A Comprehensive Landscape of Imaging Feature-Associated RNA Expression Profiles in Human Breast Tissue
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. Image Data Processiong
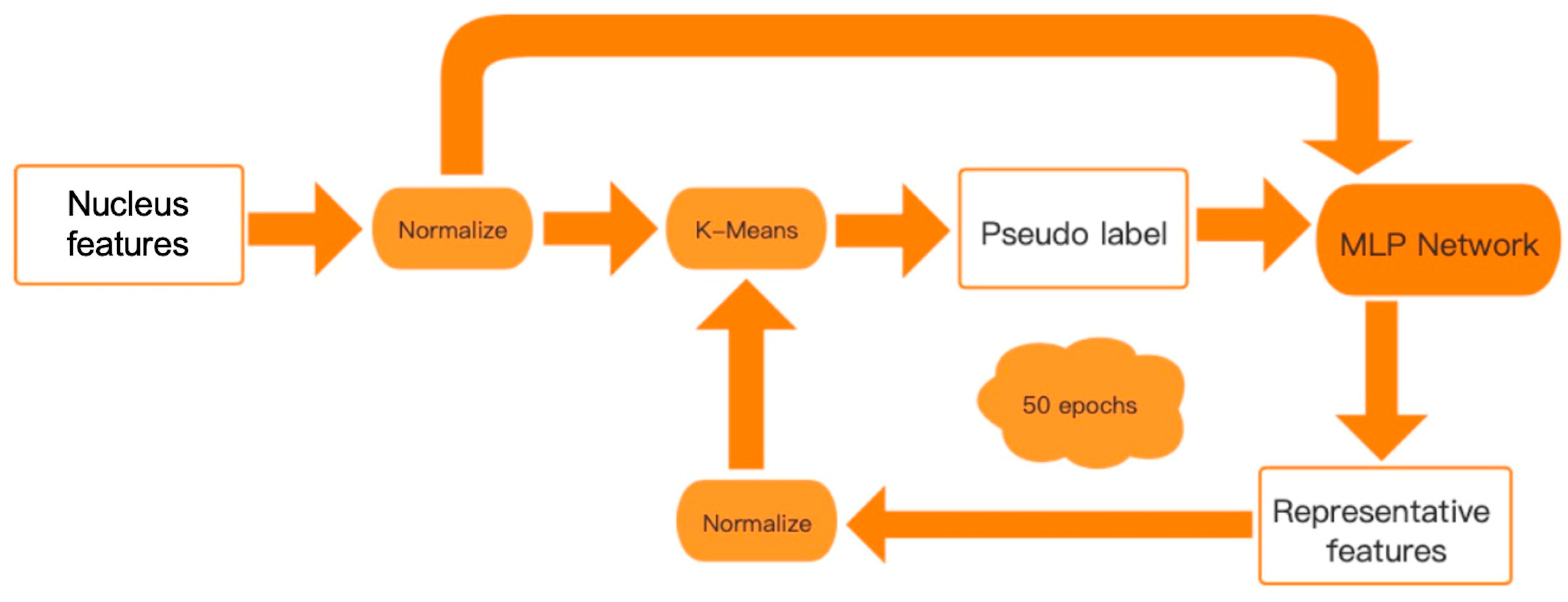
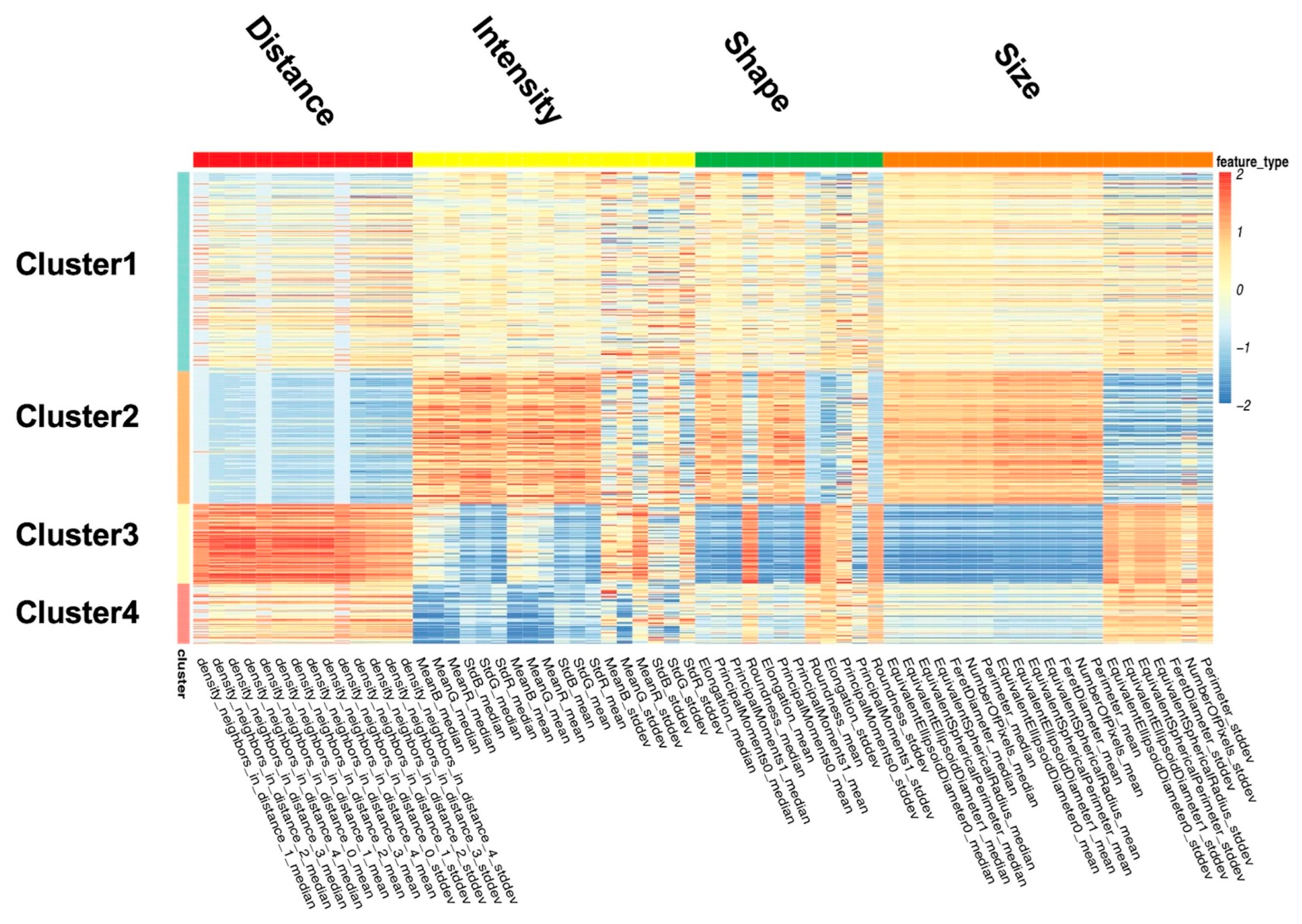
2.3. Unsupervised Analysis of Breast Images Using Nucleus Features
2.4. Identification of Feature-Specific Genes
3. Results
3.1. Image-Genetic Joint Analysis Pipeline
3.2. Glandular Tissue Segmentation
3.3. Nuclei Segmentation
3.4. Classification of Image Features
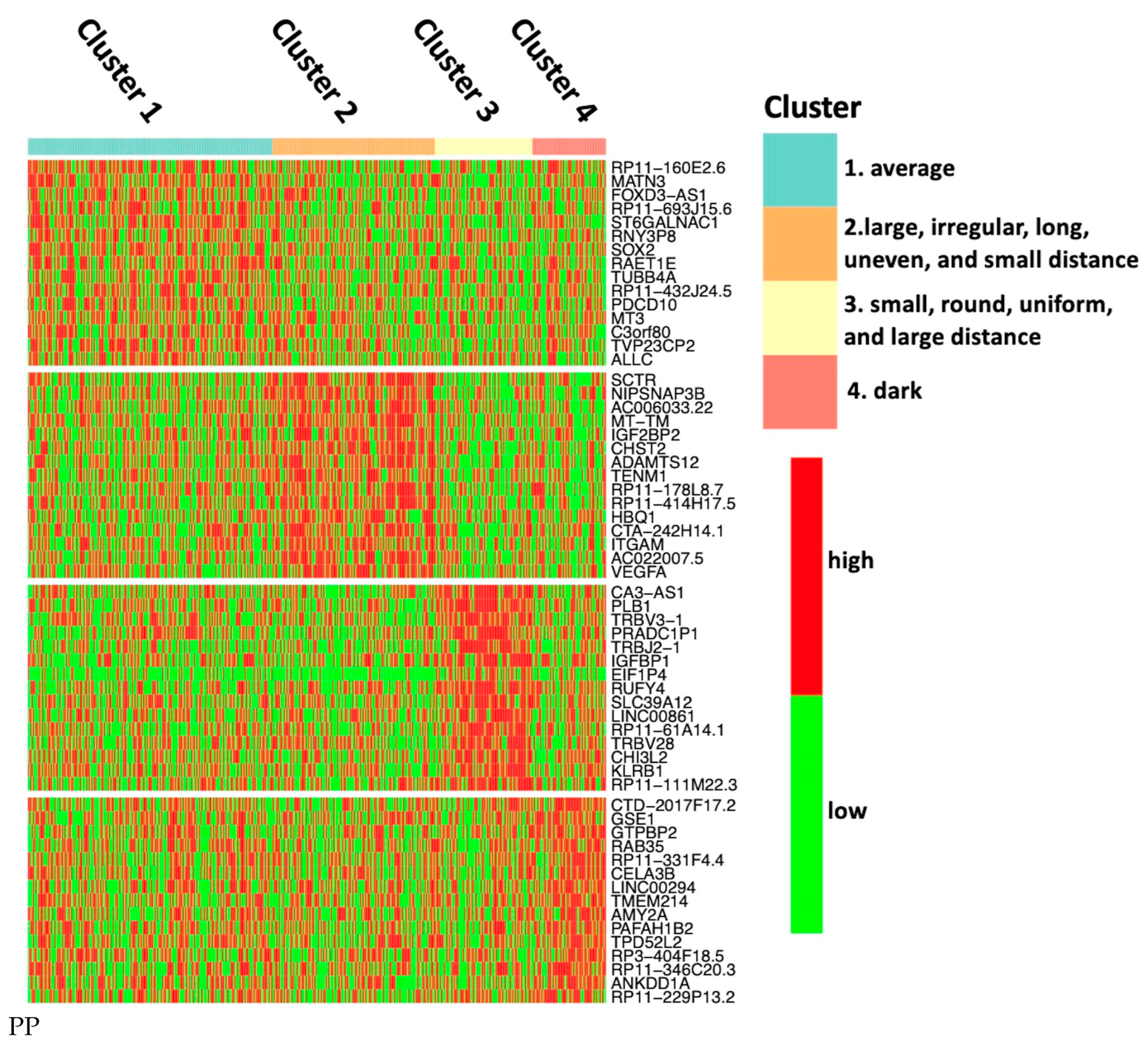
- Cluster 1: All nuclear features are close to the sample mean.
- Cluster 2: The nuclei in this cluster are large, irregular, long, and dark, with the most uneven color distribution. The distance between the nuclei is small.
- Cluster 3: The nuclei in this cluster are small and round, with uniform color distribution. The distances among the nuclei are large.
- Cluster 4: The nuclei in this cluster appear to be quite dark.
3.5. Discovery of Feature-Specific Genes
3.6. Pathway Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhan, X.; Cheng, J.; Huang, Z.; Han, Z.; Helm, B.; Liu, X.; Zhang, J.; Wang, T.-F.; Ni, D.; Huang, K. Correlation Analysis of Histopathology and Proteogenomics Data for Breast Cancer. Mol. Cell. Proteom. 2019, 18, S37–S51. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.K.; Cristofanilli, M.; Zhou, X.; Welch, K.L.; Smith, T.L.; Yang, Y.; Sneige, N.; Sahin, A.A.; Gilcrease, M.Z. β4 integrin subunit gene expression correlates with tumor size and nuclear grade in early breast cancer. Mod. Pathol. 2005, 18, 1165–1175. [Google Scholar] [CrossRef]
- Bianchini, G.; Qi, Y.; Alvarez, R.H.; Iwamoto, T.; Coutant, C.; Ibrahim, N.K.; Valero, V.; Cristofanilli, M.; Green, M.C.; Radvanyi, L.; et al. Molecular Anatomy of Breast Cancer Stroma and Its Prognostic Value in Estrogen Receptor–Positive and –Negative Cancers. J. Clin. Oncol. 2010, 28, 4316–4323. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Perou, C.M. Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 2010, 5, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.H.; Sangoi, A.R.; Leung, S.; Marinelli, R.J.; Nielsen, T.O.; van de Vijver, M.J.; West, R.B.; van de Rijn, M.; Koller, D. Systematic Analysis of Breast Cancer Morphology Uncovers Stromal Features Associated with Survival. Sci. Transl. Med. 2011, 3, 108ra113. [Google Scholar] [CrossRef] [PubMed]
- Ash, J.T.; Darnell, G.; Munro, D.; Engelhardt, B.E. Joint analysis of expression levels and histological images identifies genes associated with tissue morphology. Nat. Commun. 2021, 12, 1609. [Google Scholar] [CrossRef] [PubMed]
- Irshad, H.; Veillard, A.; Roux, L.; Racoceanu, D. Methods for Nuclei Detection, Segmentation, and Classification in Digital Histopathology: A Review—Current Status and Future Potential. IEEE Rev. Biomed. Eng. 2013, 7, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Veta, M.M.; Van Diest, P.J.; Kornegoor, R.; Huisman, A.; Viergever, M.A.; Pluim, J.P.W. Automatic Nuclei Segmentation in H&E Stained Breast Cancer Histopathology Images. PLoS ONE 2013, 8, e70221. [Google Scholar] [CrossRef]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef]
- Sirinukunwattana, K.; Raza, S.E.A.; Tsang, Y.-W.; Snead, D.R.J.; Cree, I.A.; Rajpoot, N.M. Locality Sensitive Deep Learning for Detection and Classification of Nuclei in Routine Colon Cancer Histology Images. IEEE Trans. Med. Imaging 2016, 35, 1196–1206. [Google Scholar] [CrossRef]
- Wang, P.; Hu, X.; Li, Y.; Liu, Q.; Zhu, X. Automatic cell nuclei segmentation and classification of breast cancer histopathology images. Signal Process. 2016, 122, 1–13. [Google Scholar] [CrossRef]
- Sun, D.; Li, A.; Tang, B.; Wang, M. Integrating genomic data and pathological images to effectively predict breast cancer clinical outcome. Comput. Methods Programs Biomed. 2018, 161, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, H.; Zhou, X. Nuclei Segmentation Using Marker-Controlled Watershed, Tracking Using Mean-Shift, and Kalman Filter in Time-Lapse Microscopy. IEEE Trans. Circuits Syst. I: Regul. Pap. 2006, 53, 2405–2414. [Google Scholar] [CrossRef]
- Naik, S.; Doyle, S.; Feldman, M.; Tomaszewski, J.; Madabhushi, A. Gland segmentation and computerized gleason grading of prostate histology by integrating low-, high-level and domain specific information. In MIAAB Workshop; Citeseer: Piscataway, NJ, USA, 2007; pp. 1–8. [Google Scholar]
- Isaksson, J.; Arvidsson, I.; Aastrom, K.; Heyden, A. Semantic Segmentation of Microscopic Images of H&E Stained Prostatic Tissue Using CNN; IEEE: Toulouse, France, 2017; pp. 1252–1256. [Google Scholar] [CrossRef]
- Johnson, J.W. Adapting mask-rcnn for automatic nucleus segmentation. arXiv preprint 2018, arXiv:1805.00500. [Google Scholar] [CrossRef]
- Vuola, A.O.; Akram, S.U.; Kannala, J. Mask-RCNN and U-net ensembled for nuclei segmentation. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 208–212. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]
- Abràmoff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ. Biophotonics international 2004, 11, 36–42. [Google Scholar]
- Bahlmann, C.; Patel, A.; Johnson, J.; Ni, J.; Chekkoury, A.; Khurd, P.; Kamen, A.; Grady, L.; Krupinski, E.; Graham, A.; et al. Automated detection of diagnostically relevant regions in H&E stained digital pathology slides. Prog. Biomed. Opt. Imaging 2012, 8315, 831504. [Google Scholar] [CrossRef]
- Linder, N.; Konsti, J.; Turkki, R.; Rahtu, E.; Lundin, M.; Nordling, S.; Haglund, C.; Ahonen, T.; Pietikäinen, M.; Lundin, J. Identification of tumor epithelium and stroma in tissue microarrays using texture analysis. Diagn. Pathol. 2012, 7, 22. [Google Scholar] [CrossRef]
- McKenna, S.J.; Amaral, T.; Akbar, S.; Jordan, L.; Thompson, A. Immunohistochemical analysis of breast tissue microarray images using contextual classifiers. J. Pathol. Informatics 2013, 4, 13. [Google Scholar] [CrossRef]
- Vink, J.P.; Van Leeuwen, M.B.; Van Deurzen, C.H.M.; De Haan, G.G. Efficient nucleus detector in histopathology images. J. Microsc. 2013, 249, 124–135. [Google Scholar] [CrossRef]
- Khan, A.M.; El-Daly, H.; Simmons, E.; Rajpoot, N.M. HyMaP: A hybrid magnitude-phase approach to unsupervised segmentation of tumor areas in breast cancer histology images. J. Pathol. Informatics 2013, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Kim, C.; Chae, S.W.; Oh, S. Unsupervised Segmentation of Overlapped Nuclei Using Bayesian Classification. IEEE Trans. Biomed. Eng. 2010, 57, 2825–2832. [Google Scholar] [CrossRef] [PubMed]
- Saltz, J.; Almeida, J.; Gao, Y.; Sharma, A.; Bremer, E.; DiPrima, T.; Saltz, M.; Kalpathy-Cramer, J.; Kurc, T. Towards Generation, Management, and Exploration of Combined Radiomics and Pathomics Datasets for Cancer Research. AMIA Jt. Summits Transl. Sci. proceedings. AMIA Jt. Summits Transl. Sci. 2017, 2017, 85–94. [Google Scholar]
- Wen, S.; Kurc, T.M.; Gao, Y.; Zhao, T.; Saltz, J.H.; Zhu, W. A Methodology for Texture Feature-based Quality Assessment in Nucleus Segmentation of Histopathology Image. J. Pathol. Informatics 2017, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Han, Z.; Mehra, R.; Shao, W.; Cheng, M.; Feng, Q.; Ni, D.; Huang, K.; Cheng, L.; Zhang, J. Computational analysis of pathological images enables a better diagnosis of TFE3 Xp11.2 translocation renal cell carcinoma. Nat. Commun. 2020, 11, 1778. [Google Scholar] [CrossRef]
- Blaveri, E.; Simko, J.P.; Korkola, J.E.; Brewer, J.L.; Baehner, F.; Mehta, K.; DeVries, S.; Koppie, T.; Pejavar, S.; Carroll, P.; et al. Bladder Cancer Outcome and Subtype Classification by Gene Expression. Clin. Cancer Res. 2005, 11, 4044–4055. [Google Scholar] [CrossRef] [PubMed]
- Sotiriou, C.; Pusztai, L. Gene-Expression Signatures in Breast Cancer. New Engl. J. Med. 2009, 360, 790–800. [Google Scholar] [CrossRef]
- Barry, W.T.; Kernagis, D.N.; Dressman, H.K.; Griffis, R.J.; Hunter, J.D.; Olson, J.A.; Marks, J.R.; Ginsburg, G.S.; Marcom, P.K.; Nevins, J.R.; et al. Intratumor Heterogeneity and Precision of Microarray-Based Predictors of Breast Cancer Biology and Clinical Outcome. J. Clin. Oncol. 2010, 28, 2198–2206. [Google Scholar] [CrossRef]
- Gevaert, O.; Xu, J.; Hoang, C.D.; Leung, A.N.; Xu, Y.; Quon, A.; Rubin, D.L.; Napel, S.; Plevritis, S.K. Non–Small Cell Lung Cancer: Identifying Prognostic Imaging Biomarkers by Leveraging Public Gene Expression Microarray Data—Methods and Preliminary Results. Radiology 2012, 264, 387–396. [Google Scholar] [CrossRef]
- Chen, R.J.; Lu, M.Y.; Wang, J.; Williamson, D.F.K.; Rodig, S.J.; Lindeman, N.I.; Mahmood, F. Pathomic Fusion: An Integrated Framework for Fusing Histopathology and Genomic Features for Cancer Diagnosis and Prognosis. IEEE Trans. Med. Imaging 2022, 41, 757–770. [Google Scholar] [CrossRef]
- Dolezal, J.M.; Trzcinska, A.; Liao, C.-Y.; Kochanny, S.; Blair, E.; Agrawal, N.; Keutgen, X.M.; Angelos, P.; Cipriani, N.A.; Pearson, A.T. Deep learning prediction of BRAF-RAS gene expression signature identifies noninvasive follicular thyroid neoplasms with papillary-like nuclear features. Mod. Pathol. 2021, 34, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Pineda-Cirera, L.; Cabana-Domínguez, J.; Lee, P.H.; Fernàndez-Castillo, N.; Cormand, B. Identification of genetic variants influencing methylation in brain with pleiotropic effects on psychiatric disorders. Prog. Neuro-Psychopharmacology Biol. Psychiatry 2021, 113, 110454. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Huber, P.J. Robust statistics. In International encyclopedia of statistical science; Springer: Berlin, Heidelberg, 2011; pp. 1248–1251. [Google Scholar]
- Pichon, E.; Tannenbaum, A.; Kikinis, R. A statistically based flow for image segmentation. Med. Image Anal. 2004, 8, 267–274. [Google Scholar] [CrossRef]
- Gao, Y.; Kikinis, R.; Bouix, S.; Shenton, M.; Tannenbaum, A. A 3D interactive multi-object segmentation tool using local robust statistics driven active contours. Med. Image Anal. 2012, 16, 1216–1227. [Google Scholar] [CrossRef]
- Ruifrok, A.C.; Johnston, D.A. Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 2001, 23, 291–299. [Google Scholar]
- Vese, L.A.; Chan, T.F. A Multiphase Level Set Framework for Image Segmentation Using the Mumford and Shah Model. Int. J. Comput. Vis. 2002, 50, 271–293. [Google Scholar] [CrossRef]
- Qi, X.; Xing, F.; Foran, D.J.; Yang, L. Robust Segmentation of Overlapping Cells in Histopathology Specimens Using Parallel Seed Detection and Repulsive Level Set. IEEE Trans. Biomed. Eng. 2011, 59, 754–765. [Google Scholar] [CrossRef]
- Lehmann, G. Label object representation and manipulation with ITK. Insight J. 2007. [Google Scholar] [CrossRef]
- Doyle, S.; Hwang, M.; Shah, K.; Madabhushi, A.; Feldman, M.; Tomaszeweski, J. Automated Grading Of Prostate Cancer Using Architectural And Textural Image Features; IEEE: Toulouse, France, 2007; pp. 1284–1287. [Google Scholar] [CrossRef]
- Caron, M.; Bojanowski, P.; Joulin, A.; Douze, M. Deep clustering for unsupervised learning of visual features. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 132–149. [Google Scholar] [CrossRef]
- Min, E.; Guo, X.; Liu, Q.; Zhang, G.; Cui, J.; Long, J. A Survey of Clustering With Deep Learning: From the Perspective of Network Architecture. IEEE Access 2018, 6, 39501–39514. [Google Scholar] [CrossRef]
- MacQueen, J. Classification and analysis of multivariate observations. 5th Berkeley Symp. Math. Statist. Probability 1967, 281–297. [Google Scholar]
- Vu, T.N.; Pramana, S.; Calza, S.; Suo, C.; Lee, D.; Pawitan, Y. Comprehensive landscape of subtype-specific coding and non-coding RNA transcripts in breast cancer. Oncotarget 2016, 7, 68851–68863. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Pawitan, Y.; Michiels, S.; Koscielny, S.; Gusnanto, A.; Ploner, A. False discovery rate, sensitivity and sample size for microarray studies. Bioinformatics 2005, 21, 3017–3024. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Medical Image Computing C.A.I.S., 2016. Miccai2016. Available online: http://www.miccai2016.org/en (accessed on 13 October 2021).
- Sirinukunwattana, K.; Pluim, J.P.; Chen, H.; Qi, X.; Heng, P.-A.; Guo, Y.B.; Wang, L.Y.; Matuszewski, B.J.; Bruni, E.; Sanchez, U.; et al. Gland segmentation in colon histology images: The glas challenge contest. Med. Image Anal. 2017, 35, 489–502. [Google Scholar] [CrossRef]
- Maška, M.; Ulman, V.; Svoboda, D.; Matula, P.; Matula, P.; Ederra, C.; Urbiola, A.; España, T.; Venkatesan, S.; Balak, D.M.; et al. A benchmark for comparison of cell tracking algorithms. Bioinformatics 2014, 30, 1609–1617. [Google Scholar] [CrossRef]
- Caicedo, J.C.; Goodman, A.; Karhohs, K.W.; Cimini, B.A.; Ackerman, J.; Haghighi, M.; Heng, C.; Becker, T.; Doan, M.; McQuin, C.; et al. Nucleus segmentation across imaging experiments: The 2018 Data Science Bowl. Nat. Methods 2019, 16, 1247–1253. [Google Scholar] [CrossRef]
- Mou, T.; Pawitan, Y.; Stahl, M.; Vesterlund, M.; Deng, W.; Jafari, R.; Bohlin, A.; Österroos, A.; Siavelis, L.; Bäckvall, H.; et al. The transcriptome-wide landscape of molecular subtype-specific mRNA expression profiles in acute myeloid leukemia. Am. J. Hematol. 2021, 96, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Onori, P.; Wise, C.; Gaudio, E.; Franchitto, A.; Francis, H.; Carpino, G.; Lee, V.; Lam, I.; Miller, T.; Dostal, D.E.; et al. Secretin inhibits cholangiocarcinoma growth via dysregulation of the cAMP-dependent signaling mechanisms of secretin receptor. Int. J. Cancer 2009, 127, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, L.; Fu, J.; Huang, H.; Sun, S.; Zhang, D.; Zhao, L.; Onwuka, J.U.; Zhao, Y.; Cui, B. SCTR hypermethylation is a diagnostic biomarker in colorectal cancer. Cancer Sci. 2020, 111, 4558–4566. [Google Scholar] [CrossRef]
- Kang, S.; Kim, B.; Kang, H.-S.; Jeong, G.; Bae, H.; Lee, H.; Lee, S.; Kim, S.J. SCTR regulates cell cycle-related genes toward anti-proliferation in normal breast cells while having pro-proliferation activity in breast cancer cells. Int. J. Oncol. 2015, 47, 1923–1931. [Google Scholar] [CrossRef] [PubMed]
- McMullen, E.R.; Gonzalez, M.E.; Skala, S.L.; Tran, M.; Thomas, D.; Djomehri, S.I.; Burman, B.; Kidwell, K.M.; Kleer, C.G. CCN6 regulates IGF2BP2 and HMGA2 signaling in metaplastic carcinomas of the breast. Breast Cancer Res. Treat. 2018, 172, 577–586. [Google Scholar] [CrossRef]
- Kim, H.-Y.; Thi, H.T.H.; Hong, S. IMP2 and IMP3 cooperate to promote the metastasis of triple-negative breast cancer through destabilization of progesterone receptor. Cancer Lett. 2018, 415, 30–39. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Zhuang, H.-W.; Wang, J.; Shen, Y.; Bu, Y.-Z.; Guan, B.-G.; Xu, F.; Dou, J. Long noncoding RNA CA3-AS1 suppresses gastric cancer migration and invasion by sponging miR-93-5p and targeting BTG3. Gene Ther. 2020, 29, 566–574. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Yang, Z.; Lin, B. Overexpression of long non coding RNA CA3-AS1 suppresses proliferation, invasion and promotes apoptosis via miRNA-93/PTEN axis in colorectal cancer. Gene 2018, 687, 9–15. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.A.; Clark, J.; Ireland, A.; Lomax, J.; Ashburner, M.; Foulger, R.; Eilbeck, K.; Lewis, S.; Marshall, B.; Mungall, C.; et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [CrossRef]
- Hajizadeh, F.; Maleki, L.A.; Alexander, M.; Mikhailova, M.V.; Masjedi, A.; Ahmadpour, M.; Hashemi, V.; Jadidi-Niaragh, F. Tumor-associated neutrophils as new players in immunosuppressive process of the tumor microenvironment in breast cancer. Life Sci. 2020, 264, 118699. [Google Scholar] [CrossRef]
- Mollinedo, F. Neutrophil Degranulation, Plasticity, and Cancer Metastasis. Trends Immunol. 2019, 40, 228–242. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.U.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Psyrri, A.; Kalogeras, K.; Kronenwett, R.; Wirtz, R.; Batistatou, A.; Bournakis, E.; Timotheadou, E.; Gogas, H.; Aravantinos, G.; Christodoulou, C.; et al. Prognostic significance of UBE2C mRNA expression in high-risk early breast cancer. A Hellenic Cooperative Oncology Group (HeCOG) Study. Ann. Oncol. 2012, 23, 1422–1427. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Zhou, C.; Liu, F.; Xu, G.; Zheng, R.; Zhang, X. MicroRNA-196a post-transcriptionally upregulates the UBE2C proto-oncogene and promotes cell proliferation in breast cancer. Oncol. Rep. 2015, 34, 877–883. [Google Scholar] [CrossRef] [PubMed]
- Bièche, I.; Vacher, S.; Lallemand, F.; Tozlu-Kara, S.; Bennani, H.; Beuzelin, M.; Driouch, K.; Rouleau, E.; Lerebours, F.; Ripoche, H.; et al. Expression analysis of mitotic spindle checkpoint genes in breast carcinoma: Role of NDC80/HEC1 in early breast tumorigenicity, and a two-gene signature for aneuploidy. Mol. Cancer 2011, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Wang, Y.; Wang, Y.; Lv, S.; Xu, X.; Dong, X. Screening of differentially expressed genes and identification of NUF2 as a prognostic marker in breast cancer. Int. J. Mol. Med. 2019, 44, 390–404. [Google Scholar] [CrossRef]
- Opoku, F.; Bedu-Addo, K.; Titiloye, N.A.; Manu, E.A.; Ameh-Mensah, C.; Duduyemi, B.M. Expression profile of tumour suppressor protein p53 and its regulator MDM2 in a cohort of breast cancer patients in a Tertiary Hospital in Ghana. PLoS ONE 2021, 16, e0258543. [Google Scholar] [CrossRef]
- Wege, A.K.; Rom-Jurek, E.; Jank, P.; Denkert, C.; Ugocsai, P.; Solbach, C.; Blohmer, J.; Sinn, B.; Mackelenbergh, M.; Möbus, V.; et al. mdm2 gene amplification is associated with luminal breast cancer progression in humanized PDX mice and a worse outcome of estrogen receptor positive disease. Int. J. Cancer 2021, 150, 1357–1372. [Google Scholar] [CrossRef]
- Ethier, J.-L.; Desautels, D.; Templeton, A.; Shah, P.S.; Amir, E. Prognostic role of neutrophil-to-lymphocyte ratio in breast cancer: A systematic review and meta-analysis. Breast Cancer Res. 2017, 19, 2. [Google Scholar] [CrossRef]
- Kakinuma, T. Chemokines, chemokine receptors, and cancer metastasis. J. Leukoc. Biol. 2006, 79, 639–651. [Google Scholar] [CrossRef]
- Pour, A.F.; White, B.S.; Park, J.; Sheridan, T.B.; Chuang, J.H. Deep learning features encode interpretable morphologies within histological images. Sci. Rep. 2022, 12, 9428. [Google Scholar] [CrossRef]






| DSC (mean ± std) | IoU (mean ± std) | Average HD (mean ± std) | |
|---|---|---|---|
| QuPath | 0.7002 ± 0.0904 | 0.5451 ± 0.1006 | 2.8108 ± 2.0811 |
| UNet | 0.7592 ± 0.0983 | 0.6204 ± 0.1187 | 1.3044 ± 0.7208 |
| Proposed | 0.7797 ± 0.0525 | 0.6416 ± 0.0691 | 1.2942 ± 0.6634 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mou, T.; Liang, J.; Vu, T.N.; Tian, M.; Gao, Y. A Comprehensive Landscape of Imaging Feature-Associated RNA Expression Profiles in Human Breast Tissue. Sensors 2023, 23, 1432. https://doi.org/10.3390/s23031432
Mou T, Liang J, Vu TN, Tian M, Gao Y. A Comprehensive Landscape of Imaging Feature-Associated RNA Expression Profiles in Human Breast Tissue. Sensors. 2023; 23(3):1432. https://doi.org/10.3390/s23031432
Chicago/Turabian StyleMou, Tian, Jianwen Liang, Trung Nghia Vu, Mu Tian, and Yi Gao. 2023. "A Comprehensive Landscape of Imaging Feature-Associated RNA Expression Profiles in Human Breast Tissue" Sensors 23, no. 3: 1432. https://doi.org/10.3390/s23031432
APA StyleMou, T., Liang, J., Vu, T. N., Tian, M., & Gao, Y. (2023). A Comprehensive Landscape of Imaging Feature-Associated RNA Expression Profiles in Human Breast Tissue. Sensors, 23(3), 1432. https://doi.org/10.3390/s23031432





