Beef Quality Classification with Reduced E-Nose Data Features According to Beef Cut Types
Abstract
1. Introduction
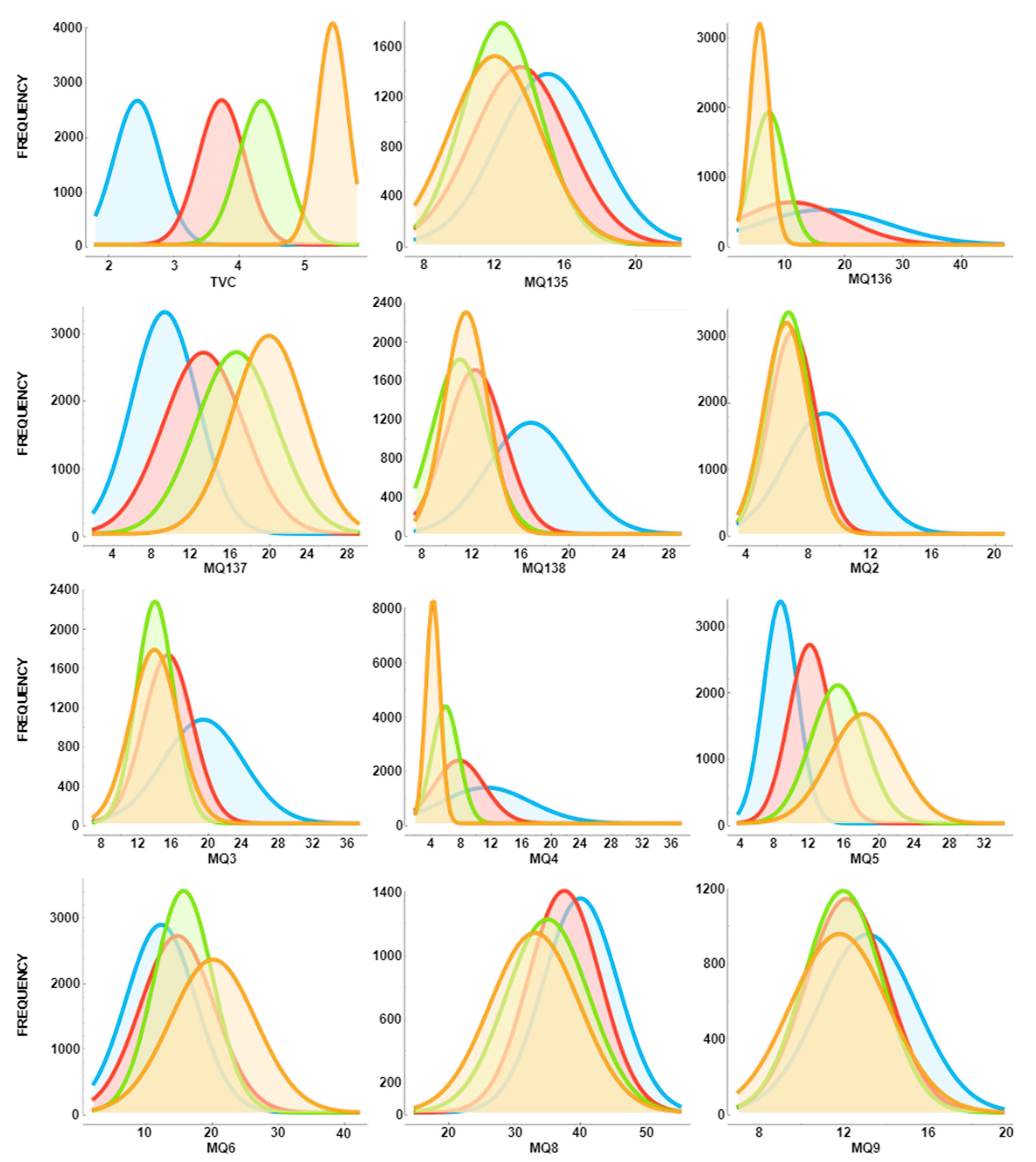
- E-nose data obtained from 12 different beef cuts was used.
- The data in the 12 tables created for each beef cut in the dataset were classified separately.
- The data in each of the 12 tables were reclassified with the active features determined by the ANOVA method.
- In order to eliminate the beef cut shape factor, all tables were combined into a single table.
- The 26,640 rows of data obtained as a result of combining all tables were classified by using the same methods.
- Classifications with 26,640 rows of data and features selected by ANOVA were performed with the same methods again.
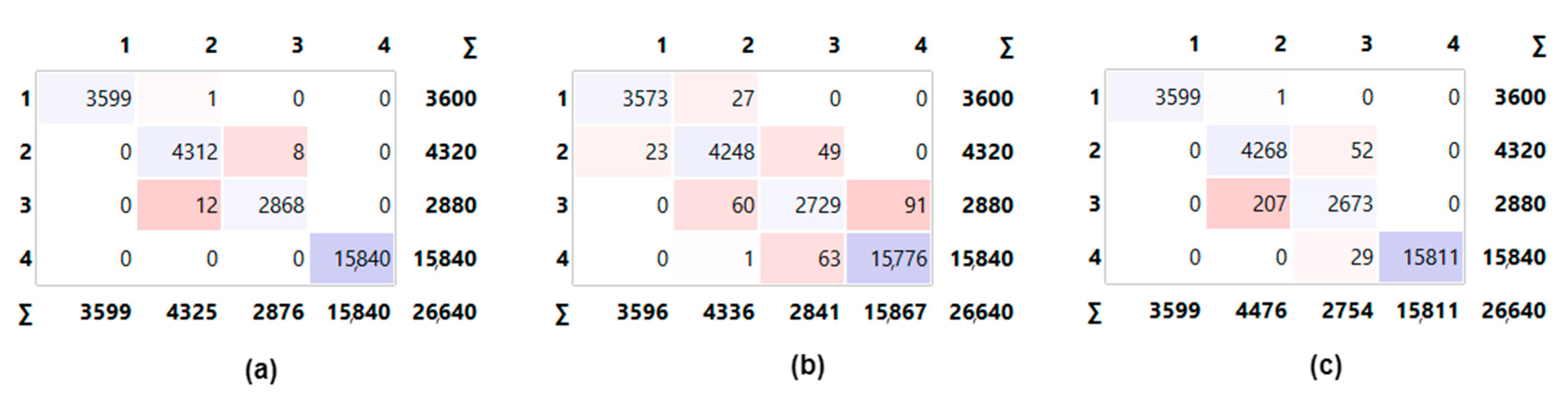
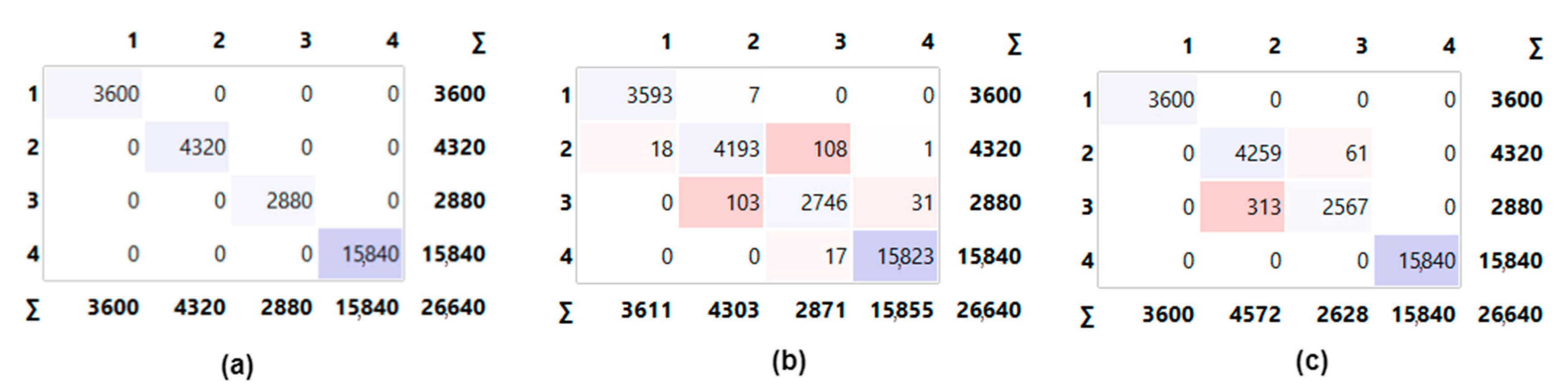
- The obtained results were analyzed with confusion matrices and performance metrics.
- The 12-table data generated for each beef cut was classified separately. In this way, the classification accuracy for each cut could be analyzed.
- Effective features were determined with ANOVA and more accurate and faster results were obtained with fewer data in each table.
- By combining all the tables in the dataset, the beef cut factor was disabled and classifications were carried out.
- With the effective features determined by ANOVA, high accuracy and high-speed prediction accuracy were achieved on all data in the dataset.
2. Material and Methods
2.1. Dataset: Electronic Nose from Various Beef Cuts (ENVBC)
2.2. Machine Learning (ML)
2.2.1. Artificial Neural Network (ANN)
2.2.2. K-Nearest Neighbor (KNN)
2.2.3. Logistic Regression (LR)
2.3. (Analysis of Variance) ANOVA
2.4. Confusion Matrix
2.5. Performance Evaluation
3. Experimental Results
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wijaya, D.R.; Afianti, F. Information-theoretic ensemble feature selection with multi-stage aggregation for sensor array optimization. IEEE Sens. J. 2020, 21, 476–489. [Google Scholar] [CrossRef]
- Huang, C.; Gu, Y. A Machine Learning Method for the Quantitative Detection of Adulterated Meat Using a MOS-Based E-Nose. Foods 2022, 11, 602. [Google Scholar] [CrossRef]
- Wijaya, D.R.; Sarno, R.; Zulaika, E. DWTLSTM for electronic nose signal processing in beef quality monitoring. Sens. Actuators B Chem. 2021, 326, 128931. [Google Scholar] [CrossRef]
- Mu, F.; Gu, Y.; Zhang, J.; Zhang, L. Milk source identification and milk quality estimation using an electronic nose and machine learning techniques. Sensors 2020, 20, 4238. [Google Scholar] [CrossRef] [PubMed]
- Hui, T.; Fang, Z.; Ma, Q.; Hamid, N.; Li, Y. Effect of cold atmospheric plasma-assisted curing process on the color, odor, volatile composition, and heterocyclic amines in beef meat roasted by charcoal and superheated steam. Meat Sci. 2023, 196, 109046. [Google Scholar] [CrossRef]
- Huang, Y.; Doh, I.-J.; Bae, E. Design and validation of a portable machine learning-based electronic nose. Sensors 2021, 21, 3923. [Google Scholar] [CrossRef]
- Hua, Z.; Yu, Y.; Zhao, C.; Zong, J.; Shi, Y.; Men, H. A feature dimensionality reduction strategy coupled with an electronic nose to identify the quality of egg. J. Food Process Eng. 2021, 44, e13873. [Google Scholar] [CrossRef]
- Chen, J.; Gu, J.; Zhang, R.; Mao, Y.; Tian, S. Freshness evaluation of three kinds of meats based on the electronic nose. Sensors 2019, 19, 605. [Google Scholar] [CrossRef]
- Stassen, I.; Bueken, B.; Reinsch, H.; Oudenhoven, J.F.M.; Wputers, D.; Hajek, J.; Van Speybroeck, V.; Stock, N.; Vereecken, P.M.; Van Schaijk, R.; et al. Towards metal–organic framework based field effect chemical sensors: UiO-66-NH2 for nerve agent detection. Chem. Sci. 2016, 7, 5827–5832. [Google Scholar] [CrossRef]
- Eamsa-Ard, T.; Seesaard, T.; Kitiyakara, T.; Kerdcharoen, T. Screening and discrimination of Hepatocellular carcinoma patients by testing exhaled breath with smart devices using composite polymer/carbon nanotube gas sensors. In Proceedings of the 2016 9th Biomedical Engineering International Conference (BMEiCON), Laung Prabang, Laos, 7–9 December 2016; IEEE: Manhattan, NY, USA, 2016. [Google Scholar]
- Adigal, S.S.; Rayaroth, N.V.; John, R.V.; Pai, K.M.; Bhandari, S.; Mohapatra, A.K.; Lukose, J.; Patil, A.; Bankapur, A.; Chidangil, S. A review on human body fluids for the diagnosis of viral infections: Scope for rapid detection of COVID-19. Expert Rev. Mol. Diagn. 2021, 21, 31–42. [Google Scholar] [CrossRef]
- Cheng, J.; Sun, J.; Yao, K.; Xu, M.; Tian, Y.; Dai, C. A decision fusion method based on hyperspectral imaging and electronic nose techniques for moisture content prediction in frozen-thawed pork. LWT 2022, 165, 113778. [Google Scholar] [CrossRef]
- Avian, C.; Leu, J.-S.; Prakosa, S.W.; Faisal, M. An Improved Classification of Pork Adulteration in Beef Based on Electronic Nose Using Modified Deep Extreme Learning with Principal Component Analysis as Feature Learning. Food Anal. Methods 2022, 15, 3020–3031. [Google Scholar] [CrossRef]
- Roy, M.; Yadav, B. Electronic nose for detection of food adulteration: A review. J. Food Sci. Technol. 2022, 59, 846–858. [Google Scholar] [CrossRef]
- Wen, R.; Kong, B.; Yin, X.; Zhang, H.; Chen, Q. Characterisation of flavour profile of beef jerky inoculated with different autochthonous lactic acid bacteria using electronic nose and gas chromatography–ion mobility spectrometry. Meat Sci. 2022, 183, 108658. [Google Scholar] [CrossRef]
- Andre, R.S.; Facure, M.H.; Mercante, L.A.; Correa, D.S. Electronic nose based on hybrid free-standing nanofibrous mats for meat spoilage monitoring. Sens. Actuators B Chem. 2022, 353, 131114. [Google Scholar] [CrossRef]
- Panigrahi, S.; Balasubramanian, S.; Gu, H.; Logue, C.; Marchello, M. Neural-network-integrated electronic nose system for identification of spoiled beef. LWT-Food Sci. Technol. 2006, 39, 135–145. [Google Scholar] [CrossRef]
- Li, C.; Heinemann, P.; Sherry, R. Neural network and Bayesian network fusion models to fuse electronic nose and surface acoustic wave sensor data for apple defect detection. Sens. Actuators B Chem. 2007, 125, 301–310. [Google Scholar] [CrossRef]
- Cevoli, C.; Cerretani, L.; Gori, A.; Caboni, M.F.; Toschi, T.G.; Fabbri, A. Classification of Pecorino cheeses using electronic nose combined with artificial neural network and comparison with GC–MS analysis of volatile compounds. Food Chem. 2011, 129, 1315–1319. [Google Scholar] [CrossRef]
- Papadopoulou, O.S.; Panagou, E.Z.; Mohareb, F.R.; Nychas, G.-J.E. Sensory and microbiological quality assessment of beef fillets using a portable electronic nose in tandem with support vector machine analysis. Food Res. Int. 2013, 50, 241–249. [Google Scholar] [CrossRef]
- Dang, L.; Tian, F.; Zhang, L.; Kadri, C.; Yin, X.; Peng, X.; Liu, S. A novel classifier ensemble for recognition of multiple indoor air contaminants by an electronic nose. Sens. Actuators A Phys. 2014, 207, 67–74. [Google Scholar] [CrossRef]
- Wijaya, D.R.; Sarno, R.; Zulaika, E.; Sabila, S.I. Development of mobile electronic nose for beef quality monitoring. Procedia Comput. Sci. 2017, 124, 728–735. [Google Scholar] [CrossRef]
- Liu, H.; Li, Q.; Yan, B.; Zhang, L.; Gu, Y. Bionic electronic nose based on MOS sensors array and machine learning algorithms used for wine properties detection. Sensors 2018, 19, 45. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi-Varnamkhasti, M.; Mohammad-Razdari, A.; Yoosefian, S.H.; Izadi, Z.; Siadat, M. Aging discrimination of French cheese types based on the optimization of an electronic nose using multivariate computational approaches combined with response surface method (RSM). LWT 2019, 111, 85–98. [Google Scholar] [CrossRef]
- Sarno, R.; Triyana, K.; Sabilla, S.I.; Wijaya, D.R.; Sunaryono, D.; Fatichah, C. Detecting pork adulteration in beef for halal authentication using an optimized electronic nose system. IEEE Access 2020, 8, 221700–221711. [Google Scholar] [CrossRef]
- Wakhid, S.; Sarno, R.; Sabilla, S.I. The effect of gas concentration on detection and classification of beef and pork mixtures using E-nose. Comput. Electron. Agric. 2022, 195, 106838. [Google Scholar] [CrossRef]
- Hibatulah, R.P.; Wijaya, D.R.; Wikusna, W. Prediction of Microbial Population in Meat Using Electronic Nose and Support Vector Regression Algorithm. In Proceedings of the 2022 1st International Conference on Information System & Information Technology (ICISIT), Yogyakarta, Indonesia, 27–28 July 2022; IEEE: Manhattan, NY, USA, 2022. [Google Scholar]
- Wijaya, D.R. Dataset for Electronic Nose from Various Beef Cuts; Harvard Dataverse: Cambridge, MA, USA, 2018. [Google Scholar]
- Wijaya, D.R.; Sarno, R.; Zulaika, E.; Afianti, F. Electronic nose homogeneous data sets for beef quality classification and microbial population prediction. BMC Res. Notes 2022, 15, 237. [Google Scholar] [CrossRef]
- Taspinar, Y.S.; Cinar, I.; Koklu, M. Prediction of Computer Type Using Benchmark Scores of Hardware Units. Selcuk University J. Eng. Sci. 2021, 20, 11–17. [Google Scholar]
- Sabanci, K.; Aslan, M.F.; Slavova, V.; Genova, S. The Use of Fluorescence Spectroscopic Data and Machine-Learning Algorithms to Discriminate Red Onion Cultivar and Breeding Line. Agriculture 2022, 12, 1652. [Google Scholar] [CrossRef]
- Ropelewska, E.; Slavova, V.; Sabanci, K.; Aslan, M.F.; Masheva, V.; Petkova, M. Differentiation of Yeast-Inoculated and Uninoculated Tomatoes Using Fluorescence Spectroscopy Combined with Machine Learning. Agriculture 2022, 12, 1887. [Google Scholar] [CrossRef]
- Kishore, B.; Yasar, A.; Taspinar, Y.S.; Kursun, R.; Cinar, I.; Shankar, V.G.; Koklu, M.; Ofori, I. Computer-aided multiclass classification of corn from corn images integrating deep feature extraction. Comput. Intell. Neurosci. 2022, 2022, 2062944. [Google Scholar] [CrossRef]
- Kaya, E.; Saritas, İ. Towards a real-time sorting system: Identification of vitreous durum wheat kernels using ANN based on their morphological, colour, wavelet and gaborlet features. Comput. Electron. Agric. 2019, 166, 105016. [Google Scholar] [CrossRef]
- Yasar, A.; Kaya, E.; Saritas, I. Classification of wheat types by artificial neural network. Int. J. Intell. Syst. Appl. Eng. 2016, 4, 12–15. [Google Scholar] [CrossRef]
- Al-Doori, S.K.S.; Taspinar, Y.S.; Koklu, M. Distracted Driving Detection with Machine Learning Methods by CNN Based Feature Extraction. Int. J. Appl. Math. Electron. Comput. 2021, 9, 116–121. [Google Scholar] [CrossRef]
- Koklu, M.; Kursun, R.; Taspinar, Y.S.; Cinar, I. Classification of date fruits into genetic varieties using image analysis. Math. Probl. Eng. 2021, 2021, 4793293. [Google Scholar] [CrossRef]
- Cinar, I.; Koklu, M. Determination of Effective and Specific Physical Features of Rice Varieties by Computer Vision In Exterior Quality Inspection. Selcuk J. Agric. Food Sci. 2021, 35, 229–243. [Google Scholar] [CrossRef]
- Sulak, S.A.; Kaplan, S.E. The Examination of Perception of Social Values of Elemantary School Students According to Different Variables. Bartin Univ. J. Fac. Educ. 2017, 6, 840–858. [Google Scholar]
- Yu, Z.; Guindani, M.; Grieco, S.F.; Chen, L.; Holmes, T.C.; Xu, X. Beyond t test and ANOVA: Applications of mixed-effects models for more rigorous statistical analysis in neuroscience research. Neuron 2021, 10, 21–35. [Google Scholar] [CrossRef]
- Taspinar, Y.S.; Koklu, M.; Altin, M. Classification of flame extinction based on acoustic oscillations using artificial intelligence methods. Case Stud. Therm. Eng. 2021, 28, 101561. [Google Scholar] [CrossRef]
- Ropelewska, E.; Slavova, V.; Sabanci, K.; Aslan, M.F.; Cai, X.; Genova, S. Discrimination of onion subjected to drought and normal watering mode based on fluorescence spectroscopic data. Comput. Electron. Agric. 2022, 196, 106916. [Google Scholar] [CrossRef]
- Singh, D.; Taspinar, Y.S.; Kursun, R.; Cinar, I.; Koklu, M.; Ozkan, I.A.; Lee, H.-N. Classification and Analysis of Pistachio Species with Pre-Trained Deep Learning Models. Electronics 2022, 11, 981. [Google Scholar] [CrossRef]
- Taspinar, Y.S.; Dogan, M.; Cinar, I.; Kursun, R.; Ozkan, I.A.; Koklu, M. Computer vision classification of dry beans (Phaseolus vulgaris L.) based on deep transfer learning techniques. Eur. Food Res. Technol. 2022, 248, 2707–2725. [Google Scholar] [CrossRef]
- Unal, Y.; Taspinar, Y.S.; Cinar, I.; Kursun, R.; Koklu, M. Application of pre-trained deep convolutional neural networks for coffee beans species detection. Food Anal. Methods 2022, 15, 3232–3243. [Google Scholar] [CrossRef]
- Kaya, A.; Keçeli, A.S.; Catal, C.; Tekinerdogan, B. Sensor failure tolerable machine learning-based food quality prediction model. Sensors 2020, 20, 3173. [Google Scholar] [CrossRef] [PubMed]
- Wijaya, D.R.; Afianti, F.; Arifianto, A.; Rahmawati, D.; Kodogiannis, V.S. Ensemble machine learning approach for electronic nose signal processing. Sens. Bio-Sens. Res. 2022, 36, 100495. [Google Scholar] [CrossRef]
- Enériz, D.; Medrano, N.; Calvo, B. An FPGA-based machine learning tool for in-situ food quality tracking using sensor fusion. Biosensors 2021, 11, 366. [Google Scholar] [CrossRef]
- Pulluri, K.K.; Kumar, V.N. Development of an Integrated Soft E-nose for Food Quality Assessment. IEEE Sens. J. 2022, 22, 15111–15122. [Google Scholar] [CrossRef]

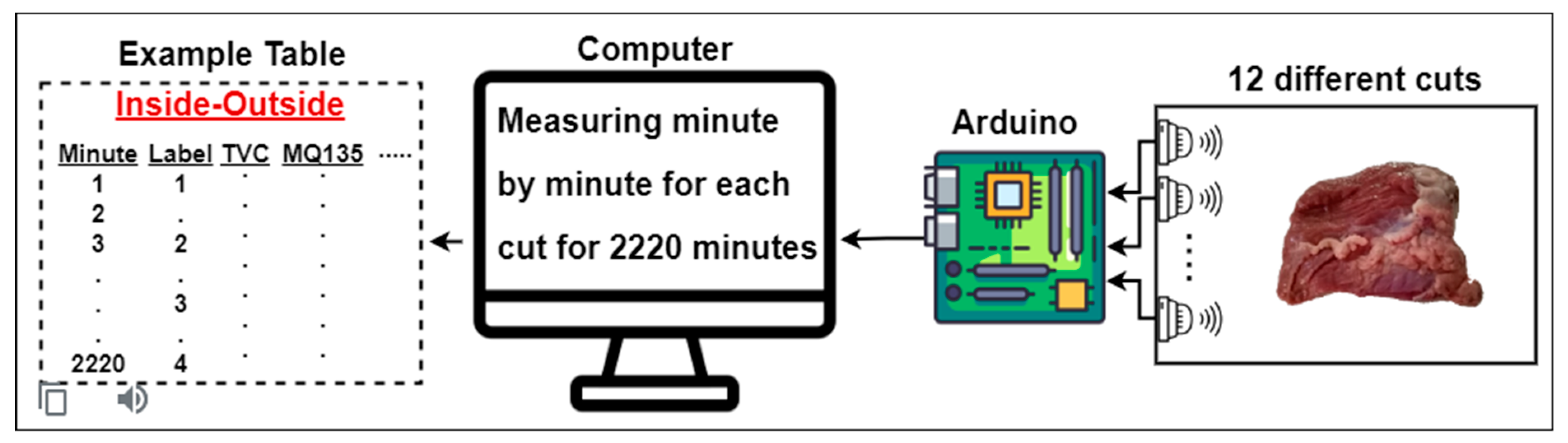
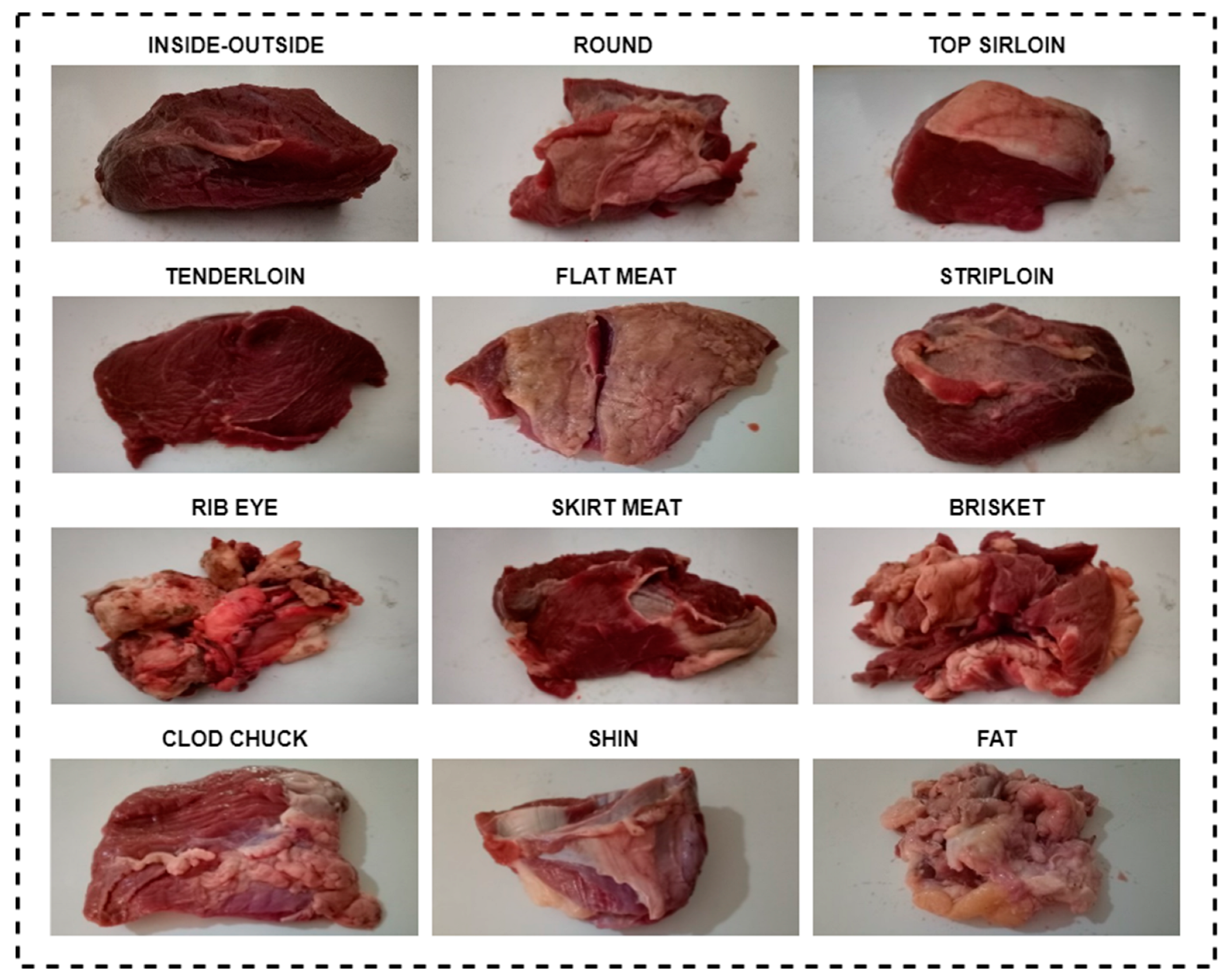



| Gas Sensor | What Gases Does it Detect? |
|---|---|
| MQ2 | Alcohol, LPG, smoke, propane, methane, butane, hydrogen |
| MQ3 | Alcohol, carbon monoxide, methane, LPG, hexane |
| MQ4 | Methane |
| MQ5 | Alcohol, carbon monoxide, hydrogen, LPG, methane |
| MQ6 | LPG, Propane, Iso-butane |
| MQ8 | Hydrogen |
| MQ9 | Methane, propane and carbon monoxide |
| MQ135 | Nox, Alcohol, carbon dioxide, smoke, ammonia, benzene |
| MQ136 | Hydrogen sulfide |
| MQ137 | Ammonia |
| MQ138 | Alcohols, aldehydes, ketones |
| Metrics | Formula |
|---|---|
| Accuracy (AC) | |
| F1 Score (F1) | |
| Precision (PR) | |
| Recall (RE) | |
| Specificity (SP) |
| AC (%) | F1 | PR | RE | SP | Train Time (s) | Test Time (s) | ||
|---|---|---|---|---|---|---|---|---|
| Inside-Outside | ANN | 98.9 | 0.989 | 0.989 | 0.989 | 0.997 | 19.616 | 0.038 |
| KNN | 98.5 | 0.985 | 0.985 | 0.985 | 0.995 | 0.104 | 0.124 | |
| LR | 98.7 | 0.987 | 0.988 | 0.987 | 0.996 | 11.948 | 0.012 | |
| Round | ANN | 99.3 | 0.993 | 0.993 | 0.993 | 0.997 | 19.133 | 0.037 |
| KNN | 98.8 | 0.988 | 0.988 | 0.988 | 0.994 | 0.106 | 0.11 | |
| LR | 98.9 | 0.989 | 0.989 | 0.989 | 0.995 | 6.114 | 0.012 | |
| Top Sirloin | ANN | 99.3 | 0.993 | 0.993 | 0.993 | 0.998 | 19.668 | 0.039 |
| KNN | 98.2 | 0.983 | 0.983 | 0.982 | 0.993 | 0.105 | 0.116 | |
| LR | 98.8 | 0.988 | 0.988 | 0.988 | 0.993 | 19.397 | 0.014 | |
| Tenderloin | ANN | 99.4 | 0.994 | 0.994 | 0.994 | 0.998 | 18.746 | 0.036 |
| KNN | 98.6 | 0.986 | 0.986 | 0.986 | 0.994 | 0.084 | 0.119 | |
| LR | 99.5 | 0.996 | 0.996 | 0.995 | 0.998 | 12.456 | 0.018 | |
| Flap Meat | ANN | 99.5 | 0.995 | 0.995 | 0.995 | 0.998 | 18.143 | 0.036 |
| KNN | 98.2 | 0.982 | 0.982 | 0.982 | 0.989 | 0.089 | 0.108 | |
| LR | 98.6 | 0.986 | 0.986 | 0.986 | 0.991 | 11.547 | 0.013 | |
| Striploin | ANN | 99.3 | 0.993 | 0.993 | 0.993 | 0.999 | 18.604 | 0.039 |
| KNN | 99 | 0.99 | 0.99 | 0.99 | 0.992 | 0.096 | 0.12 | |
| LR | 99.3 | 0.993 | 0.993 | 0.993 | 0.996 | 5.253 | 0.014 | |
| Rib Eye | ANN | 98.7 | 0.987 | 0.987 | 0.987 | 0.997 | 18.53 | 0.038 |
| KNN | 99.4 | 0.994 | 0.994 | 0.994 | 0.999 | 0.092 | 0.102 | |
| LR | 99.2 | 0.992 | 0.992 | 0.992 | 0.999 | 9.098 | 0.015 | |
| Skirt Meat | ANN | 99.7 | 0.997 | 0.997 | 0.997 | 0.999 | 21.204 | 0.071 |
| KNN | 99.6 | 0.996 | 0.996 | 0.996 | 0.999 | 0.106 | 0.114 | |
| LR | 99.5 | 0.995 | 0.995 | 0.995 | 0.997 | 9.772 | 0.016 | |
| Brisket | ANN | 99 | 0.99 | 0.99 | 0.99 | 0.997 | 18.479 | 0.035 |
| KNN | 98.5 | 0.985 | 0.985 | 0.985 | 0.991 | 0.103 | 0.115 | |
| LR | 98.9 | 0.989 | 0.989 | 0.989 | 0.996 | 11.804 | 0.011 | |
| Clod Chuck | ANN | 99.7 | 0.997 | 0.997 | 0.997 | 0.999 | 20.717 | 0.041 |
| KNN | 98.9 | 0.989 | 0.989 | 0.989 | 0.992 | 0.099 | 0.116 | |
| LR | 99.5 | 0.995 | 0.995 | 0.995 | 0.997 | 14.857 | 0.017 | |
| Shin | ANN | 99.3 | 0.993 | 0.993 | 0.993 | 0.998 | 19.588 | 0.035 |
| KNN | 98.7 | 0.987 | 0.987 | 0.987 | 0.994 | 0.114 | 0.126 | |
| LR | 99.2 | 0.992 | 0.992 | 0.992 | 0.999 | 11.946 | 0.016 | |
| Fat | ANN | 99.5 | 0.995 | 0.995 | 0.995 | 0.997 | 19.733 | 0.037 |
| KNN | 98.6 | 0.986 | 0.986 | 0.986 | 0.992 | 0.098 | 0.135 | |
| LR | 99.1 | 0.991 | 0.991 | 0.991 | 0.994 | 22.757 | 0.013 |
| AC (%) | F1 | PR | RE | SP | Train Time (s) | Test Time (s) | ||
|---|---|---|---|---|---|---|---|---|
| Inside-Outside | ANN | 99.2 | 0.992 | 0.992 | 0.992 | 0.999 | 18.745 | 0.02 |
| KNN | 99.5 | 0.995 | 0.995 | 0.995 | 0.999 | 0.058 | 0.083 | |
| LR | 96.6 | 0.965 | 0.966 | 0.966 | 0.96 | 6.83 | 0.01 | |
| Round | ANN | 98.8 | 0.988 | 0.988 | 0.988 | 0.997 | 18.154 | 0.016 |
| KNN | 99.2 | 0.992 | 0.992 | 0.992 | 0.998 | 0.067 | 0.082 | |
| LR | 99 | 0.99 | 0.99 | 0.99 | 0.997 | 4.818 | 0.007 | |
| Top Sirloin | ANN | 98.7 | 0.987 | 0.987 | 0.987 | 0.998 | 23.533 | 0.018 |
| KNN | 99.1 | 0.991 | 0.991 | 0.991 | 0.998 | 0.084 | 0.091 | |
| LR | 96 | 0.959 | 0.959 | 0.96 | 0.966 | 9.906 | 0.008 | |
| Tenderloin | ANN | 99.6 | 0.996 | 0.996 | 0.996 | 0.999 | 17.073 | 0.022 |
| KNN | 99.7 | 0.997 | 0.997 | 0.997 | 0.999 | 0.07 | 0.081 | |
| LR | 99.1 | 0.991 | 0.991 | 0.991 | 0.993 | 5.886 | 0.007 | |
| Flap Meat | ANN | 99.6 | 0.996 | 0.996 | 0.996 | 0.999 | 17.085 | 0.018 |
| KNN | 99.9 | 0.999 | 0.999 | 0.999 | 0.999 | 0.062 | 0.076 | |
| LR | 96.8 | 0.966 | 0.969 | 0.968 | 0.961 | 5.464 | 0.005 | |
| Striploin | ANN | 99.9 | 0.999 | 0.999 | 0.999 | 1 | 18.249 | 0.021 |
| KNN | 99.9 | 0.999 | 0.999 | 0.999 | 1 | 0.064 | 0.088 | |
| LR | 99.7 | 0.997 | 0.997 | 0.997 | 0.998 | 5.683 | 0.008 | |
| Rib Eye | ANN | 99.5 | 0.995 | 0.995 | 0.995 | 0.999 | 17.062 | 0.02 |
| KNN | 99.6 | 0.996 | 0.996 | 0.996 | 0.999 | 0.063 | 0.078 | |
| LR | 98.4 | 0.984 | 0.984 | 0.984 | 0.989 | 5.216 | 0.006 | |
| Skirt Meat | ANN | 99.8 | 0.998 | 0.998 | 0.998 | 1 | 19.406 | 0.019 |
| KNN | 99.5 | 0.995 | 0.996 | 0.995 | 0.999 | 0.073 | 0.091 | |
| LR | 99.4 | 0.994 | 0.994 | 0.994 | 0.998 | 9.594 | 0.01 | |
| Brisket | ANN | 99.2 | 0.992 | 0.992 | 0.992 | 0.999 | 18.411 | 0.02 |
| KNN | 98.7 | 0.987 | 0.987 | 0.987 | 0.993 | 0.081 | 0.09 | |
| LR | 97.6 | 0.976 | 0.976 | 0.976 | 0.986 | 7.232 | 0.009 | |
| Clod Chuck | ANN | 99.9 | 0.999 | 0.999 | 0.999 | 0.999 | 19.008 | 0.021 |
| KNN | 99.7 | 0.997 | 0.997 | 0.997 | 0.997 | 0.076 | 0.091 | |
| LR | 99 | 0.989 | 0.99 | 0.99 | 0.986 | 6.641 | 0.01 | |
| Shin | ANN | 99.6 | 0.996 | 0.996 | 0.996 | 0.999 | 18.005 | 0.02 |
| KNN | 99.9 | 0.999 | 0.999 | 0.999 | 1 | 0.081 | 0.088 | |
| LR | 97.7 | 0.977 | 0.977 | 0.977 | 0.977 | 4.342 | 0.006 | |
| Fat | ANN | 99.3 | 0.993 | 0.993 | 0.993 | 0.999 | 18.875 | 0.019 |
| KNN | 99.5 | 0.995 | 0.995 | 0.995 | 0.999 | 0.065 | 0.089 | |
| LR | 98.6 | 0.986 | 0.986 | 0.986 | 0.991 | 7.323 | 0.008 |
| AC (%) | F1 | PR | RE | SP | Train Time (s) | Test Time (s) | |
|---|---|---|---|---|---|---|---|
| ANN | 99.9 | 0.999 | 0.999 | 0.999 | 1 | 205.253 | 0.154 |
| KNN | 98.8 | 0.988 | 0.988 | 0.988 | 0.994 | 0.493 | 1.368 |
| LR | 98.9 | 0.989 | 0.989 | 0.989 | 0.998 | 406.085 | 0.024 |
| AC (%) | F1 | PR | RE | SP | Train Time (s) | Test Time (s) | |
|---|---|---|---|---|---|---|---|
| ANN | 100 | 1 | 1 | 1 | 1 | 137.617 | 0.129 |
| KNN | 98.9 | 0.989 | 0.989 | 0.989 | 0.997 | 0.205 | 0.0866 |
| LR | 98.6 | 0.986 | 0.986 | 0.986 | 0.997 | 45.535 | 0.011 |
| Study | Methods | Max. Performance |
|---|---|---|
| Kaya et al. [46] | KNN, Linear Discriminant, Decision Tree | Accuracy: 98% |
| Wijaya et al. [47] | Adaboost, Random Forest, Support Vector Machine (SVM) and Decision Tree | Accuracy: 99.9% |
| Wijaya et al. [1] | Theoretic Ensemble Feature Selection and KNN and SVM | F1 Score: 99% |
| Enériz et al. [48] | ANN on FPGA | Accuracy: 93.73% |
| Pulluri et al. [49] | KNN, Extreme Learning Machine (ELM), SVM, ANN, Deep Neural Network (DNN) | Accuracy: 98% |
| Hibatulah et al. [27] | Support Vector Regression | R2: 0.977 and RMSE: 0.026 |
| This Study | ANN, KNN, SVM | Accuracy: 100% Precision: 100% Recall: 100% F1 Score: 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feyzioglu, A.; Taspinar, Y.S. Beef Quality Classification with Reduced E-Nose Data Features According to Beef Cut Types. Sensors 2023, 23, 2222. https://doi.org/10.3390/s23042222
Feyzioglu A, Taspinar YS. Beef Quality Classification with Reduced E-Nose Data Features According to Beef Cut Types. Sensors. 2023; 23(4):2222. https://doi.org/10.3390/s23042222
Chicago/Turabian StyleFeyzioglu, Ahmet, and Yavuz Selim Taspinar. 2023. "Beef Quality Classification with Reduced E-Nose Data Features According to Beef Cut Types" Sensors 23, no. 4: 2222. https://doi.org/10.3390/s23042222
APA StyleFeyzioglu, A., & Taspinar, Y. S. (2023). Beef Quality Classification with Reduced E-Nose Data Features According to Beef Cut Types. Sensors, 23(4), 2222. https://doi.org/10.3390/s23042222







