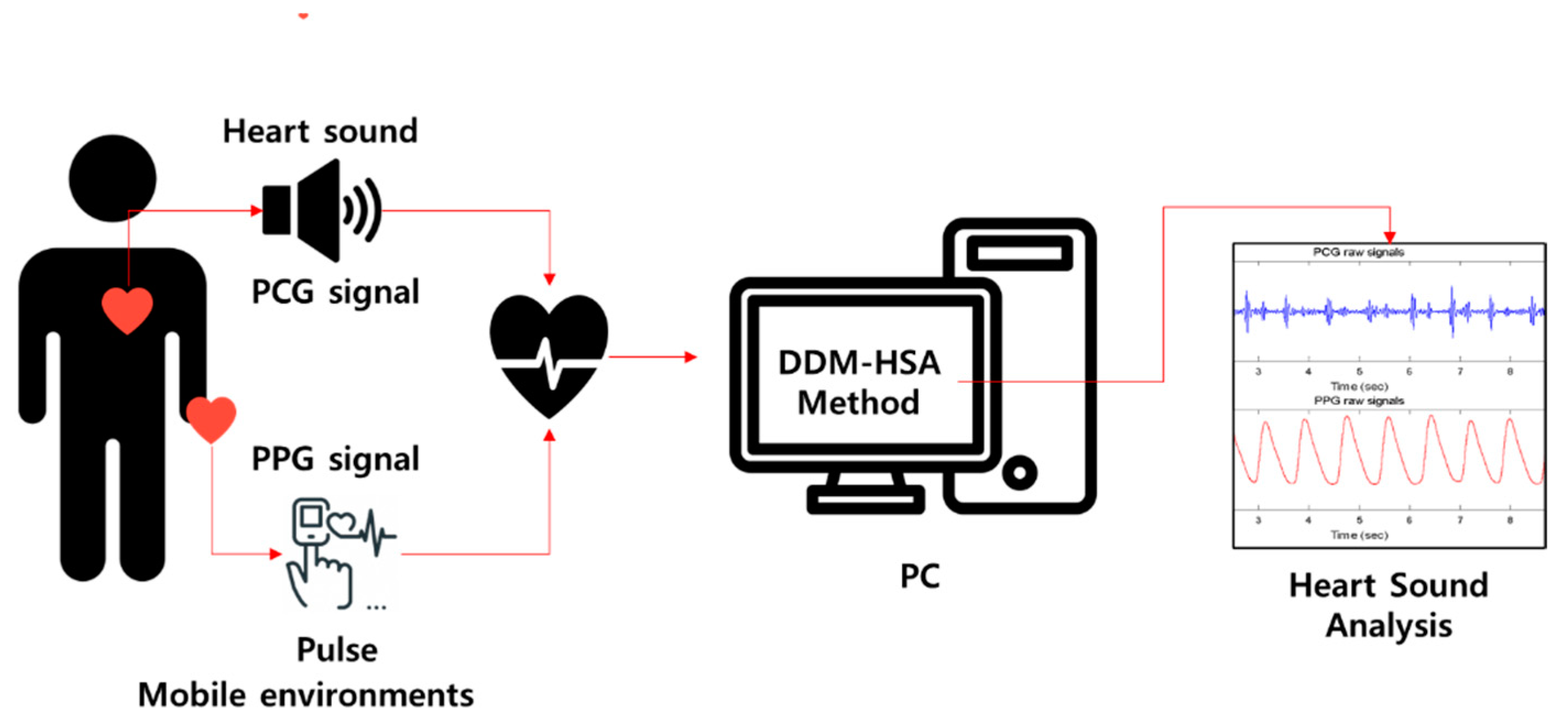
2.3. DDM-HSA
2.3.1. Overview
2.3.2. PCG Analysis
2.3.3. PPG Analysis
2.3.4. VTT Calculation and S2 Detection
2.4. Performance Measures
- True positive (TP): actual heartbeat correctly detected as an actual heartbeat.
- False negative (FN): not heartbeat detected as not heartbeat.
- True negative (TN): not heartbeat correctly detected as an actual heartbeat.
- False positive (FP): actual heartbeat detected as not heartbeat.
3. Results
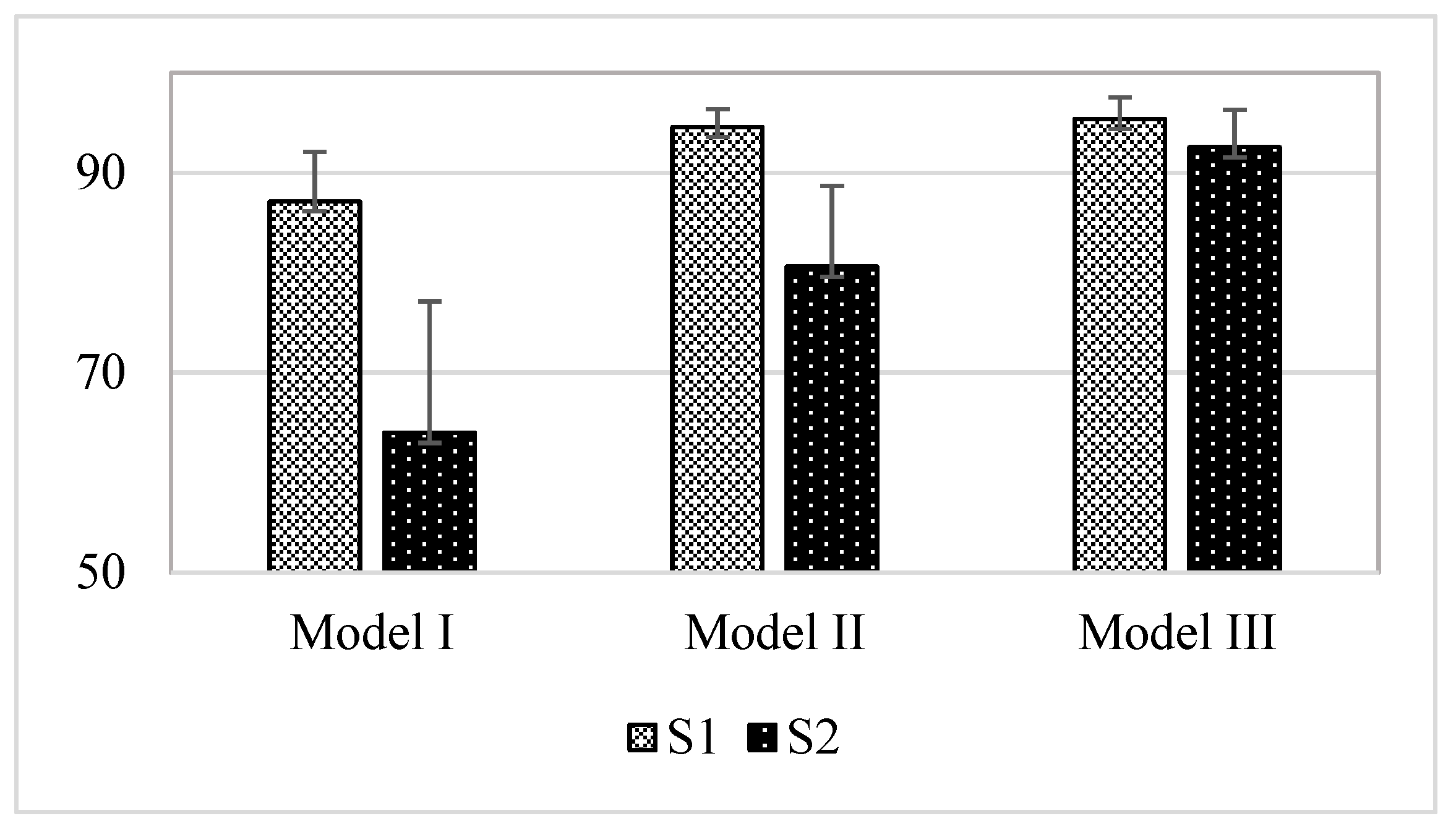
3.1. Comparison Result of Models
3.2. Result of Model I (DDM-HSA)
3.3. Effect of the Window (Model II)
3.4. Effect of the Envelope Filtering (Model III)
4. Discussion
4.1. Comparison with Other Approaches for Heart Sounds Analysis
4.2. Limitations
5. Conclusions
- We contributed to the analysis of heart sounds in daily life by presenting a DDM-HSA that can utilize PPG and PCG, which that can be measured using wearable devices.
- We proposed an envelope filtering method to improve the performance of S2 detection. By applying it to DDM-HSA (Model III), the performance of S2 improved by about 28.59% compared with that of the existing method (Model I).
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, S.; Wei, Q.; Park, H.; Na, Y.; Jeong, D.; Lim, H. Development of a finger-ring-shaped hybrid smart stethoscope for automatic s1 and s2 heart sound identification. Sensors 2021, 21, 6294. [Google Scholar] [CrossRef]
- Lee, M.; Song, T.G.; Lee, J.H. Heartbeat classification using local transform pattern feature and hybrid neural fuzzy-logic system based on self-organizing map. Biomed. Signal Process. Control 2020, 57, 101960. [Google Scholar] [CrossRef]
- Lee, M.; Lee, J.H. A robust fusion algorithm of LBP and IMF with recursive feature elimination-based ECG processing for QRS and arrhythmia detection. Appl. Intell. 2022, 52, 939–953. [Google Scholar] [CrossRef]
- Achenbach, S.; Daniel, W.G. Noninvasive coronary angiography—An acceptable alternative? N. Engl. J. Med. 2001, 345, 1909–1910. [Google Scholar] [CrossRef]
- Yıldırım, Ö.; Pławiak, P.; Tan, R.S.; Acharya, U.R. Arrhythmia detection using deep convolutional neural network with long duration ECG signals. Comput. Biol. Med. 2018, 102, 411–420. [Google Scholar] [CrossRef]
- Bharti, R.; Khamparia, A.; Shabaz, M.; Dhiman, G.; Pande, S.; Singh, P. Prediction of Heart Disease Using a Combination of Machine Learning and Deep Learning. Comput. Intell. Neurosci. 2021, 2021, 8387680. [Google Scholar] [CrossRef]
- Sraitih, M.; Jabrane, Y.; el Hassani, A.H. An automated system for ECG arrhythmia detection using machine learning techniques. J. Clin. Med. 2021, 10, 5450. [Google Scholar] [CrossRef]
- The Electronic Stethoscope for Telemedicine—Ekuore Smart Stethoscope. Available online: https://www.ekuore.com/electronicstethoscope-ld-coronavirus/ (accessed on 28 July 2022).
- A Smart Way of Asthma Monitoring—StethoMe Stethoscope. Available online: https://www.stethome.com/en-gb/ (accessed on 28 July 2022).
- A Smart Way of PPG Monitoring—Galaxy Watch Active. Available online: https://www.samsung.com/sec/watches/galaxy-watch-active2-r820/SM-R820NSSAKOO/ (accessed on 28 July 2022).
- Park, H.; Dong, S.-Y.; Lee, M.; Youn, I. The Role of Heart-Rate Variability Parameters in Activity Recognition and Energy-Expenditure Estimation Using Wearable Sensors. Sensors 2017, 17, 1698. [Google Scholar] [CrossRef]
- Nieman, D.C.; Austin, M.D.; Benezra, L.; Pearce, S.; McInnis, T.; Unick, J.; Gross, S.J. Validation of cosmed’s fitmate in measuring oxygen consumption and estimating resting metabolic rate. Res. Sports Med. 2006, 14, 89–96. [Google Scholar] [CrossRef]
- Melia, U.; Clariá, F.; Vallverdú, M.; Caminal, P. Filtering and thresholding the analytic signal envelope in order to improve peak and spike noise reduction in EEG signals. Med. Eng. Phys. 2014, 36, 547–553. [Google Scholar] [CrossRef]
- Abdulmunem, M.E.; Badr, A.A. Hilbert Transform and its Applications: A survey. Int. J. Sci. Eng. Res. 2017, 8, 699–704. [Google Scholar]
- Lee, M.; Park, D.; Dong, S.Y.; Youn, I. A novel R peak detection method for mobile environments. IEEE Access 2018, 6, 51227–51237. [Google Scholar] [CrossRef]
- Rastogi, N.; Mehra, R. Analysis of Savitzky-Golay filter for baseline wander cancellation in ECG using wavelets. Int. J. Eng. Sci. Emerg. Technol. 2013, 6, 15–23. [Google Scholar]
- Agrawal, S.; Gupta, A. Fractal and EMD based removal of baseline wander and powerline interference from ECG signals. Comput. Biol. Med. 2013, 43, 1889–1899. [Google Scholar] [CrossRef]
- Giordano, N.; Knaflitz, M. A novel method for measuring the timing of heart sound components through digital phonocardiography. Sensors 2019, 19, 1868. [Google Scholar] [CrossRef]
- Allen, J. Photoplethysmography and its Application in Clinical Physiological Measurement. Physiol. Meas. 2007, 28, R1. [Google Scholar] [CrossRef]
- Shelley, K.H. Photoplethysmography: Beyond the Calculation of Arterial Oxygen Saturation and Heart Rate. Anesth. Analg. 2007, 105, S31–S36. [Google Scholar] [CrossRef]
- Nam, D.H.; Lee, W.B.; Hong, Y.S.; Lee, S.S. Measurement of Spatial Pulse Wave Velocity by Using a Clip-Type Pulsimeter Equipped with a Hall Sensor and Photoplethysmography. J. Sens. Sci. Technol. 2013, 13, 4714–4723. [Google Scholar] [CrossRef]
- Elgendi, M.; Fletcher, R.; Liang, Y.; Howard, N.; Lovell, N.H.; Abbott, D.; Lim, K.; Ward, R. The use of photoplethysmography for assessing hypertension. NPJ Digit. Med. 2019, 2, 1–11. [Google Scholar] [CrossRef]
- Varghees, V.N.; Ramachandran, K. A novel heart sound activity detection framework for automated heart sound analysis. Biomed. Signal Process. Control 2014, 13, 174–188. [Google Scholar] [CrossRef]
- Babu, K.A.; Ramkumar, B.; Manikandan, M.S. Automatic identification of S1 and S2 heart sounds using simultaneous PCG and PPG recordings. IEEE Sens. J. 2018, 18, 9430–9440. [Google Scholar] [CrossRef]
- Huang, C.; Chen, H.; Yang, L.; Zhang, Q. BreathLive: Liveness detection for heart sound authentication with deep breathing. Proc. ACM Interact. Mob. Wearable Ubiquitous Technol. 2018, 2, 12. [Google Scholar] [CrossRef]
- Park, H.; Wei, Q.; Lee, S.; Lee, M. Novel Design of a Multimodal Technology-Based Smart Stethoscope for Personal Cardiovascular Health Monitoring. Sensors 2022, 22, 6465. [Google Scholar] [CrossRef]






| Subject ID | Sex | Age |
|---|---|---|
| S1 | F | 32 |
| S2 | F | 24 |
| S3 | F | 31 |
| S4 | F | 28 |
| S5 | F | 26 |
| S6 | F | 30 |
| S7 | F | 28 |
| S8 | F | 28 |
| S9 | F | 28 |
| S10 | M | 29 |
| S11 | M | 30 |
| S12 | M | 28 |
| S13 | M | 34 |
| S14 | M | 24 |
| S15 | M | 25 |
| S16 | M | 31 |
| S17 | M | 26 |
| S18 | M | 25 |
| S19 | M | 27 |
| S20 | M | 22 |
| TOTAL | M = 10, F = 10 | 27.4 (3.2) |
| Estimation Models | Description |
|---|---|
| Model I | DDM-HSA |
| Model II | DDM-HSA + window |
| Model III | DDM-HSA + window + envelope filter (EF) |
| Sub. No. | Mean HR [bpm] | TP | TN | FP | FN | SEN | PRE | SPE | ACC |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 61.50 | 353.00 | 255.00 | 114.00 | 16.00 | 95.66 | 75.59 | 69.11 | 82.38 |
| 2 | 77.50 | 453.00 | 385.00 | 80.00 | 12.00 | 97.42 | 84.99 | 82.80 | 90.11 |
| 3 | 76.67 | 386.00 | 322.00 | 138.00 | 74.00 | 83.91 | 73.66 | 70.00 | 76.96 |
| 4 | 77.17 | 376.00 | 363.00 | 100.00 | 87.00 | 81.21 | 78.99 | 78.40 | 79.81 |
| 5 | 73.33 | 419.00 | 366.00 | 74.00 | 21.00 | 95.23 | 84.99 | 83.18 | 89.20 |
| 6 | 70.50 | 355.00 | 300.00 | 123.00 | 68.00 | 83.92 | 74.27 | 70.92 | 77.42 |
| 7 | 59.67 | 342.00 | 318.00 | 40.00 | 16.00 | 95.53 | 89.53 | 88.83 | 92.18 |
| 8 | 72.33 | 411.00 | 379.00 | 55.00 | 23.00 | 94.70 | 88.20 | 87.33 | 91.01 |
| 9 | 68.33 | 388.00 | 315.00 | 95.00 | 22.00 | 94.63 | 80.33 | 76.83 | 85.73 |
| 10 | 59.17 | 343.00 | 251.00 | 104.00 | 12.00 | 96.62 | 76.73 | 70.70 | 83.66 |
| 11 | 61.17 | 318.00 | 300.00 | 67.00 | 49.00 | 86.65 | 82.60 | 81.74 | 84.20 |
| 12 | 67.33 | 375.00 | 362.00 | 42.00 | 29.00 | 92.82 | 89.93 | 89.60 | 91.21 |
| 13 | 63.17 | 355.00 | 315.00 | 64.00 | 24.00 | 93.67 | 84.73 | 83.11 | 88.39 |
| 14 | 67.00 | 399.00 | 289.00 | 113.00 | 3.00 | 99.25 | 77.93 | 71.89 | 85.57 |
| 15 | 73.00 | 414.00 | 367.00 | 71.00 | 24.00 | 94.52 | 85.36 | 83.79 | 89.16 |
| 16 | 63.67 | 355.00 | 327.00 | 55.00 | 27.00 | 92.93 | 86.59 | 85.60 | 89.27 |
| 17 | 86.67 | 488.00 | 486.00 | 34.00 | 32.00 | 93.85 | 93.49 | 93.46 | 93.65 |
| 18 | 76.67 | 422.00 | 438.00 | 22.00 | 38.00 | 91.74 | 95.05 | 95.22 | 93.48 |
| 19 | 80.17 | 413.00 | 448.00 | 33.00 | 68.00 | 85.86 | 92.60 | 93.14 | 89.50 |
| 20 | 62.33 | 355.00 | 314.00 | 60.00 | 19.00 | 94.92 | 85.54 | 83.96 | 89.44 |
| AVG | 70.00 | 386.00 | 345.00 | 74.20 | 33.20 | 92.25 | 84.05 | 81.98 | 87.12 |
| STD | 7.75 | 41.77 | 61.52 | 33.18 | 23.47 | 5.07 | 6.43 | 8.24 | 4.99 |
| Sub. No. | Mean HR [bpm] | TP | TN | FP | FN | SEN | PRE | SPE | ACC |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 61.50 | 310 | 268 | 27 | 59 | 84.01 | 91.99 | 90.85 | 87.05 |
| 2 | 77.50 | 226 | 187 | 161 | 239 | 48.60 | 58.40 | 53.74 | 50.80 |
| 3 | 76.67 | 251 | 199 | 111 | 209 | 54.57 | 69.34 | 64.19 | 58.44 |
| 4 | 77.17 | 267 | 243 | 131 | 196 | 57.67 | 67.09 | 64.97 | 60.93 |
| 5 | 73.33 | 386 | 300 | 40 | 54 | 87.73 | 90.61 | 88.24 | 87.95 |
| 6 | 70.50 | 281 | 138 | 58 | 142 | 66.43 | 82.89 | 70.41 | 67.69 |
| 7 | 59.67 | 260 | 111 | 43 | 98 | 72.63 | 85.81 | 72.08 | 72.46 |
| 8 | 72.33 | 226 | 188 | 99 | 208 | 52.07 | 69.54 | 65.51 | 57.42 |
| 9 | 68.33 | 109 | 311 | 58 | 301 | 26.59 | 65.27 | 84.28 | 53.92 |
| 10 | 59.17 | 170 | 297 | 24 | 185 | 47.89 | 87.63 | 92.52 | 69.08 |
| 11 | 61.17 | 252 | 50 | 57 | 115 | 68.66 | 81.55 | 46.73 | 63.71 |
| 12 | 67.33 | 83 | 243 | 95 | 321 | 20.54 | 46.63 | 71.89 | 43.94 |
| 13 | 63.17 | 105 | 211 | 101 | 274 | 27.70 | 50.97 | 67.63 | 45.73 |
| 14 | 67.00 | 194 | 298 | 56 | 208 | 48.26 | 77.60 | 84.18 | 65.08 |
| 15 | 73.00 | 252 | 136 | 94 | 186 | 57.53 | 72.83 | 59.13 | 58.08 |
| 16 | 63.67 | 190 | 241 | 93 | 192 | 49.74 | 67.14 | 72.16 | 60.20 |
| 17 | 86.67 | 424 | 255 | 59 | 96 | 81.54 | 87.78 | 81.21 | 81.41 |
| 18 | 76.67 | 375 | 168 | 43 | 85 | 81.52 | 89.71 | 79.62 | 80.92 |
| 19 | 80.17 | 299 | 213 | 54 | 182 | 62.16 | 84.70 | 79.78 | 68.45 |
| 20 | 62.33 | 124 | 197 | 128 | 250 | 33.16 | 49.21 | 60.62 | 45.92 |
| AVG | 70.00 | 239.20 | 212.70 | 76.60 | 180.00 | 56.45 | 73.83 | 72.49 | 63.96 |
| STD | 7.75 | 94.14 | 69.26 | 37.74 | 77.33 | 19.68 | 14.47 | 12.52 | 13.18 |
| Sub. No. | Mean HR [bpm] | TP | TN | FP | FN | SEN | PRE | SPE | ACC |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 61.50 | 366 | 310 | 59 | 3 | 99.19 | 86.12 | 84.01 | 91.60 |
| 2 | 77.50 | 451 | 427 | 38 | 14 | 96.99 | 92.23 | 91.83 | 94.41 |
| 3 | 76.67 | 434 | 403 | 57 | 26 | 94.35 | 88.39 | 87.61 | 90.98 |
| 4 | 77.17 | 424 | 420 | 43 | 39 | 91.58 | 90.79 | 90.71 | 91.14 |
| 5 | 73.33 | 431 | 412 | 28 | 9 | 97.95 | 93.90 | 93.64 | 95.80 |
| 6 | 70.50 | 411 | 368 | 55 | 12 | 97.16 | 88.20 | 87.00 | 92.08 |
| 7 | 59.67 | 350 | 332 | 26 | 8 | 97.77 | 93.09 | 92.74 | 95.25 |
| 8 | 72.33 | 422 | 401 | 33 | 12 | 97.24 | 92.75 | 92.40 | 94.82 |
| 9 | 68.33 | 404 | 367 | 43 | 6 | 98.54 | 90.38 | 89.51 | 94.02 |
| 10 | 59.17 | 351 | 317 | 38 | 4 | 98.87 | 90.23 | 89.30 | 94.08 |
| 11 | 61.17 | 363 | 340 | 27 | 4 | 98.91 | 93.08 | 92.64 | 95.78 |
| 12 | 67.33 | 388 | 386 | 18 | 16 | 96.04 | 95.57 | 95.54 | 95.79 |
| 13 | 63.17 | 371 | 346 | 33 | 8 | 97.89 | 91.83 | 91.29 | 94.59 |
| 14 | 67.00 | 400 | 359 | 43 | 2 | 99.50 | 90.29 | 89.30 | 94.40 |
| 15 | 73.00 | 434 | 400 | 38 | 4 | 99.09 | 91.95 | 91.32 | 95.21 |
| 16 | 63.67 | 377 | 357 | 25 | 5 | 98.69 | 93.78 | 93.46 | 96.07 |
| 17 | 86.67 | 500 | 507 | 13 | 20 | 96.15 | 97.47 | 97.50 | 96.83 |
| 18 | 76.67 | 438 | 449 | 11 | 22 | 95.22 | 97.55 | 97.61 | 96.41 |
| 19 | 80.17 | 464 | 464 | 17 | 17 | 96.47 | 96.47 | 96.47 | 96.47 |
| 20 | 62.33 | 366 | 348 | 26 | 8 | 97.86 | 93.37 | 93.05 | 95.45 |
| AVG | 70.00 | 407.25 | 385.65 | 33.55 | 11.95 | 97.27 | 92.37 | 91.85 | 94.56 |
| STD | 7.75 | 40.82 | 51.19 | 13.94 | 9.32 | 1.93 | 3.03 | 3.50 | 1.79 |
| Sub. No. | Mean HR [bpm] | TP | TN | FP | FN | SEN | PRE | SPE | ACC |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 61.50 | 333 | 268 | 11 | 36 | 90.24 | 96.80 | 96.06 | 92.75 |
| 2 | 77.50 | 400 | 187 | 26 | 65 | 86.02 | 93.90 | 87.79 | 86.58 |
| 3 | 76.67 | 388 | 199 | 43 | 72 | 84.35 | 90.02 | 82.23 | 83.62 |
| 4 | 77.17 | 300 | 243 | 86 | 163 | 64.79 | 77.72 | 73.86 | 68.56 |
| 5 | 73.33 | 411 | 300 | 11 | 29 | 93.41 | 97.39 | 96.46 | 94.67 |
| 6 | 70.50 | 355 | 138 | 36 | 68 | 83.92 | 90.79 | 79.31 | 82.58 |
| 7 | 59.67 | 311 | 111 | 16 | 47 | 86.87 | 95.11 | 87.40 | 87.01 |
| 8 | 72.33 | 343 | 188 | 34 | 91 | 79.03 | 90.98 | 84.68 | 80.95 |
| 9 | 68.33 | 352 | 311 | 24 | 58 | 85.85 | 93.62 | 92.84 | 88.99 |
| 10 | 59.17 | 264 | 297 | 24 | 91 | 74.37 | 91.67 | 92.52 | 82.99 |
| 11 | 61.17 | 311 | 50 | 16 | 56 | 84.74 | 95.11 | 75.76 | 83.37 |
| 12 | 67.33 | 274 | 243 | 55 | 130 | 67.82 | 83.28 | 81.54 | 73.65 |
| 13 | 63.17 | 255 | 211 | 60 | 124 | 67.28 | 80.95 | 77.86 | 71.69 |
| 14 | 67.00 | 294 | 298 | 56 | 108 | 73.13 | 84.00 | 84.18 | 78.31 |
| 15 | 73.00 | 294 | 136 | 88 | 144 | 67.12 | 76.96 | 60.71 | 64.95 |
| 16 | 63.67 | 264 | 241 | 78 | 118 | 69.11 | 77.19 | 75.55 | 72.04 |
| 17 | 86.67 | 434 | 255 | 54 | 86 | 83.46 | 88.93 | 82.52 | 83.11 |
| 18 | 76.67 | 399 | 168 | 23 | 61 | 86.74 | 94.55 | 87.96 | 87.10 |
| 19 | 80.17 | 344 | 213 | 54 | 137 | 71.52 | 86.43 | 79.78 | 74.47 |
| 20 | 62.33 | 254 | 197 | 34 | 120 | 67.91 | 88.19 | 85.28 | 74.55 |
| AVG | 70.00 | 329.00 | 212.70 | 41.45 | 90.20 | 78.39 | 88.68 | 83.22 | 80.60 |
| STD | 7.75 | 55.88 | 69.26 | 24.09 | 38.51 | 9.14 | 6.64 | 8.45 | 8.07 |
| Sub. No. | Mean HR [bpm] | TP | TN | FP | FN | SEN | PRE | SPE | ACC |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 61.50 | 359 | 349 | 20 | 10 | 97.29 | 94.72 | 94.58 | 95.93 |
| 2 | 77.50 | 457 | 464 | 1 | 8 | 98.28 | 99.78 | 99.78 | 99.03 |
| 3 | 76.67 | 439 | 427 | 33 | 21 | 95.43 | 93.01 | 92.83 | 94.13 |
| 4 | 77.17 | 433 | 425 | 38 | 30 | 93.52 | 91.93 | 91.79 | 92.66 |
| 5 | 73.33 | 413 | 422 | 18 | 27 | 93.86 | 95.82 | 95.91 | 94.89 |
| 6 | 70.50 | 423 | 386 | 37 | 0 | 100.00 | 91.96 | 91.25 | 95.63 |
| 7 | 59.67 | 323 | 336 | 22 | 35 | 90.22 | 93.62 | 93.85 | 92.04 |
| 8 | 72.33 | 433 | 403 | 31 | 1 | 99.77 | 93.32 | 92.86 | 96.31 |
| 9 | 68.33 | 384 | 370 | 40 | 26 | 93.66 | 90.57 | 90.24 | 91.95 |
| 10 | 59.17 | 336 | 319 | 36 | 19 | 94.65 | 90.32 | 89.86 | 92.25 |
| 11 | 61.17 | 361 | 347 | 20 | 6 | 98.37 | 94.75 | 94.55 | 96.46 |
| 12 | 67.33 | 401 | 388 | 16 | 3 | 99.26 | 96.16 | 96.04 | 97.65 |
| 13 | 63.17 | 362 | 342 | 37 | 17 | 95.51 | 90.73 | 90.24 | 92.88 |
| 14 | 67.00 | 401 | 360 | 42 | 1 | 99.75 | 90.52 | 89.55 | 94.65 |
| 15 | 73.00 | 432 | 419 | 19 | 6 | 98.63 | 95.79 | 95.66 | 97.15 |
| 16 | 63.67 | 376 | 360 | 22 | 6 | 98.43 | 94.47 | 94.24 | 96.34 |
| 17 | 86.67 | 506 | 511 | 9 | 14 | 97.31 | 98.25 | 98.27 | 97.79 |
| 18 | 76.67 | 434 | 446 | 14 | 26 | 94.35 | 96.88 | 96.96 | 95.65 |
| 19 | 80.17 | 478 | 463 | 18 | 3 | 99.38 | 96.37 | 96.26 | 97.82 |
| 20 | 62.33 | 374 | 349 | 25 | 0 | 100.00 | 93.73 | 93.32 | 96.66 |
| AVG | 70.00 | 406.25 | 394.30 | 24.90 | 12.95 | 96.88 | 94.14 | 93.90 | 95.39 |
| STD | 7.75 | 47.32 | 51.57 | 11.30 | 11.31 | 2.79 | 2.67 | 2.88 | 2.14 |
| Sub. No. | Mean HR [bpm] | TP | TN | FP | FN | SEN | PRE | SPE | ACC |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 61.50 | 365 | 268 | 22 | 4 | 98.92 | 94.32 | 92.41 | 96.05 |
| 2 | 77.50 | 445 | 187 | 27 | 20 | 95.70 | 94.28 | 87.38 | 93.08 |
| 3 | 76.67 | 446 | 199 | 43 | 14 | 96.96 | 91.21 | 82.23 | 91.88 |
| 4 | 77.17 | 457 | 243 | 37 | 6 | 98.70 | 92.51 | 86.79 | 94.21 |
| 5 | 73.33 | 411 | 300 | 22 | 29 | 93.41 | 94.92 | 93.17 | 93.31 |
| 6 | 70.50 | 365 | 138 | 15 | 58 | 86.29 | 96.05 | 90.20 | 87.33 |
| 7 | 59.67 | 345 | 111 | 21 | 13 | 96.37 | 94.26 | 84.09 | 93.06 |
| 8 | 72.33 | 346 | 188 | 30 | 88 | 79.72 | 92.02 | 86.24 | 81.90 |
| 9 | 68.33 | 369 | 311 | 24 | 41 | 90.00 | 93.89 | 92.84 | 91.28 |
| 10 | 59.17 | 305 | 297 | 18 | 50 | 85.92 | 94.43 | 94.29 | 89.85 |
| 11 | 61.17 | 366 | 50 | 12 | 1 | 99.73 | 96.83 | 80.65 | 96.97 |
| 12 | 67.33 | 374 | 243 | 18 | 30 | 92.57 | 95.41 | 93.10 | 92.78 |
| 13 | 63.17 | 355 | 211 | 13 | 24 | 93.67 | 96.47 | 94.20 | 93.86 |
| 14 | 67.00 | 365 | 298 | 23 | 37 | 90.80 | 94.07 | 92.83 | 91.70 |
| 15 | 73.00 | 394 | 136 | 18 | 44 | 89.95 | 95.63 | 88.31 | 89.53 |
| 16 | 63.67 | 351 | 241 | 9 | 31 | 91.88 | 97.50 | 96.40 | 93.67 |
| 17 | 86.67 | 519 | 255 | 12 | 1 | 99.81 | 97.74 | 95.51 | 98.35 |
| 18 | 76.67 | 423 | 168 | 13 | 37 | 91.96 | 97.02 | 92.82 | 92.20 |
| 19 | 80.17 | 432 | 213 | 9 | 49 | 89.81 | 97.96 | 95.95 | 91.75 |
| 20 | 62.33 | 372 | 197 | 16 | 2 | 99.47 | 95.88 | 92.49 | 96.93 |
| AVG | 70.00 | 390.25 | 212.70 | 20.10 | 28.95 | 93.08 | 95.12 | 90.59 | 92.48 |
| STD | 7.75 | 50.26 | 69.26 | 8.95 | 22.60 | 5.35 | 1.89 | 4.64 | 3.64 |
| Publication | Database | Approach | Highest Performance | Key Contributions |
|---|---|---|---|---|
| Giordano et al. [18] | Own database (ECG and PCG) | Measuring the timing of heart sound components | S1: 99.6 (a) S2: 98.9 (a) (a) Sensitivity | Robust Performance |
| Babu et al. [24] | Own database (PPG and PCG) | Variational mode decomposition-based heart sound endpoint determination | S1: 100 (b) S2: 100 (b) (b) Accuracy | Robust Performance |
| Huang et al. [25] | Own database (Gyroscope and Acoustic signal) | Deep-breath detection and various feature extraction methods (duration ratio, amplitude ration, correlation coefficient) | S1: N/R S2: N/R | Liveness detection |
| This study | Own database (PPG and PCG) | Dual deterministic model with window, envelope filtering | S1: 95.39 (c) S2: 92.55 (c) (c) Accuracy | Mobile Environments |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, M.; Wei, Q.; Lee, S.; Park, H. DDM-HSA: Dual Deterministic Model-Based Heart Sound Analysis for Daily Life Monitoring. Sensors 2023, 23, 2423. https://doi.org/10.3390/s23052423
Lee M, Wei Q, Lee S, Park H. DDM-HSA: Dual Deterministic Model-Based Heart Sound Analysis for Daily Life Monitoring. Sensors. 2023; 23(5):2423. https://doi.org/10.3390/s23052423
Chicago/Turabian StyleLee, Miran, Qun Wei, Soomin Lee, and Heejoon Park. 2023. "DDM-HSA: Dual Deterministic Model-Based Heart Sound Analysis for Daily Life Monitoring" Sensors 23, no. 5: 2423. https://doi.org/10.3390/s23052423
APA StyleLee, M., Wei, Q., Lee, S., & Park, H. (2023). DDM-HSA: Dual Deterministic Model-Based Heart Sound Analysis for Daily Life Monitoring. Sensors, 23(5), 2423. https://doi.org/10.3390/s23052423









