Data Augmentation Effects on Highly Imbalanced EEG Datasets for Automatic Detection of Photoparoxysmal Responses
Abstract
1. Introduction
- Type-1: spikes in the occipital region.
- Type-2: spikes followed by a biphasic slow wave in the occipital and parietal regions.
- Type-3: spikes followed by a biphasic slow wave in the occipital and parietal regions, spreading to the frontal regions.
- Type-4: generalized poly-spikes and waves.
Virtual Reality and Artificial Intelligence for PPR Detection
2. Materials and Methods
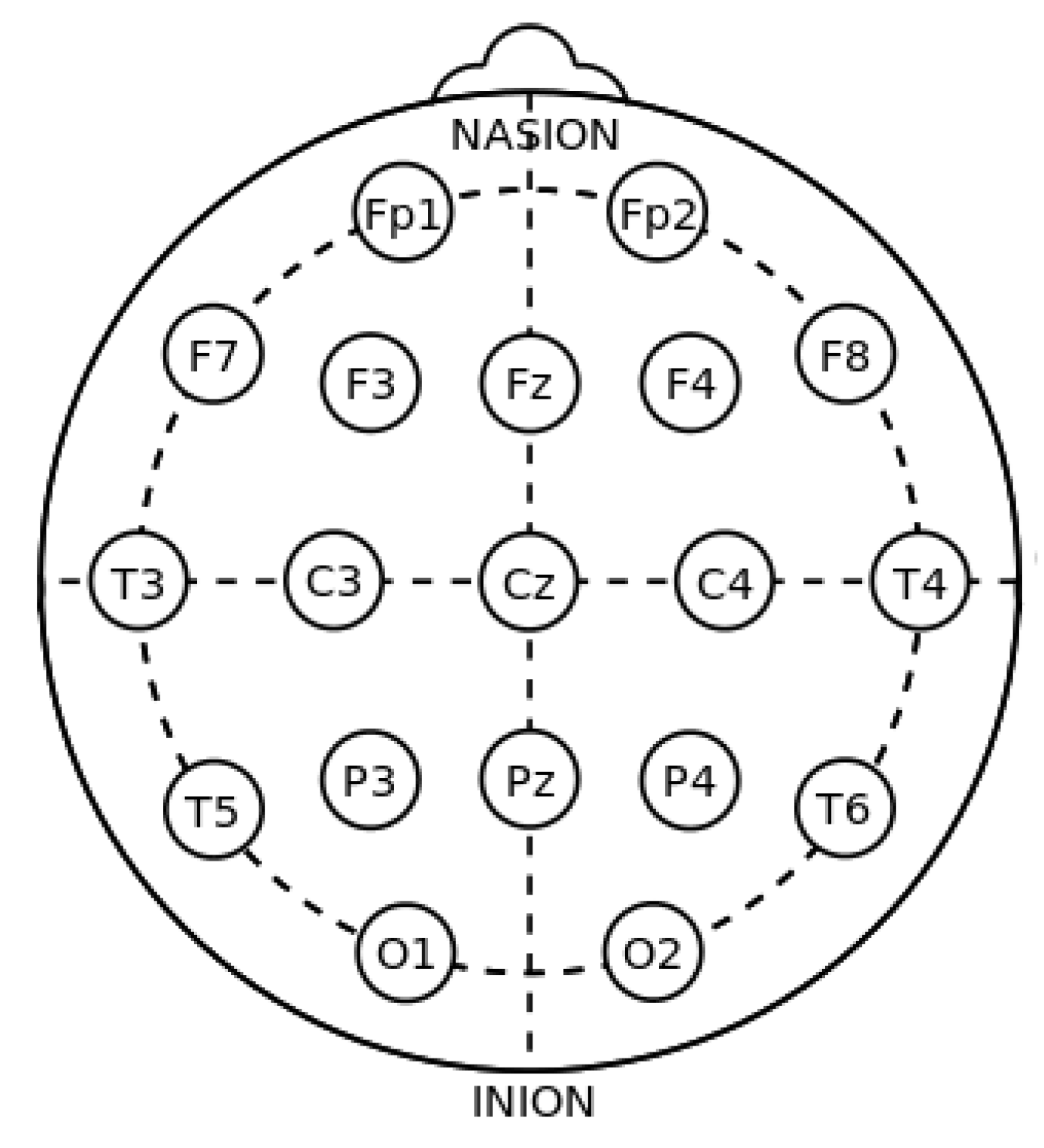
2.1. Dataset
- Total number of EEG windows: 29,190:
- –
- Number of non-PPR windows: 27,968 (95.81%).
- –
- Number of PPR windows: 1222 (4.19%).
2.2. The Data Augmentation Strategy
2.3. Preprocessing and Dimensional Reduction
2.4. Cross-Validation Scheme
- There were 3000 non-PPR and 500 PPR windows randomly selected.
- There were 2500 synthetic PPR windows created from the 500 selected, up to a total of 3000 PPR instances.
- The training set was formed by these 6000 instances with a perfect balance of 50%.
- Two test sets were formed:
- The first one (Test 1) was made from the rest of the non-PPR and only the real PPR windows.
- The test synthetic PPR windows were created up until another 3000 PPR instances again.
- The second test set (Test 2) was created from the rest of the non-PPR and the synthetic PPR windows.
2.5. Modeling and Evaluation
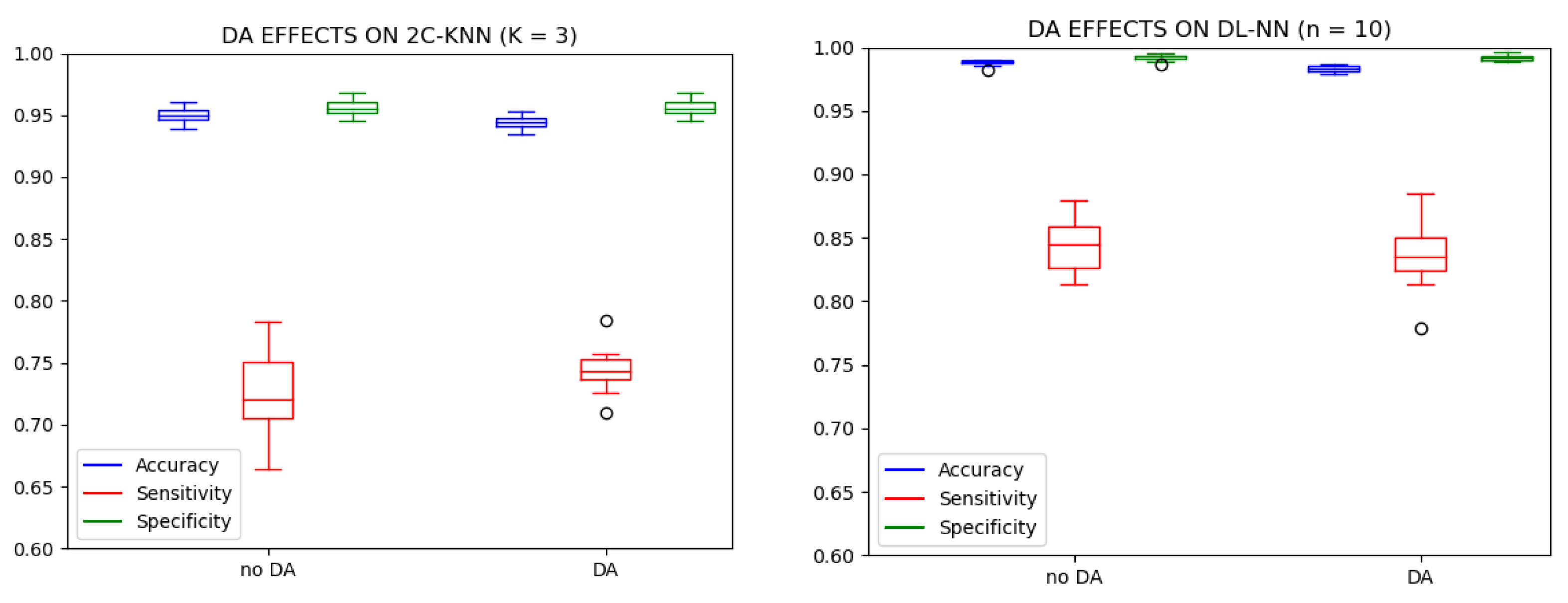
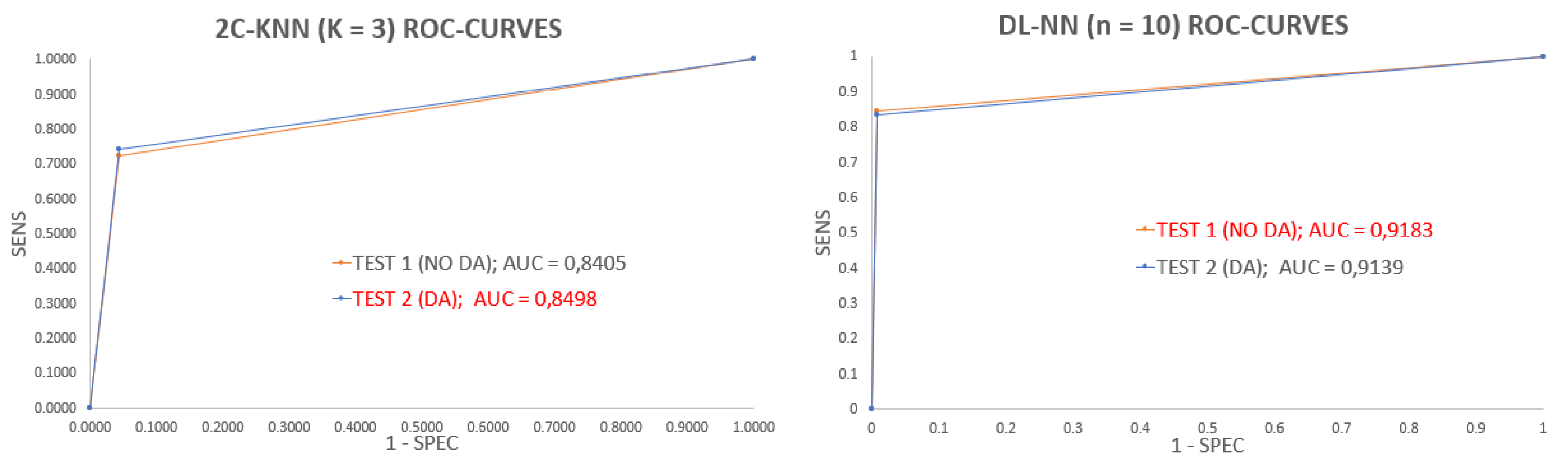
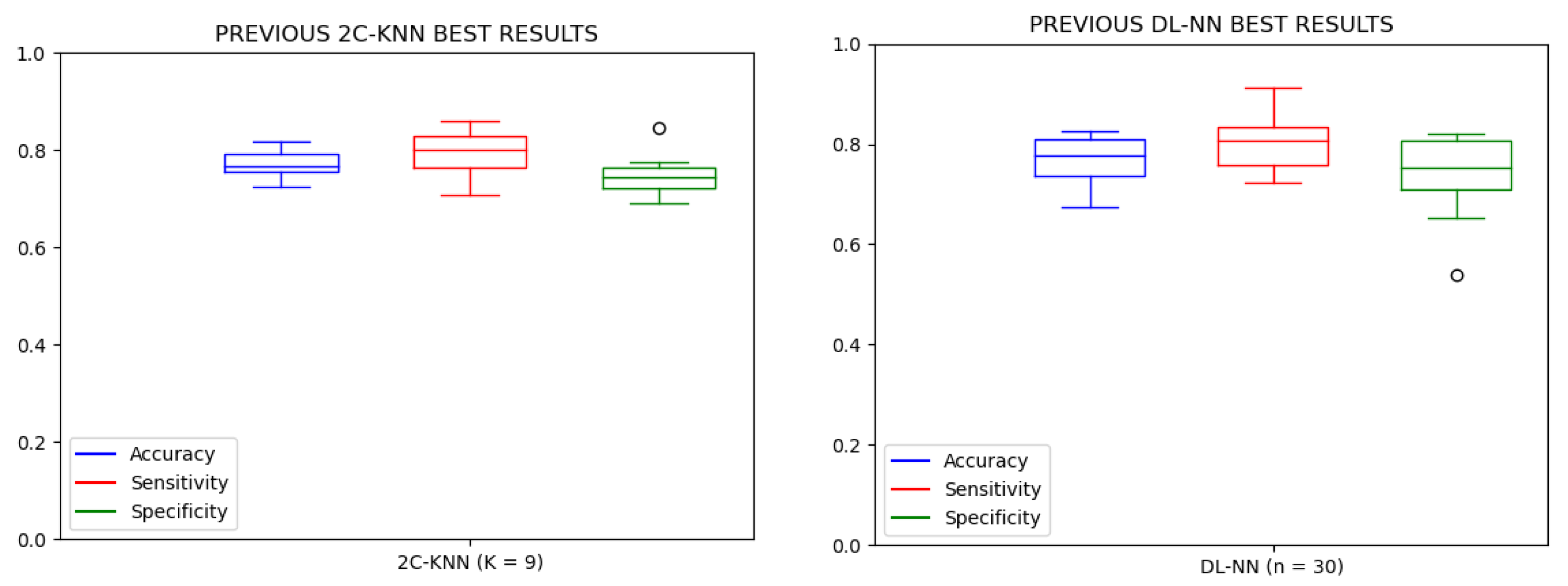
3. Results and Discussion
- To determine if all the 2C-KNN models obtained using Test 1 belong to the same population or not.
- To determine if all the 2C-KNN models trained using Test 2 belong to the same population or not.
- To determine if all the DL-NN models obtained using Test 1 belong to the same population or not.
- To determine if all the DL-NN models trained using Test 2 belong to the same population or not.
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| EEG | Electroencephalogram |
| IPS | Intermittent Photic Stimulation |
| PPR | Photoparoxysmal Response |
| ML | Machine Learning |
| DL | Deep Learning |
| DA | Data Augmentation |
| VR | Virtual Reality |
| AI | Artificial Intelligence |
| PCA | Principal Component Analysis |
| 2C-KNN | Two-Class K-Nearest-Neighbors |
| DL-NN | Dense-Layer Neural Network |
| ACC | Accuracy |
| SENS | Sensitivity |
| SPEC | Specificity |
| ROC | Receiver Operating Characteristic |
| AUC | Area Under the Curve |
References
- Waltz, S.; Christen, H.J.; Doose, H. The different patterns of the photoparoxysmal response—A genetic study. Electroencephalogr. Clin. Neurophysiol. 1992, 83, 138–145. [Google Scholar] [CrossRef]
- Trenité, D.K.N. Photosensitivity in epilepsy. Electrophisiological and clinical correlates. Acta Neurol. Scand. 1989, 125, 3–149. [Google Scholar]
- Hughes, J. The photoparoxysmal response: The probable cause of attacks during video games. Clin. EEG Neurosci. 2008, 39, 1–7. [Google Scholar] [CrossRef]
- Staff, R. London 2012 Logo Footage Withdrawn Amid Epilepsy Fears. 2007. Available online: https://www.reuters.com/article/us-britain-olympics-logo-idUSHO63873520070606 (accessed on 9 December 2022).
- Fisher, R.S.; Acharya, J.N.; Baumer, F.M.; French, J.A.; Parisi, P.; Solodar, J.H.; Szaflarski, J.P.; Thio, L.L.; Tolchin, B.; Wilkins, A.J.; et al. Visually sensitive seizures: An updated review by the Epilepsy Foundation. Epilepsia 2022, 63, 739–768. [Google Scholar] [CrossRef]
- Trenité, D.G.N.; Binnie, C.D.; Harding, G.F.; Wilkins, A. Photic stimulation: Standardization of screening methods. Epilepsia 1999, 40, 75–79. [Google Scholar] [CrossRef]
- Trenite, D.K.N. Photosensitivity and Epilepsy. In Clinical Electroencephalography; Springer: Cham, Switzerland, 2019; pp. 487–495. [Google Scholar] [CrossRef]
- Rubboli, G.; Parra, J.; Seri, S.; Takahashi, T.; Thomas, P. EEG Diagnostic Procedures and Special Investigations in the Assessment of Photosensitivity. Epilepsia 2004, 45, 35–39. [Google Scholar] [CrossRef]
- Shepherd, A.J.; Siniatchkin, M. Visual Pattern Adaptation in Subjects with Photoparoxysmal EEG Response: Evidence for Increased Visual Cortical Excitability. Investig. Ophthalmol. Vis. Sci. 2009, 50, 1470–1476. [Google Scholar] [CrossRef]
- Strigaro, G.; Gori, B.; Varrasi, C.; Fleetwood, T.; Cantello, G.; Cantello, R. Flash-evoked high-frequency EEG oscillations in photosensitive epilepsies. Epilepsy Res. 2021, 172, 106597. [Google Scholar] [CrossRef]
- Dong, H.; Supratak, A.; Pan, W.; Wu, C.; Matthews, P.M.; Guo, Y. Mixed Neural Network Approach for Temporal Sleep Stage Classification. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 324–333. [Google Scholar] [CrossRef]
- Rathore, C.; Prakash, S.; Makwana, P. Prevalence of photoparoxysmal response in patients with epilepsy: Effect of the underlying syndrome and treatment status. Seizure 2020, 82, 39–43. [Google Scholar] [CrossRef]
- Meritam Larsen, P.; Wüstenhagen, S.; Terney, D.; Gardella, E.; Alving, J.; Aurlien, H.; Beniczky, S. Photoparoxysmal response and its characteristics in a large EEG database using the SCORE system. Clin. Neurophysiol. 2021, 132, 365–371. [Google Scholar] [CrossRef]
- Kalitzin, S.; Parra, J.; Velis, D.; Lopes da Silva, F. Enhancement of phase clustering in the EEG/MEG gamma frequency band anticipates transitions to paroxysmal epileptiform activity in epileptic patients with know visual sensitivity. IEEE Trans. Bio-Med. Eng. 2002, 49, 1279–1286. [Google Scholar] [CrossRef]
- Moncada, F.; Martín, S.; González, V.M.; Álvarez, V.M.; García-López, B.; Gómez-Menéndez, A.I.; Villar, J.R. Virtual reality and Machine Learning in the automatic photoparoxysmal response detection. Neural Comput. Appl. 2022, 1–17. [Google Scholar] [CrossRef]
- Martins, F.M.; González, V.M.; García, B.; Álvarez, V.; Villar, J.R. A Comparison of Machine Learning Techniques for the Detection o Type-4 PhotoParoxysmal Responses in Electroencephalographic Signals. In Proceedings of the Hybrid Artificial Intelligent Systems; Springer International Publishing: Cham, Switzerland, 2022; pp. 3–13. [Google Scholar] [CrossRef]
- Vanabelle, P.; Handschutter, P.D.; Tahry, R.E.; Benjelloun, M.; Boukhebouze, M. Epileptic seizure detection using EEG signals and extreme gradient boosting. J. Biomed. Res. 2020, 34, 228–239. [Google Scholar] [CrossRef]
- Omidvarnia, A.; Warren, A.E.; Dalic, L.J.; Pedersen, M.; Jackson, G. Automatic detection of generalized paroxysmal fast activity in interictal EEG using time-frequency analysis. Comput. Biol. Med. 2021, 133, 104287. [Google Scholar] [CrossRef]
- Mammone, N.; Duun-Henriksen, J.; Kjaer, T.W.; Morabito, F.C. Differentiating Interictal and Ictal States in Childhood Absence Epilepsy through Permutation Rényi Entropy. Entropy 2015, 17, 4627–4643. [Google Scholar] [CrossRef]
- Choubey, H.; Pandey, A. A combination of statistical parameters for the detection of epilepsy and EEG classification using ANN and KNN classifier. Signal Image Video Process. 2021, 15, 475–483. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Swetapadma, A.; Pattnaik, P.K. A channel independent generalized seizure detection method for pediatric epileptic seizures. Comput. Methods Programs Biomed. 2021, 209, 106335. [Google Scholar] [CrossRef]
- Jahanbekam, A.; Baumann, J.; Nass, R.D.; Bauckhage, C.; Hill, H.; Elger, C.E.; Surges, R. Performance of ECG-based seizure detection algorithms strongly depends on training and test conditions. Epilepsia Open 2021, 6, 597–606. [Google Scholar] [CrossRef]
- Jeppesen, J.; Fuglsang-Frederiksen, A.; Johansen, P.; Christensen, J.; Wüstenhagen, S.; Tankisi, H.; Qerama, E.; Hess, A.; Beniczky, S. Seizure detection based on heart rate variability using a wearable electrocardiography device. Epilepsia 2019, 60, 2105–2113. [Google Scholar] [CrossRef]
- Ufongene, C.; Atrache, R.E.; Loddenkemper, T.; Meisel, C. Electrocardiographic changes associated with epilepsy beyond heart rate and their utilization in future seizure detection and forecasting methods. Clin. Neurophysiol. 2020, 131, 866–879. [Google Scholar] [CrossRef]
- Yang, Y.; Truong, N.; Maher, C.; Kavehei, O.; Truong, N.D.; Eshraghian, J.K.; Nikpour, A. A multimodal AI system for out-of-distribution generalization of seizure detection. IEEE J. Biomed. Health Inform. 2021. [Google Scholar] [CrossRef]
- Beniczky, S.; Conradsen, I.; Henning, O.; Fabricius, M.; Wolf, P. Automated real-time detection of tonic-clonic seizures using a wearable EMG device. Neurology 2018, 90, 428–434. [Google Scholar] [CrossRef]
- Zibrandtsen, I.C.; Kidmose, P.; Kjaer, T.W. Detection of generalized tonic-clonic seizures from ear-EEG based on EMG analysis. Seizure 2018, 59, 54–59. [Google Scholar] [CrossRef]
- Soriano, M.C.; Niso, G.; Clements, J.; Ortín, S.; Carrasco, S.; Gudín, M.; Mirasso, C.R.; Pereda, E. Automated Detection of Epileptic Biomarkers in Resting-State Interictal MEG Data. Front. Neuroinform. 2017, 11, 43. [Google Scholar] [CrossRef]
- Rasheed, K.; Qayyum, A.; Qadir, J.; Sivathamboo, S.; Kwan, P.; Kuhlmann, L.; O’Brien, T.; Razi, A. Machine Learning for Predicting Epileptic Seizures Using EEG Signals: A Review. IEEE Rev. Biomed. Eng. 2021, 14, 139–155. [Google Scholar] [CrossRef]
- Iwana, B.K.; Uchida, S. An empirical survey of data augmentation for time series classification with neural networks. PLoS ONE 2021, 16, e0254841. [Google Scholar] [CrossRef]
- Bandara, K.; Hewamalage, H.; Liu, Y.H.; Kang, Y.; Bergmeir, C. Improving the accuracy of global forecasting models using time series data augmentation. Pattern Recognit. 2021, 120, 108148. [Google Scholar] [CrossRef]
- Demir, S.; Mincev, K.; Kok, K.; Paterakis, N.G. Data augmentation for time series regression: Applying transformations, autoencoders and adversarial networks to electricity price forecasting. Appl. Energy 2021, 304, 117695. [Google Scholar] [CrossRef]
- Miller, E.; MacFarlane, Z.; Martin, S.; Banerjee, N.; Zhu, T. Radar-based monitoring system for medication tampering using data augmentation and multivariate time series classification. Smart Health 2022, 23, 100245. [Google Scholar] [CrossRef]
- García, E.; Villar, M.; Fáñez, M.; Villar, J.R.; de la Cal, E.; Cho, S.B. Towards effective detection of elderly falls with CNN-LSTM neural networks. Neurocomputing 2022, 500, 231–240. [Google Scholar] [CrossRef]
- Jacaruso, L.C. Accuracy improvement for Fully Convolutional Networks via selective augmentation with applications to electrocardiogram data. Informatics Med. Unlocked 2021, 26, 100729. [Google Scholar] [CrossRef]
- Lashgari, E.; Liang, D.; Maoz, U. Data augmentation for deep-learning-based electroencephalography. J. Neurosci. Methods 2020, 346, 108885. [Google Scholar] [CrossRef]
- He, C.; Liu, J.; Zhu, Y.; Du, W. Data Augmentation for Deep Neural Networks Model in EEG Classification Task: A Review. Front. Hum. Neurosci. 2021, 15, 765525. [Google Scholar] [CrossRef]
- Wang, F.; Zhong, S.H.; Peng, J.; Jiang, J.; Liu, Y. Data augmentation for eeg-based emotion recognition with deep convolutional neural networks. In Proceedings of the MMM 2018: International Conference on MultiMedia Modeling, Bangkok, Thailand, 5–7 February 2018; Lecture Notes in Computer Science. Springer: Cham, Switzerland, 2018; Volume 10705. [Google Scholar] [CrossRef]
- Luo, Y.; Zhu, L.Z.; Wan, Z.Y.; Lu, B.L. Data augmentation for enhancing EEG-based emotion recognition with deep generative models. J. Neural Eng. 2020, 17, 5. [Google Scholar] [CrossRef]
- George, O.; Smith, R.; Madiraju, P.; Yahyasoltani, N.; Ahamed, S.I. Data augmentation strategies for EEG-based motor imagery decoding. Heliyon 2022, 8, e10240. [Google Scholar] [CrossRef]
- He, Z.; Du, L.; Wang, P.; Xia, P.; Liu, Z.; Song, Y.; Chen, X.; Fang, Z. Single-channel EEG sleep staging based on data augmentation and cross-subject discrepancy alleviation. Comput. Biol. Med. 2022, 149, 106044. [Google Scholar] [CrossRef]
- Jasper, H. Report of the committee on methods of clinical examination in electroencephalography. Electroencephalogr. Clin. Neurophysiol. 1958, 10, 370–375. [Google Scholar] [CrossRef]
- Barandas, M.; Folgado, D.; Fernandes, L.; Santos, S.; Abreu, M.; Bota, P.; Liu, H.; Schultz, T.; Gamboa, H. TSFEL: Time Series Feature Extraction Library. SoftwareX 2020, 11, 100456. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Principal Component Analysis, Second Edition. Encycl. Stat. Behav. Sci. 2002, 30, 351–352. [Google Scholar] [CrossRef]
- Zwillinger, D.; Kokoska, S. CRC Standard Probability and Statistics Tables and Formulae; CRC Press: Boca Raton, FL, USA, 1999. [Google Scholar] [CrossRef]
- Welch, P.D. The Use of Fast Fourier Transform for the Estimation of Power Spectra: A Method Based on Time Averaging Over Short, Modified Periodograms. IEEE Trans. Audio Electroacoust. 1967, 15, 70–73. [Google Scholar] [CrossRef]
- Peeters, G.; Giordano, B.L.; Susini, P.; Misdariis, N.; McAdams, S. The Timbre Toolbox: Extracting audio descriptors from musical signals. J. Acoust. Soc. Am. 2011, 130, 2902. [Google Scholar] [CrossRef]
- Pan, Y.N.; Chen, J.; Li, X.L. Spectral entropy: A complementary index for rolling element bearing performance degradation assessment. Proc. Inst. Mech. Eng. Part C J. Mech. Eng. Sci. 2009, 223, 1223–1231. [Google Scholar] [CrossRef]
- Henriksson, U. Power Spectrum and Bandwidth; Tech. Rep.; Linköping University, 2003; Available online: https://www.commsys.isy.liu.se/TSDT45/Material/UlfsSpectrum2003.pdf (accessed on 9 December 2022).
- Fernandes, L.; Barandas, M.; Gamboa, H. Learning Human Behaviour Patterns by Trajectory and Activity Recognition. In Proceedings of the 13th International Joint Conference on Biomedical Engineering Systems and Technologies—Biosignals, Valletta, Malta, 24–26 February 2020; INSTICC, SciTePress: Setúbal, Portugal, 2020; pp. 220–227. [Google Scholar] [CrossRef]
- Polat, E.I. Hypothesis Testing with Python: Step by Step Hands-on Tutorial with Practical Examples. 2022. Available online: https://towardsdatascience.com/hypothesis-testing-with-python-step-by-step-hands-on-tutorial-with-practical-examples-e805975ea96e (accessed on 9 December 2022).






| Statistical Domain |
|---|
| Kurtosis [45], Skewness [45], Standard Deviation, Maximum, |
| Minimum, Mean of Absolute Deviation, Root Mean Square. |
| Temporal Domain |
| Sum of Absolute Values, Maximum Amplitude, Sum of Absolute Differences, |
| Total Energy, Absolute Energy, Area Under the Curve, Entropy, Autocorrelation. |
| Spectral Domain |
| Fundamental Frequency, Maximum Frequency, Median Frequency, Maximum Power |
| Spectrum [46], Spectral Centroid [47], Spectral Decrease [47], Spread [47], |
| Spectral Distance, Spectral Kurtosis [47], Spectral Skewness [47], Spectral Entropy [48], |
| Positive Turning Points, Roll-Off [46], Roll-On [46], Variation [47], |
| Power Bandwidth [49], Human Range Energy [50]. |
| K = 3 | K = 5 | K = 7 | K = 9 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACC | SENS | SPEC | ACC | SENS | SPEC | ACC | SENS | SPEC | ACC | SENS | SPEC | |
| 0.9521 | 0.6639 | 0.9603 | 0.9647 | 0.6681 | 0.9731 | 0.9699 | 0.6639 | 0.9786 | 0.9728 | 0.6610 | 0.9817 | |
| 0.9464 | 0.7496 | 0.9520 | 0.9569 | 0.7398 | 0.9631 | 0.9616 | 0.7398 | 0.9679 | 0.9660 | 0.7271 | 0.9728 | |
| 0.9607 | 0.7159 | 0.9677 | 0.9686 | 0.7032 | 0.9761 | 0.9750 | 0.6990 | 0.9828 | 0.9783 | 0.6962 | 0.9863 | |
| 0.9541 | 0.7511 | 0.9599 | 0.9657 | 0.7215 | 0.9727 | 0.9705 | 0.7342 | 0.9772 | 0.9729 | 0.7145 | 0.9803 | |
| 0.9502 | 0.7243 | 0.9567 | 0.9599 | 0.7117 | 0.9669 | 0.9659 | 0.7103 | 0.9732 | 0.9695 | 0.7117 | 0.9769 | |
| 0.9492 | 0.7834 | 0.9539 | 0.9579 | 0.7722 | 0.9632 | 0.9638 | 0.7623 | 0.9695 | 0.9690 | 0.7665 | 0.9748 | |
| 0.9543 | 0.7032 | 0.9614 | 0.9641 | 0.7018 | 0.9716 | 0.9687 | 0.6906 | 0.9766 | 0.9726 | 0.6934 | 0.9805 | |
| 0.9394 | 0.7103 | 0.9459 | 0.9523 | 0.7131 | 0.9591 | 0.9577 | 0.6948 | 0.9652 | 0.9620 | 0.6779 | 0.9701 | |
| 0.9426 | 0.7637 | 0.9477 | 0.9556 | 0.7426 | 0.9617 | 0.9602 | 0.7370 | 0.9666 | 0.9651 | 0.7229 | 0.972 | |
| 0.9463 | 0.6864 | 0.9537 | 0.9553 | 0.6624 | 0.9636 | 0.9612 | 0.6667 | 0.9696 | 0.9641 | 0.6667 | 0.9725 | |
| mean | 0.9495 | 0.7252 | 0.9559 | 0.9601 | 0.7136 | 0.9671 | 0.9654 | 0.7098 | 0.9727 | 0.9692 | 0.7038 | 0.9768 |
| median | 0.9497 | 0.7201 | 0.9553 | 0.9589 | 0.7124 | 0.9653 | 0.9648 | 0.7046 | 0.9714 | 0.9693 | 0.7039 | 0.9758 |
| std | 0.0062 | 0.0368 | 0.0066 | 0.0054 | 0.0333 | 0.0058 | 0.0055 | 0.0328 | 0.0059 | 0.0050 | 0.0317 | 0.0052 |
| K = 11 | K = 13 | K = 15 | ||||||||||
| ACC | SENS | SPEC | ACC | SENS | SPEC | ACC | SENS | SPEC | ||||
| 0.9739 | 0.6498 | 0.9832 | 0.9752 | 0.6456 | 0.9846 | 0.9764 | 0.6414 | 0.9859 | ||||
| 0.9683 | 0.7201 | 0.9753 | 0.9696 | 0.7103 | 0.9770 | 0.9707 | 0.7060 | 0.9783 | ||||
| 0.9794 | 0.6821 | 0.9878 | 0.9802 | 0.6864 | 0.9886 | 0.9810 | 0.6779 | 0.9896 | ||||
| 0.9763 | 0.7173 | 0.9837 | 0.9777 | 0.7131 | 0.9852 | 0.9787 | 0.7089 | 0.9864 | ||||
| 0.9725 | 0.7089 | 0.9800 | 0.9742 | 0.7060 | 0.9819 | 0.9750 | 0.6948 | 0.9829 | ||||
| 0.9730 | 0.7595 | 0.9791 | 0.9752 | 0.7581 | 0.9814 | 0.9773 | 0.7511 | 0.9837 | ||||
| 0.9741 | 0.6821 | 0.9825 | 0.9756 | 0.6751 | 0.9842 | 0.9771 | 0.6821 | 0.9855 | ||||
| 0.9642 | 0.6765 | 0.9724 | 0.9664 | 0.6779 | 0.9746 | 0.9674 | 0.6765 | 0.9757 | ||||
| 0.9671 | 0.7201 | 0.9741 | 0.9681 | 0.7131 | 0.9754 | 0.9707 | 0.7117 | 0.9781 | ||||
| 0.9663 | 0.6624 | 0.9749 | 0.9683 | 0.6540 | 0.9773 | 0.9698 | 0.6498 | 0.9789 | ||||
| mean | 0.9715 | 0.6979 | 0.9793 | 0.9731 | 0.6940 | 0.9810 | 0.9744 | 0.6900 | 0.9825 | |||
| median | 0.9728 | 0.6955 | 0.9796 | 0.9747 | 0.6962 | 0.9816 | 0.9757 | 0.6885 | 0.9833 | |||
| std | 0.0048 | 0.0330 | 0.0050 | 0.0046 | 0.0331 | 0.0047 | 0.0045 | 0.0320 | 0.0045 | |||
| K = 3 | K = 5 | K = 7 | K = 9 | |||||||||
| ACC | SENS | SPEC | ACC | SENS | SPEC | ACC | SENS | SPEC | ACC | SENS | SPEC | |
| 0.9470 | 0.7260 | 0.9603 | 0.9587 | 0.7190 | 0.9731 | 0.9635 | 0.7120 | 0.9786 | 0.9659 | 0.7033 | 0.9817 | |
| 0.9410 | 0.7573 | 0.9520 | 0.9507 | 0.7433 | 0.9631 | 0.9550 | 0.7410 | 0.9679 | 0.9593 | 0.7333 | 0.9728 | |
| 0.9531 | 0.7100 | 0.9677 | 0.9604 | 0.6983 | 0.9761 | 0.9662 | 0.6897 | 0.9828 | 0.9689 | 0.6790 | 0.9863 | |
| 0.9476 | 0.7437 | 0.9599 | 0.9588 | 0.7270 | 0.9727 | 0.9627 | 0.7207 | 0.9772 | 0.9649 | 0.7097 | 0.9803 | |
| 0.9453 | 0.7553 | 0.9567 | 0.9542 | 0.7433 | 0.9669 | 0.9600 | 0.7400 | 0.9732 | 0.9632 | 0.7360 | 0.9769 | |
| 0.9443 | 0.7840 | 0.9539 | 0.9527 | 0.7770 | 0.9632 | 0.9578 | 0.7627 | 0.9695 | 0.9627 | 0.7620 | 0.9748 | |
| 0.9490 | 0.7417 | 0.9614 | 0.9579 | 0.7303 | 0.9716 | 0.9620 | 0.7183 | 0.9766 | 0.9656 | 0.7170 | 0.9805 | |
| 0.9345 | 0.7450 | 0.9459 | 0.9464 | 0.7353 | 0.9591 | 0.9515 | 0.7227 | 0.9652 | 0.9557 | 0.7170 | 0.9701 | |
| 0.9357 | 0.7350 | 0.9477 | 0.9486 | 0.7300 | 0.9617 | 0.9528 | 0.7230 | 0.9666 | 0.9576 | 0.7180 | 0.9720 | |
| 0.9415 | 0.7393 | 0.9537 | 0.9506 | 0.7343 | 0.9636 | 0.9554 | 0.7197 | 0.9696 | 0.9581 | 0.7180 | 0.9725 | |
| mean | 0.9439 | 0.7437 | 0.9559 | 0.9539 | 0.7338 | 0.9671 | 0.9587 | 0.7250 | 0.9727 | 0.9622 | 0.7193 | 0.9768 |
| median | 0.9448 | 0.7427 | 0.9553 | 0.9535 | 0.7323 | 0.9653 | 0.9589 | 0.7217 | 0.9714 | 0.9630 | 0.7175 | 0.9758 |
| std | 0.0058 | 0.0197 | 0.0066 | 0.0049 | 0.0200 | 0.0058 | 0.0049 | 0.0195 | 0.0059 | 0.0043 | 0.0218 | 0.0052 |
| K = 11 | K = 13 | K = 15 | ||||||||||
| ACC | SENS | SPEC | ACC | SENS | SPEC | ACC | SENS | SPEC | ||||
| 0.9672 | 0.7010 | 0.9832 | 0.9682 | 0.6947 | 0.9846 | 0.9693 | 0.6923 | 0.9859 | ||||
| 0.9615 | 0.7310 | 0.9753 | 0.9626 | 0.7227 | 0.9770 | 0.9637 | 0.7217 | 0.9783 | ||||
| 0.9698 | 0.6703 | 0.9878 | 0.9706 | 0.6707 | 0.9886 | 0.9713 | 0.6677 | 0.9896 | ||||
| 0.9678 | 0.7037 | 0.9837 | 0.9691 | 0.7013 | 0.9852 | 0.9703 | 0.7023 | 0.9864 | ||||
| 0.9657 | 0.7280 | 0.9800 | 0.9673 | 0.7250 | 0.9819 | 0.9680 | 0.7193 | 0.9829 | ||||
| 0.9667 | 0.7607 | 0.9791 | 0.9688 | 0.7580 | 0.9814 | 0.9708 | 0.7550 | 0.9837 | ||||
| 0.9671 | 0.7117 | 0.9825 | 0.9688 | 0.7120 | 0.9842 | 0.9699 | 0.7100 | 0.9855 | ||||
| 0.9580 | 0.7177 | 0.9724 | 0.9600 | 0.7160 | 0.9746 | 0.9608 | 0.7137 | 0.9757 | ||||
| 0.9596 | 0.7183 | 0.9741 | 0.9605 | 0.7130 | 0.9754 | 0.9630 | 0.7113 | 0.9781 | ||||
| 0.9602 | 0.7147 | 0.9749 | 0.9619 | 0.7060 | 0.9773 | 0.9632 | 0.7017 | 0.9789 | ||||
| mean | 0.9644 | 0.7157 | 0.9793 | 0.9658 | 0.7119 | 0.9810 | 0.9670 | 0.7095 | 0.9825 | |||
| median | 0.9662 | 0.7162 | 0.9796 | 0.9677 | 0.7125 | 0.9816 | 0.9687 | 0.7107 | 0.9833 | |||
| std | 0.0041 | 0.0232 | 0.0050 | 0.0040 | 0.0225 | 0.0047 | 0.0039 | 0.0223 | 0.0045 | |||
| n = 10 | n = 20 | n = 30 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ACC | SENS | SPEC | ACC | SENS | SPEC | ACC | SENS | SPEC | |
| 0.9871 | 0.8467 | 0.9911 | 0.9859 | 0.8326 | 0.9903 | 0.9893 | 0.8312 | 0.9938 | |
| 0.9903 | 0.8284 | 0.9950 | 0.9861 | 0.8172 | 0.9909 | 0.9836 | 0.7947 | 0.9890 | |
| 0.9879 | 0.8594 | 0.9916 | 0.9844 | 0.8594 | 0.9879 | 0.9892 | 0.8312 | 0.9937 | |
| 0.9820 | 0.8256 | 0.9865 | 0.9885 | 0.8242 | 0.9932 | 0.9879 | 0.8340 | 0.9923 | |
| 0.9893 | 0.8129 | 0.9944 | 0.9879 | 0.8256 | 0.9926 | 0.9887 | 0.8200 | 0.9936 | |
| 0.9851 | 0.8439 | 0.9891 | 0.9827 | 0.8425 | 0.9867 | 0.9851 | 0.8565 | 0.9887 | |
| 0.9899 | 0.8790 | 0.9930 | 0.9883 | 0.8523 | 0.9922 | 0.9889 | 0.8551 | 0.9928 | |
| 0.9877 | 0.8158 | 0.9926 | 0.9864 | 0.8101 | 0.9914 | 0.9890 | 0.8129 | 0.9940 | |
| 0.9892 | 0.8579 | 0.9929 | 0.9877 | 0.8523 | 0.9915 | 0.9908 | 0.8608 | 0.9946 | |
| 0.9902 | 0.8776 | 0.9934 | 0.9906 | 0.8734 | 0.9939 | 0.9901 | 0.8945 | 0.9928 | |
| mean | 0.9879 | 0.8447 | 0.9920 | 0.9868 | 0.8390 | 0.9911 | 0.9883 | 0.8391 | 0.9925 |
| median | 0.9886 | 0.8453 | 0.9928 | 0.9870 | 0.8376 | 0.9914 | 0.9890 | 0.8326 | 0.9932 |
| std | 0.0026 | 0.0239 | 0.0025 | 0.0023 | 0.0203 | 0.0023 | 0.0022 | 0.0284 | 0.0020 |
| n = 40 | n = 50 | ||||||||
| ACC | SENS | SPEC | ACC | SENS | SPEC | ||||
| 0.9879 | 0.8312 | 0.9924 | 0.9898 | 0.8200 | 0.9946 | ||||
| 0.9896 | 0.8031 | 0.9949 | 0.9888 | 0.8214 | 0.9936 | ||||
| 0.9919 | 0.8594 | 0.9957 | 0.9812 | 0.8031 | 0.9862 | ||||
| 0.9893 | 0.8481 | 0.9934 | 0.9905 | 0.8298 | 0.9950 | ||||
| 0.9876 | 0.8270 | 0.9922 | 0.9872 | 0.8101 | 0.9922 | ||||
| 0.9851 | 0.8298 | 0.9895 | 0.9891 | 0.8158 | 0.9941 | ||||
| 0.9874 | 0.8565 | 0.9911 | 0.9900 | 0.8495 | 0.9940 | ||||
| 0.9875 | 0.8214 | 0.9923 | 0.9870 | 0.8186 | 0.9917 | ||||
| 0.9919 | 0.8383 | 0.9962 | 0.9879 | 0.8523 | 0.9917 | ||||
| 0.9906 | 0.8636 | 0.9942 | 0.9839 | 0.8819 | 0.9868 | ||||
| mean | 0.9889 | 0.8378 | 0.9932 | 0.9875 | 0.8302 | 0.9920 | |||
| median | 0.9886 | 0.8347 | 0.9929 | 0.9883 | 0.8207 | 0.9929 | |||
| std | 0.0022 | 0.0191 | 0.0021 | 0.0029 | 0.0240 | 0.0031 | |||
| n = 10 | n = 20 | n = 30 | |||||||
| ACC | SENS | SPEC | ACC | SENS | SPEC | ACC | SENS | SPEC | |
| 0.9812 | 0.8453 | 0.9894 | 0.9835 | 0.8463 | 0.9917 | 0.9839 | 0.8447 | 0.9923 | |
| 0.9836 | 0.7793 | 0.9959 | 0.9808 | 0.7913 | 0.9921 | 0.9797 | 0.7790 | 0.9918 | |
| 0.9814 | 0.8277 | 0.9907 | 0.9791 | 0.8163 | 0.9888 | 0.9816 | 0.8050 | 0.9922 | |
| 0.9787 | 0.8137 | 0.9886 | 0.9837 | 0.8263 | 0.9932 | 0.9802 | 0.8077 | 0.9906 | |
| 0.9846 | 0.8397 | 0.9933 | 0.9840 | 0.8480 | 0.9922 | 0.9845 | 0.8383 | 0.9933 | |
| 0.9808 | 0.8310 | 0.9897 | 0.9751 | 0.8140 | 0.9848 | 0.9783 | 0.8273 | 0.9873 | |
| 0.9852 | 0.8517 | 0.9933 | 0.9831 | 0.8473 | 0.9912 | 0.9830 | 0.8423 | 0.9915 | |
| 0.9827 | 0.8227 | 0.9923 | 0.9815 | 0.8200 | 0.9912 | 0.9851 | 0.8220 | 0.9949 | |
| 0.9854 | 0.8630 | 0.9927 | 0.9854 | 0.8597 | 0.9930 | 0.9879 | 0.8647 | 0.9953 | |
| 0.9866 | 0.8847 | 0.9928 | 0.9880 | 0.8787 | 0.9945 | 0.9844 | 0.8793 | 0.9907 | |
| mean | 0.9830 | 0.8359 | 0.9919 | 0.9824 | 0.8348 | 0.9913 | 0.9829 | 0.8310 | 0.9920 |
| median | 0.9831 | 0.8353 | 0.9925 | 0.9833 | 0.8363 | 0.9919 | 0.9835 | 0.8328 | 0.9920 |
| std | 0.0025 | 0.0287 | 0.0022 | 0.0036 | 0.0257 | 0.0027 | 0.0029 | 0.0295 | 0.0023 |
| ACC | SENS | SPEC | ACC | SENS | SPEC | ||||
| 0.9827 | 0.8330 | 0.9917 | 0.9847 | 0.8437 | 0.9932 | ||||
| 0.9819 | 0.7700 | 0.9947 | 0.9824 | 0.7867 | 0.9942 | ||||
| 0.9855 | 0.8290 | 0.9950 | 0.9782 | 0.8113 | 0.9883 | ||||
| 0.9842 | 0.8353 | 0.9932 | 0.9826 | 0.7947 | 0.9939 | ||||
| 0.9830 | 0.8327 | 0.9920 | 0.9847 | 0.8437 | 0.9932 | ||||
| 0.9786 | 0.8043 | 0.9891 | 0.9836 | 0.8170 | 0.9936 | ||||
| 0.9810 | 0.8337 | 0.9898 | 0.9839 | 0.8247 | 0.9935 | ||||
| 0.9830 | 0.8280 | 0.9924 | 0.9815 | 0.8220 | 0.9911 | ||||
| 0.9877 | 0.8603 | 0.9954 | 0.9857 | 0.8613 | 0.9932 | ||||
| 0.9876 | 0.8717 | 0.9945 | 0.9839 | 0.8817 | 0.9900 | ||||
| mean | 0.9835 | 0.8298 | 0.9928 | 0.9831 | 0.8287 | 0.9924 | |||
| median | 0.9830 | 0.8328 | 0.9928 | 0.9837 | 0.8233 | 0.9932 | |||
| std | 0.0028 | 0.0278 | 0.0022 | 0.0021 | 0.0293 | 0.0020 | |||
| 2C-KNN → K = 3 | DL-NN → n = 10 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Test 1 (No DA) | Test 2 (DA) | Test 1 (No DA) | Test 2 (DA) | |||||||||
| Pi | ACC | SENS | SPEC | ACC | SENS | SPEC | ACC | SENS | SPEC | ACC | SENS | SPEC |
| 0.9521 | 0.6639 | 0.9603 | 0.9470 | 0.7260 | 0.9603 | 0.9871 | 0.8467 | 0.9911 | 0.9812 | 0.8453 | 0.9894 | |
| 0.9464 | 0.7496 | 0.9520 | 0.9410 | 0.7573 | 0.9520 | 0.9903 | 0.8284 | 0.9950 | 0.9836 | 0.7793 | 0.9959 | |
| 0.9607 | 0.7159 | 0.9677 | 0.9531 | 0.7100 | 0.9677 | 0.9879 | 0.8594 | 0.9916 | 0.9814 | 0.8277 | 0.9907 | |
| 0.9541 | 0.7511 | 0.9599 | 0.9476 | 0.7437 | 0.9599 | 0.9820 | 0.8256 | 0.9865 | 0.9787 | 0.8137 | 0.9886 | |
| 0.9502 | 0.7243 | 0.9567 | 0.9453 | 0.7553 | 0.9567 | 0.9893 | 0.8129 | 0.9944 | 0.9846 | 0.8397 | 0.9933 | |
| 0.9492 | 0.7834 | 0.9539 | 0.9443 | 0.7840 | 0.9539 | 0.9851 | 0.8439 | 0.9891 | 0.9808 | 0.8310 | 0.9897 | |
| 0.9543 | 0.7032 | 0.9614 | 0.9490 | 0.7417 | 0.9614 | 0.9899 | 0.8790 | 0.9930 | 0.9852 | 0.8517 | 0.9933 | |
| 0.9394 | 0.7103 | 0.9459 | 0.9345 | 0.7450 | 0.9459 | 0.9877 | 0.8158 | 0.9926 | 0.9827 | 0.8227 | 0.9923 | |
| 0.9426 | 0.7637 | 0.9477 | 0.9357 | 0.7350 | 0.9477 | 0.9892 | 0.8579 | 0.9929 | 0.9854 | 0.8630 | 0.9927 | |
| 0.9463 | 0.6864 | 0.9537 | 0.9415 | 0.7393 | 0.9537 | 0.9902 | 0.8776 | 0.9934 | 0.9866 | 0.8847 | 0.9928 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martins, F.M.; Suárez, V.M.G.; Flecha, J.R.V.; López, B.G. Data Augmentation Effects on Highly Imbalanced EEG Datasets for Automatic Detection of Photoparoxysmal Responses. Sensors 2023, 23, 2312. https://doi.org/10.3390/s23042312
Martins FM, Suárez VMG, Flecha JRV, López BG. Data Augmentation Effects on Highly Imbalanced EEG Datasets for Automatic Detection of Photoparoxysmal Responses. Sensors. 2023; 23(4):2312. https://doi.org/10.3390/s23042312
Chicago/Turabian StyleMartins, Fernando Moncada, Víctor Manuel González Suárez, José Ramón Villar Flecha, and Beatriz García López. 2023. "Data Augmentation Effects on Highly Imbalanced EEG Datasets for Automatic Detection of Photoparoxysmal Responses" Sensors 23, no. 4: 2312. https://doi.org/10.3390/s23042312
APA StyleMartins, F. M., Suárez, V. M. G., Flecha, J. R. V., & López, B. G. (2023). Data Augmentation Effects on Highly Imbalanced EEG Datasets for Automatic Detection of Photoparoxysmal Responses. Sensors, 23(4), 2312. https://doi.org/10.3390/s23042312






