Emerging Microfluidic Tools for Simultaneous Exosomes and Cargo Biosensing in Liquid Biopsy: New Integrated Miniaturized FFF-Assisted Approach for Colon Cancer Diagnosis
Abstract
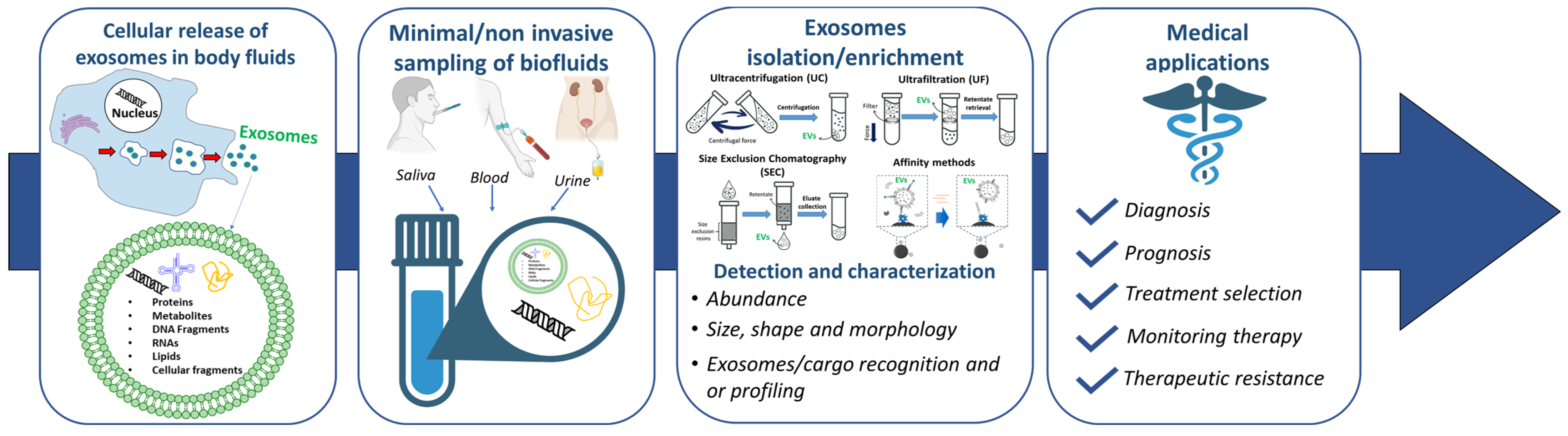
:1. Introduction
2. Exosomes for the Early Diagnosis of Cancer
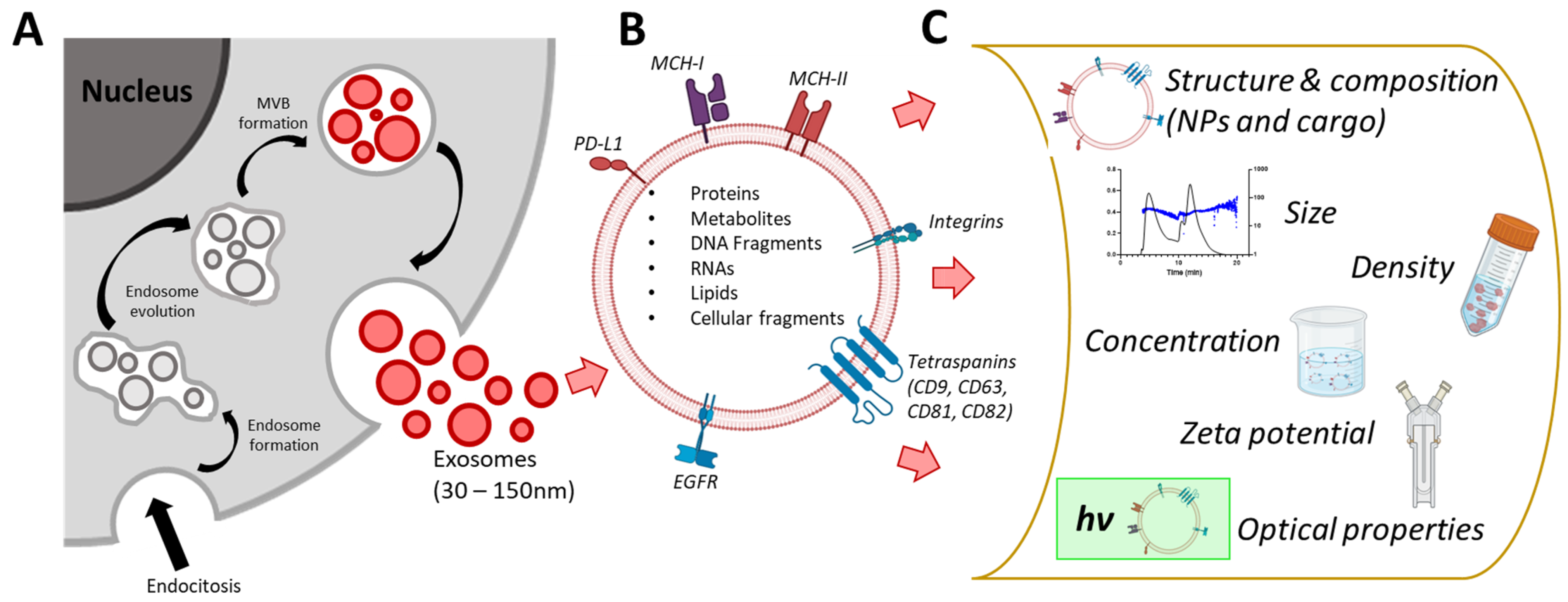
2.1. Exosome Biogenesis and Biophysics
2.2. Exosomes for the Early Diagnosis of Colorectal Cancer
3. Integrated Microfluidic System for Exosomes Analysis
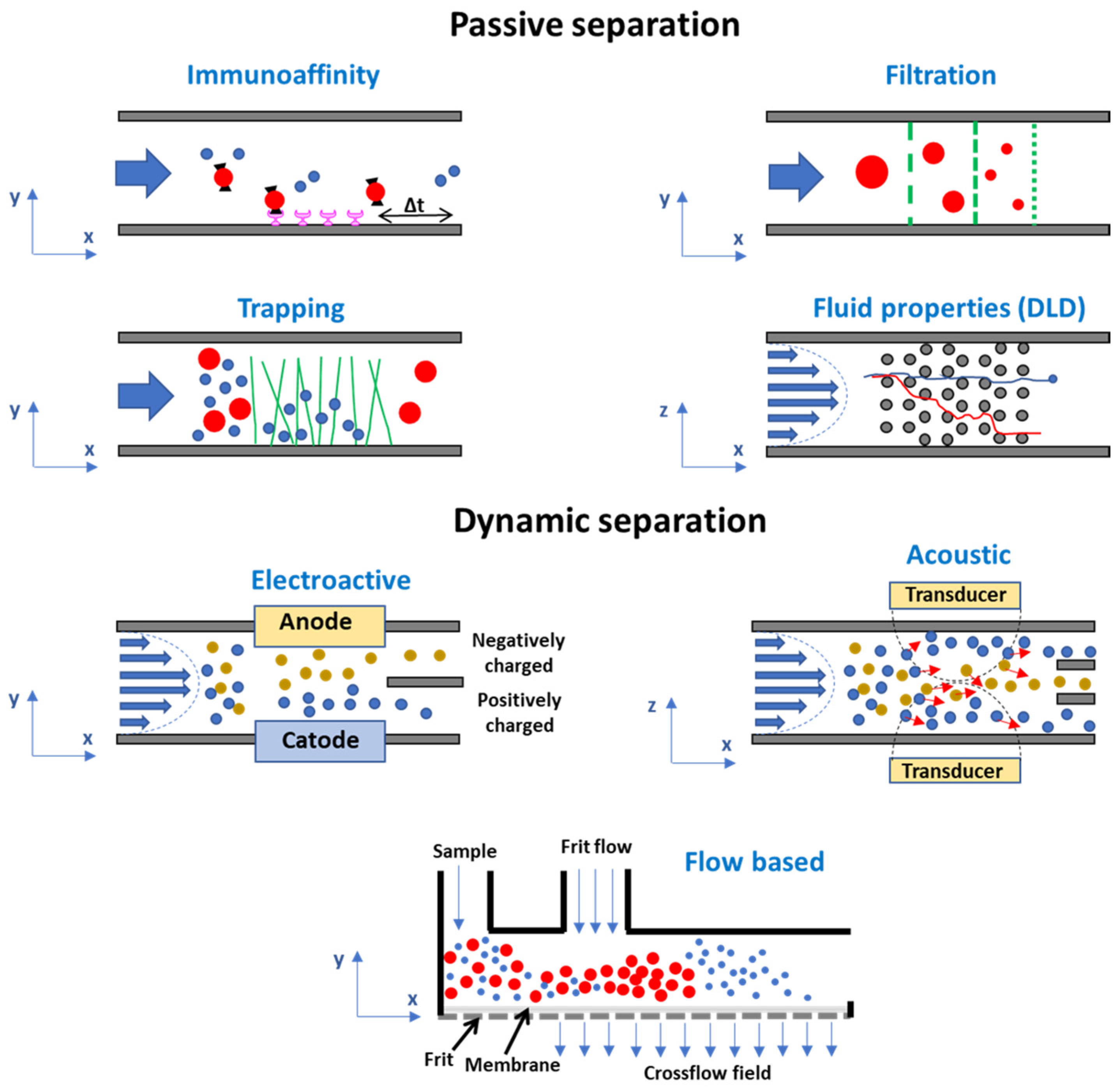
3.1. Microfluidic Systems for Exosome Isolation
3.1.1. Microfluidic Systems Based on the Intrinsic Properties of Exosomes
Immunoaffinity-Based Exosome Isolation
Label-Free Microfluidic Separation of Exosomes: Filtration and Trapping Separation; Fluid-Based Separation
3.1.2. Microfluidic Systems Based on Dynamic Separation
Electroactive and Acoustic Separation
Flow-Based Separation
3.2. Biosensing Approaches
3.2.1. Reagent-Based Systems
3.2.2. Reagent-Less-Based Systems
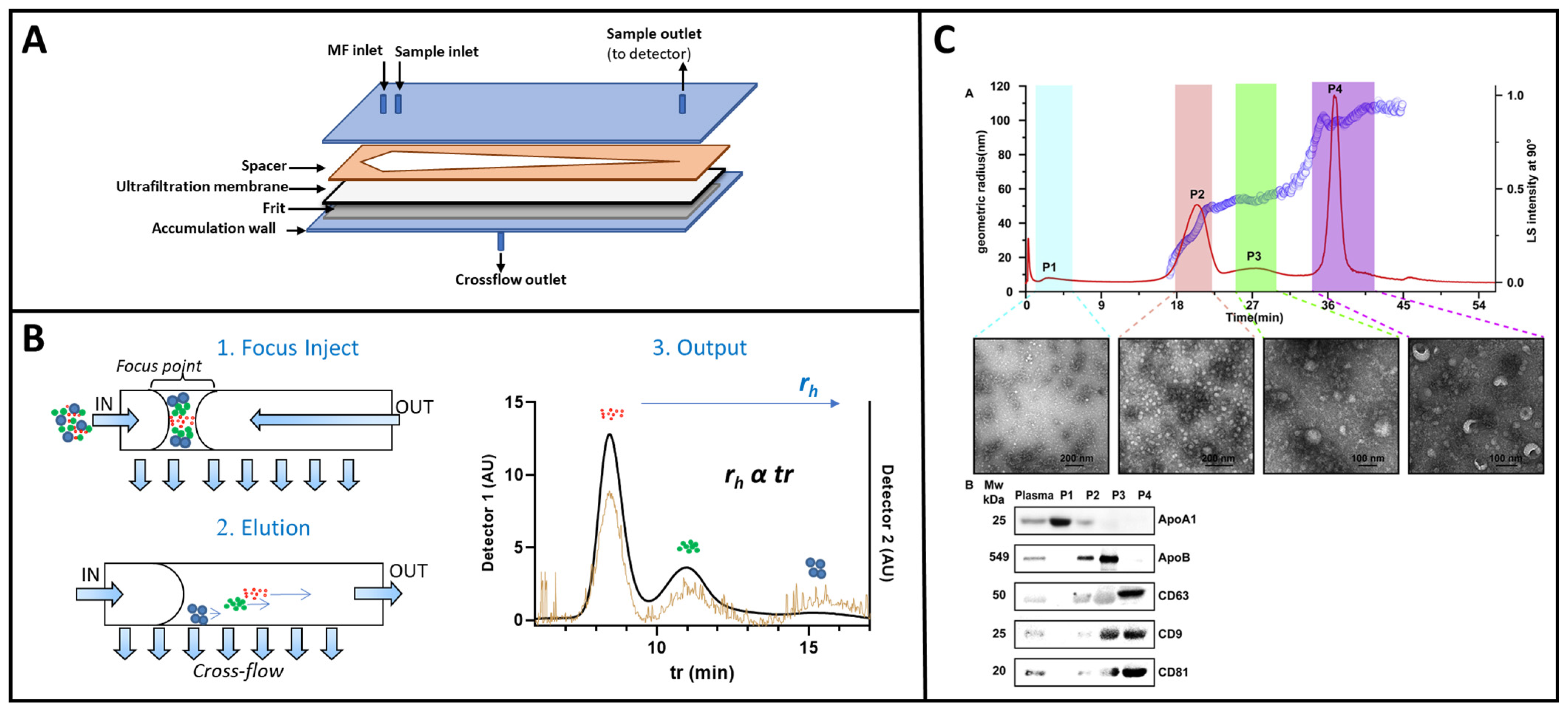
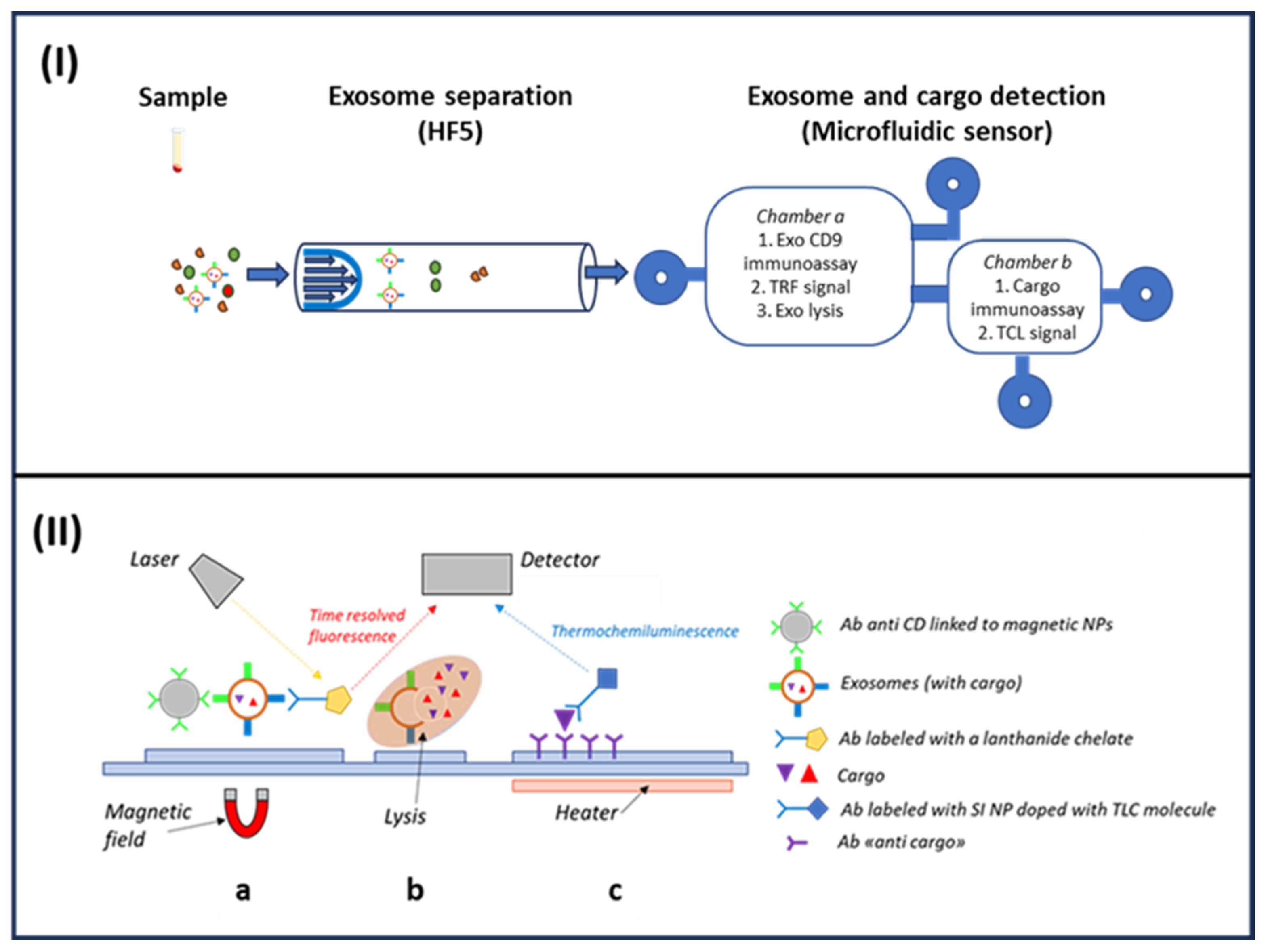
4. Towards a New FFF-Based Multiplex Biosensor
4.1. Exosomes Isolation and Cargo Analysis
4.1.1. Study Design and Serum Sample Collection
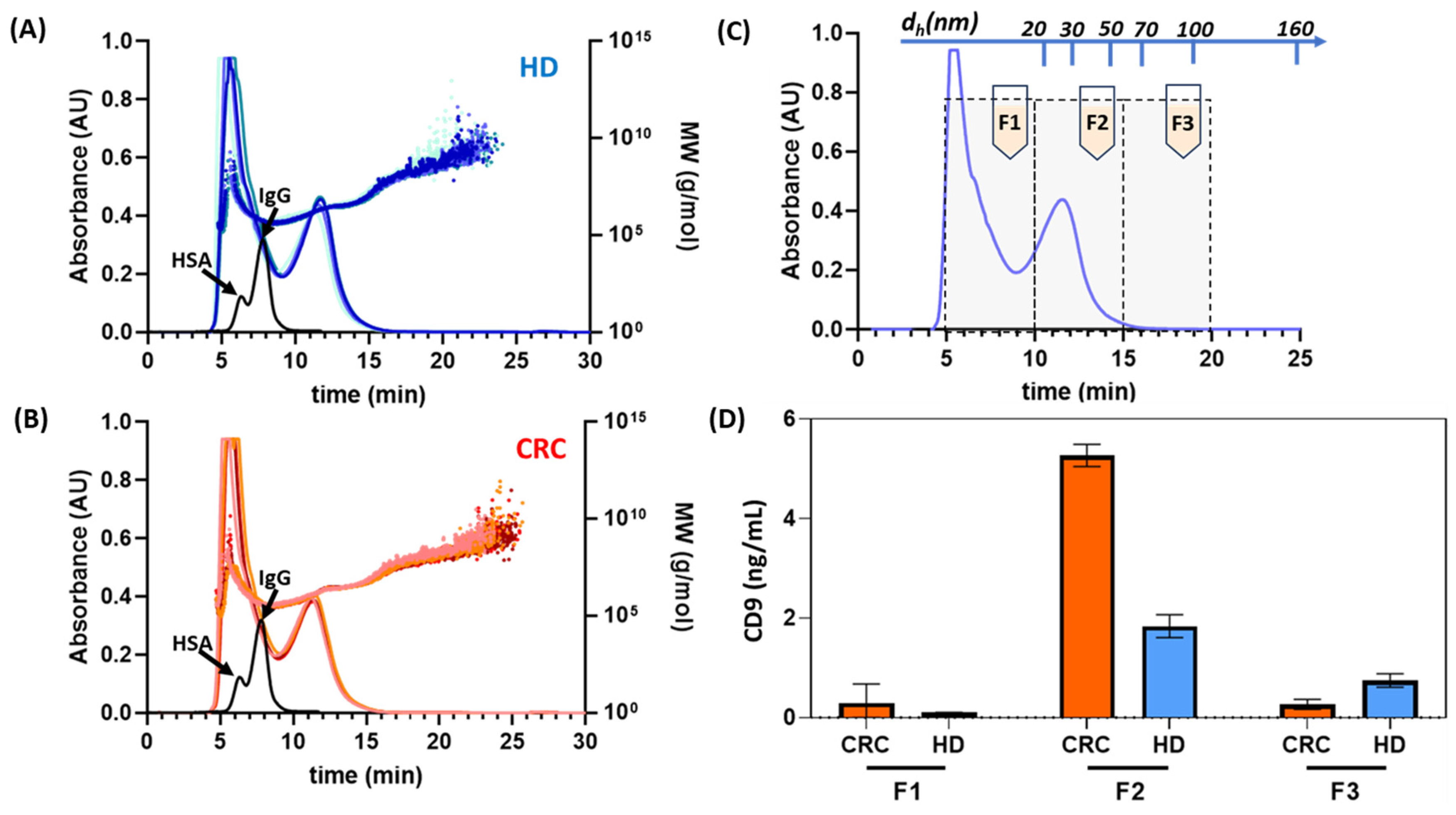
4.1.2. HF5 Instrumental Setup and Exosomes Isolation Performances
4.1.3. HF5-Based Microfluidic Tool: Simultaneous Exosomes CD9 Membrane Protein and IL6 Quantification
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lone, S.N.; Nisar, S.; Masoodi, T.; Singh, M.; Rizwan, A.; Hashem, S.; El-Rifai, W.; Bedognetti, D.; Batra, S.K.; Haris, M. Liquid biopsy: A step closer to transform diagnosis, prognosis and future of cancer treatments. Mol. Cancer 2022, 21, 79. [Google Scholar] [CrossRef] [PubMed]
- Poulet, G.; Massias, J.; Taly, V. Liquid biopsy: General concepts. Acta Cytol. 2019, 63, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Macías, M.; Alegre, E.; Díaz-Lagares, A.; Patiño, A.; Perez-Gracia, J.L.; Sanmamed, M.; López-López, R.; Varo, N.; González, A. Liquid biopsy: From basic research to clinical practice. Adv. Clin. Chem. 2018, 83, 73–119. [Google Scholar] [PubMed]
- Ferrara, F.; Zoupanou, S.; Primiceri, E.; Ali, Z.; Chiriacò, M.S. Beyond liquid biopsy: Toward non-invasive assays for distanced cancer diagnostics in pandemics. Biosens. Bioelectron. 2022, 196, 113698. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, D. Exosomes in cancer development, metastasis, and immunity. Biochim. Biophys. Acta BBA-Rev. Cancer 2019, 1871, 455–468. [Google Scholar] [CrossRef]
- Neviani, P.; Fabbri, M. Exosomic microRNAs in the tumor microenvironment. Front. Med. 2015, 2, 47. [Google Scholar] [CrossRef]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef]
- Bahrami, A.; Binabaj, M.M.; Ferns, G.A. Exosomes: Emerging modulators of signal transduction in colorectal cancer from molecular understanding to clinical application. Biomed. Pharmacother. 2021, 141, 111882. [Google Scholar] [CrossRef]
- Yu, D.; Li, Y.; Wang, M.; Gu, J.; Xu, W.; Cai, H.; Fang, X.; Zhang, X. Exosomes as a new frontier of cancer liquid biopsy. Mol. Cancer 2022, 21, 56. [Google Scholar] [CrossRef]
- Yu, W.; Hurley, J.; Roberts, D.; Chakrabortty, S.; Enderle, D.; Noerholm, M.; Breakefield, X.; Skog, J. Exosome-based liquid biopsies in cancer: Opportunities and challenges. Ann. Oncol. 2021, 32, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Tutrone, R.; Donovan, M.J.; Torkler, P.; Tadigotla, V.; McLain, T.; Noerholm, M.; Skog, J.; McKiernan, J. Clinical utility of the exosome based ExoDx Prostate (IntelliScore) EPI test in men presenting for initial Biopsy with a PSA 2–10 ng/mL. Prostate Cancer Prostatic Dis. 2020, 23, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Mora, E.M.; Álvarez-Cubela, S.; Oltra, E. Biobanking of exosomes in the era of precision medicine: Are we there yet? Int. J. Mol. Sci. 2015, 17, 13. [Google Scholar] [CrossRef] [PubMed]
- Shao, H.; Chung, J.; Issadore, D. Diagnostic technologies for circulating tumour cells and exosomes. Biosci. Rep. 2016, 36, e00292. [Google Scholar] [CrossRef] [PubMed]
- Jablonska, J.; Pietrowska, M.; Ludwig, S.; Lang, S.; Thakur, B.K. Challenges in the isolation and proteomic analysis of cancer exosomes—Implications for translational research. Proteomes 2019, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Abels, E.R.; Breakefield, X.O. Introduction to extracellular vesicles: Biogenesis, RNA cargo selection, content, release, and uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef]
- Osaki, M.; Okada, F. Exosomes and their role in cancer progression. Yonago Acta Med. 2019, 62, 182–190. [Google Scholar] [CrossRef]
- McAndrews, K.M.; Kalluri, R. Mechanisms associated with biogenesis of exosomes in cancer. Mol. Cancer 2019, 18, 52. [Google Scholar] [CrossRef]
- Wiklander, O.P.; Nordin, J.Z.; O’Loughlin, A.; Gustafsson, Y.; Corso, G.; Mäger, I.; Vader, P.; Lee, Y.; Sork, H.; Seow, Y. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J. Extracell. Vesicles 2015, 4, 26316. [Google Scholar] [CrossRef]
- Hu, Q.; Su, H.; Li, J.; Lyon, C.; Tang, W.; Wan, M.; Hu, T.Y. Clinical applications of exosome membrane proteins. Precis. Clin. Med. 2020, 3, 54–66. [Google Scholar] [CrossRef]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Yuana, Y.; Koning, R.I.; Kuil, M.E.; Rensen, P.C.; Koster, A.J.; Bertina, R.M.; Osanto, S. Cryo-electron microscopy of extracellular vesicles in fresh plasma. J. Extracell. Vesicles 2013, 2, 21494. [Google Scholar] [CrossRef] [PubMed]
- Midekessa, G.; Godakumara, K.; Ord, J.; Viil, J.; Lättekivi, F.; Dissanayake, K.; Kopanchuk, S.; Rinken, A.; Andronowska, A.; Bhattacharjee, S. Zeta potential of extracellular vesicles: Toward understanding the attributes that determine colloidal stability. ACS Omega 2020, 5, 16701–16710. [Google Scholar] [CrossRef] [PubMed]
- Mendivil-Alvarado, H.; Limon-Miro, A.T.; Carvajal-Millan, E.; Lizardi-Mendoza, J.; Mercado-Lara, A.; Coronado-Alvarado, C.D.; Rascón-Durán, M.L.; Anduro-Corona, I.; Talamás-Lara, D.; Rascón-Careaga, A. Extracellular Vesicles and Their Zeta Potential as Future Markers Associated with Nutrition and Molecular Biomarkers in Breast Cancer. Int. J. Mol. Sci. 2023, 24, 6810. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Rietjens, I.M.; Singh, M.P.; Atkins, T.M.; Purkait, T.K.; Xu, Z.; Regli, S.; Shukaliak, A.; Clark, R.J.; Mitchell, B.S. Cytotoxicity of surface-functionalized silicon and germanium nanoparticles: The dominant role of surface charges. Nanoscale 2013, 5, 4870–4883. [Google Scholar] [CrossRef] [PubMed]
- Akagi, T.; Kato, K.; Hanamura, N.; Kobayashi, M.; Ichiki, T. Evaluation of desialylation effect on zeta potential of extracellular vesicles secreted from human prostate cancer cells by on-chip microcapillary electrophoresis. Jpn. J. Appl. Phys. 2014, 53, 06JL01. [Google Scholar] [CrossRef]
- Baran, J.; Baj-Krzyworzeka, M.; Weglarczyk, K.; Szatanek, R.; Zembala, M.; Barbasz, J.; Czupryna, A.; Szczepanik, A.; Zembala, M. Circulating tumour-derived microvesicles in plasma of gastric cancer patients. Cancer Immunol. Immunother. 2010, 59, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef]
- Ruiz-López, L.; Blancas, I.; Garrido, J.M.; Mut-Salud, N.; Moya-Jódar, M.; Osuna, A.; Rodríguez-Serrano, F. The role of exosomes on colorectal cancer: A review. J. Gastroenterol. Hepatol. 2018, 33, 792–799. [Google Scholar] [CrossRef]
- Zhou, J.; Li, X.-L.; Chen, Z.-R.; Chng, W.-J. Tumor-derived exosomes in colorectal cancer progression and their clinical applications. Oncotarget 2017, 8, 100781. [Google Scholar] [CrossRef] [PubMed]
- Caliceti, C.; Punzo, A.; Silla, A.; Simoni, P.; Roda, G.; Hrelia, S. New Insights into Bile Acids Related Signaling Pathways in the Onset of Colorectal Cancer. Nutrients 2022, 14, 2964. [Google Scholar] [CrossRef]
- Mannavola, F.; Salerno, T.; Passarelli, A.; Tucci, M.; Internò, V.; Silvestris, F. Revisiting the role of exosomes in colorectal cancer: Where are we now? Front. Oncol. 2019, 9, 521. [Google Scholar] [CrossRef] [PubMed]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef] [PubMed]
- Nabariya, D.K.; Pallu, R.; Yenuganti, V.R. Exosomes: The protagonists in the tale of colorectal cancer? Biochim. Biophys. Acta BBA-Rev. Cancer 2020, 1874, 188426. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhong, J.; Zhong, B.; Huang, J.; Jiang, L.; Jiang, Y.; Yuan, J.; Sun, J.; Dai, L.; Yang, C. Exosomes as potential sources of biomarkers in colorectal cancer. Cancer Lett. 2020, 476, 13–22. [Google Scholar] [CrossRef]
- Dang, Y.; Zhang, S.; Wang, Y.; Zhao, G.; Chen, C.; Jiang, W. State-of-the-Art: Exosomes in Colorectal Cancer. Curr. Cancer Drug Targets 2022, 22, 2–17. [Google Scholar] [CrossRef]
- Pretzsch, E.; Bösch, F.; Neumann, J.; Ganschow, P.; Bazhin, A.; Guba, M.; Werner, J.; Angele, M. Mechanisms of metastasis in colorectal cancer and metastatic organotropism: Hematogenous versus peritoneal spread. J. Oncol. 2019, 2019, 7407190. [Google Scholar] [CrossRef]
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 2014, 9, e92921. [Google Scholar] [CrossRef]
- Wang, J.; Yan, F.; Zhao, Q.; Zhan, F.; Wang, R.; Wang, L.; Zhang, Y.; Huang, X. Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci. Rep. 2017, 7, 4150. [Google Scholar] [CrossRef]
- Umwali, Y.; Yue, C.-B.; Gabriel, A.N.A.; Zhang, Y.; Zhang, X. Roles of exosomes in diagnosis and treatment of colorectal cancer. World J. Clin. Cases 2021, 9, 4467. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; He, B.; Gao, T.; Pan, Y.; Sun, H.; Xu, Y.; Li, R.; Ying, H.; Wang, F.; Liu, X. Up-regulation of 91H promotes tumor metastasis and predicts poor prognosis for patients with colorectal cancer. PLoS ONE 2014, 9, e103022. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.-x.; Liu, H.-s.; Wang, F.-w.; Xiong, L.; Zhou, C.; Hu, T.; He, X.-w.; Wu, X.-j.; Xie, D.; Wu, X.-r. LncRNA RPPH1 promotes colorectal cancer metastasis by interacting with TUBB3 and by promoting exosomes-mediated macrophage M2 polarization. Cell Death Dis. 2019, 10, 829. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Lin, W.; Qi, P.; Xu, M.-d.; Wu, X.; Ni, S.; Huang, D.; Weng, W.-w.; Tan, C.; Sheng, W. Circulating long RNAs in serum extracellular vesicles: Their characterization and potential application as biomarkers for diagnosis of colorectal cancer. Cancer Epidemiol. Biomark. Prev. 2016, 25, 1158–1166. [Google Scholar] [CrossRef]
- Titu, S.; Grapa, C.M.; Mocan, T.; Balacescu, O.; Irimie, A. Tetraspanins: Physiology, Colorectal Cancer Development, and Nanomediated Applications. Cancers 2021, 13, 5662. [Google Scholar] [CrossRef]
- Abi Zamer, B.; El-Huneidi, W.; Eladl, M.A.; Muhammad, J.S. Ins and outs of heat shock proteins in colorectal carcinoma: Its role in carcinogenesis and therapeutic perspectives. Cells 2021, 10, 2862. [Google Scholar] [CrossRef] [PubMed]
- Vukobrat-Bijedic, Z.; Husic-Selimovic, A.; Sofic, A.; Bijedic, N.; Bjelogrlic, I.; Gogov, B.; Mehmedovic, A. Cancer antigens (CEA and CA 19-9) as markers of advanced stage of colorectal carcinoma. Med. Arch. 2013, 67, 397. [Google Scholar] [CrossRef]
- Cardeñes, B.; Clares, I.; Toribio, V.; Pascual, L.; López-Martín, S.; Torres-Gomez, A.; Sainz de la Cuesta, R.; Lafuente, E.M.; López-Cabrera, M.; Yáñez-Mó, M. Cellular integrin α5β1 and exosomal ADAM17 mediate the binding and uptake of exosomes produced by colorectal carcinoma cells. Int. J. Mol. Sci. 2021, 22, 9938. [Google Scholar] [CrossRef]
- Chen, Y.; Xie, Y.; Xu, L.; Zhan, S.; Xiao, Y.; Gao, Y.; Wu, B.; Ge, W. Protein content and functional characteristics of serum-purified exosomes from patients with colorectal cancer revealed by quantitative proteomics. Int. J. Cancer 2017, 140, 900–913. [Google Scholar] [CrossRef]
- Campanella, C.; Rappa, F.; Sciumè, C.; Marino Gammazza, A.; Barone, R.; Bucchieri, F.; David, S.; Curcurù, G.; Caruso Bavisotto, C.; Pitruzzella, A. Heat shock protein 60 levels in tissue and circulating exosomes in human large bowel cancer before and after ablative surgery. Cancer 2015, 121, 3230–3239. [Google Scholar] [CrossRef]
- Calderwood, S.K.; Khaleque, M.A.; Sawyer, D.B.; Ciocca, D.R. Heat shock proteins in cancer: Chaperones of tumorigenesis. Trends Biochem. Sci. 2006, 31, 164–172. [Google Scholar] [CrossRef]
- Vafaei, S.; Fattahi, F.; Ebrahimi, M.; Janani, L.; Shariftabrizi, A.; Madjd, Z. Common molecular markers between circulating tumor cells and blood exosomes in colorectal cancer: A systematic and analytical review. Cancer Manag. Res. 2019, 2019, 8669–8698. [Google Scholar] [CrossRef]
- Wan, Y.H.; Liu, Q.S.; Wan, S.S.; Wang, R.W. Colorectal cancer-derived exosomes and modulation KRAS signaling. Clin. Transl. Oncol. 2022, 24, 2074–2080. [Google Scholar] [CrossRef]
- Beckler, M.D.; Higginbotham, J.N.; Franklin, J.L.; Ham, A.-J.; Halvey, P.J.; Imasuen, I.E.; Whitwell, C.; Li, M.; Liebler, D.C.; Coffey, R.J. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol. Cell. Proteom. 2013, 12, 343–355. [Google Scholar] [CrossRef]
- Shao, Y.; Chen, T.; Zheng, X.; Yang, S.; Xu, K.; Chen, X.; Xu, F.; Wang, L.; Shen, Y.; Wang, T. Colorectal cancer-derived small extracellular vesicles establish an inflammatory premetastatic niche in liver metastasis. Carcinogenesis 2018, 39, 1368–1379. [Google Scholar] [CrossRef]
- Deng, Z.-B.; Zhuang, X.; Ju, S.; Xiang, X.; Mu, J.; Liu, Y.; Jiang, H.; Zhang, L.; Mobley, J.; McClain, C. Exosome-like nanoparticles from intestinal mucosal cells carry prostaglandin E2 and suppress activation of liver NKT cells. J. Immunol. 2013, 190, 3579–3589. [Google Scholar] [CrossRef] [PubMed]
- Koga, Y.; Yasunaga, M.; Moriya, Y.; Akasu, T.; Fujita, S.; Yamamoto, S.; Matsumura, Y. Exosome can prevent RNase from degrading microRNA in feces. J. Gastrointest. Oncol. 2011, 2, 215. [Google Scholar] [PubMed]
- Yang, C.; Zhang, M.; Sung, J.; Wang, L.; Jung, Y.; Merlin, D. Isolation and characterization of exosomes from mouse feces. Bio-Protoc. 2020, 10, e3584. [Google Scholar] [CrossRef] [PubMed]
- Byts, N.; Makieieva, O.; Zhyvolozhnyi, A.; Bart, G.; Korvala, J.; Hekkala, J.; Salmi, S.; Samoylenko, A.; Reunanen, J. Purification of Bacterial-Enriched Extracellular Vesicle Samples from Feces by Density Gradient Ultracentrifugation. In Cell-Secreted Vesicles: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2023; pp. 211–226. [Google Scholar]
- Kumar, A.; Ren, Y.; Sundaram, K.; Mu, J.; Sriwastva, M.K.; Dryden, G.W.; Lei, C.; Zhang, L.; Yan, J.; Zhang, X. miR-375 prevents high-fat diet-induced insulin resistance and obesity by targeting the aryl hydrocarbon receptor and bacterial tryptophanase (tnaA) gene. Theranostics 2021, 11, 4061. [Google Scholar] [CrossRef]
- Martins, T.S.; Vaz, M.; Henriques, A.G. A review on comparative studies addressing exosome isolation methods from body fluids. Anal. Bioanal. Chem. 2023, 415, 1239–1263. [Google Scholar] [CrossRef] [PubMed]
- Baranyai, T.; Herczeg, K.; Onódi, Z.; Voszka, I.; Módos, K.; Marton, N.; Nagy, G.; Maeger, I.; Wood, M.J.; El Andaloussi, S. Isolation of exosomes from blood plasma: Qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS ONE 2015, 10, e0145686. [Google Scholar] [CrossRef]
- Huang, S.; Ji, X.; Jackson, K.K.; Lubman, D.M.; Ard, M.B.; Bruce, T.F.; Marcus, R.K. Rapid separation of blood plasma exosomes from low-density lipoproteins via a hydrophobic interaction chromatography method on a polyester capillary-channeled polymer fiber phase. Anal. Chim. Acta 2021, 1167, 338578. [Google Scholar] [CrossRef]
- Choi, D.S.; Kim, D.K.; Kim, Y.K.; Gho, Y.S. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics 2013, 13, 1554–1571. [Google Scholar] [CrossRef]
- Chuo, S.T.-Y.; Chien, J.C.-Y.; Lai, C.P.-K. Imaging extracellular vesicles: Current and emerging methods. J. Biomed. Sci. 2018, 25, 91. [Google Scholar] [CrossRef]
- Zabeo, D.; Cvjetkovic, A.; Lässer, C.; Schorb, M.; Lötvall, J.; Höög, J.L. Exosomes purified from a single cell type have diverse morphology. J. Extracell. Vesicles 2017, 6, 1329476. [Google Scholar] [CrossRef]
- Maas, S.L.; Broekman, M.L.; de Vrij, J. Tunable resistive pulse sensing for the characterization of extracellular vesicles. In Exosomes Microvesicles: Methods and Protocols; Springer New York: New York, NY, USA, 2017; pp. 21–33. [Google Scholar]
- Gercel-Taylor, C.; Atay, S.; Tullis, R.H.; Kesimer, M.; Taylor, D.D. Nanoparticle analysis of circulating cell-derived vesicles in ovarian cancer patients. Anal. Biochem. 2012, 428, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Belotti, Y.; Lim, C.T. Microfluidics for liquid biopsies: Recent advances, current challenges, and future directions. Anal. Chem. 2021, 93, 4727–4738. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Xia, L.; Zou, Z.; Yin, J.; Lin, N.; Mu, Y. Recent advances in integrated microfluidics for liquid biopsies and future directions. Biosens. Bioelectron. 2022, 2022, 114715. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, Y.; Lu, Y.; Luo, X.; Huang, Y.; Xie, T.; Pilarsky, C.; Dang, Y.; Zhang, J. Microfluidic Technology for the Isolation and Analysis of Exosomes. Micromachines 2022, 13, 1571. [Google Scholar] [CrossRef] [PubMed]
- Hassanpour Tamrin, S.; Sanati Nezhad, A.; Sen, A. Label-free isolation of exosomes using microfluidic technologies. ACS Nano 2021, 15, 17047–17079. [Google Scholar] [CrossRef]
- Havers, M.; Broman, A.; Lenshof, A.; Laurell, T. Advancement and obstacles in microfluidics-based isolation of extracellular vesicles. Anal. Bioanal. Chem. 2023, 415, 1265–1285. [Google Scholar] [CrossRef] [PubMed]
- Vergauwen, G.; Tulkens, J.; Pinheiro, C.; Avila Cobos, F.; Dedeyne, S.; De Scheerder, M.A.; Vandekerckhove, L.; Impens, F.; Miinalainen, I.; Braems, G. Robust sequential biophysical fractionation of blood plasma to study variations in the biomolecular landscape of systemically circulating extracellular vesicles across clinical conditions. J. Extracell. Vesicles 2021, 10, e12122. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.K.; Whiteside, T.L. Immunoaffinity-based isolation of melanoma cell-derived and T cell-derived exosomes from plasma of melanoma patients. In Melanoma: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2021; pp. 305–321. [Google Scholar]
- Han, Z.; Peng, C.; Yi, J.; Zhang, D.; Xiang, X.; Peng, X.; Su, B.; Liu, B.; Shen, Y.; Qiao, L. Highly efficient exosome purification from human plasma by tangential flow filtration based microfluidic chip. Sens. Actuators B Chem. 2021, 333, 129563. [Google Scholar] [CrossRef]
- Correll, V.L.; Otto, J.J.; Risi, C.M.; Main, B.P.; Boutros, P.C.; Kislinger, T.; Galkin, V.E.; Nyalwidhe, J.O.; Semmes, O.J.; Yang, L. Optimization of small extracellular vesicle isolation from expressed prostatic secretions in urine for in-depth proteomic analysis. J. Extracell. Vesicles 2022, 11, e12184. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.; Yu, H.; Yan, G.; Gao, M.; Sun, W.; Zhang, X. Characterization of urinary exosomes purified with size exclusion chromatography and ultracentrifugation. J. Proteome Res. 2020, 19, 2217–2225. [Google Scholar] [CrossRef] [PubMed]
- Shu, S.L.; Allen, C.L.; Benjamin-Davalos, S.; Koroleva, M.; MacFarland, D.; Minderman, H.; Ernstoff, M.S. A rapid exosome isolation using ultrafiltration and size exclusion chromatography (REIUS) method for exosome isolation from melanoma cell lines. In Melanoma: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2021; pp. 289–304. [Google Scholar]
- Kang, Y.-T.; Kim, Y.J.; Bu, J.; Cho, Y.-H.; Han, S.-W.; Moon, B.-I. High-purity capture and release of circulating exosomes using an exosome-specific dual-patterned immunofiltration (ExoDIF) device. Nanoscale 2017, 9, 13495–13505. [Google Scholar] [CrossRef]
- Iliuk, A.; Wu, X.; Li, L.; Sun, J.; Hadisurya, M.; Boris, R.S.; Tao, W.A. Plasma-derived extracellular vesicle phosphoproteomics through chemical affinity purification. J. Proteome Res. 2020, 19, 2563–2574. [Google Scholar] [CrossRef]
- Fang, X.; Duan, Y.; Adkins, G.B.; Pan, S.; Wang, H.; Liu, Y.; Zhong, W. Highly efficient exosome isolation and protein analysis by an integrated nanomaterial-based platform. Anal. Chem. 2018, 90, 2787–2795. [Google Scholar] [CrossRef]
- Lou, D.; Wang, Y.; Yang, Q.; Hu, L.; Zhu, Q. Ultrafiltration combing with phospholipid affinity-based isolation for metabolomic profiling of urinary extracellular vesicles. J. Chromatogr. A 2021, 1640, 461942. [Google Scholar] [CrossRef]
- Tayebi, M.; Zhou, Y.; Tripathi, P.; Chandramohanadas, R.; Ai, Y. Exosome purification and analysis using a facile microfluidic hydrodynamic trapping device. Anal. Chem. 2020, 92, 10733–10742. [Google Scholar] [CrossRef]
- Su, J.; Chen, S.; Dou, Y.; Zhao, Z.; Jia, X.; Ding, X.; Song, S. Smartphone-based electrochemical biosensors for directly detecting serum-derived exosomes and monitoring their secretion. Anal. Chem. 2022, 94, 3235–3244. [Google Scholar] [CrossRef] [PubMed]
- Sancho-Albero, M.; Sebastián, V.; Sesé, J.; Pazo-Cid, R.; Mendoza, G.; Arruebo, M.; Martín-Duque, P.; Santamaría, J. Isolation of exosomes from whole blood by a new microfluidic device: Proof of concept application in the diagnosis and monitoring of pancreatic cancer. J. Nanobiotechnol. 2020, 18, 150. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Cheng, L.; Hu, L.; Lou, D.; Zhang, T.; Li, J.; Zhu, Q.; Liu, F. An integrative microfluidic device for isolation and ultrasensitive detection of lung cancer-specific exosomes from patient urine. Biosens. Bioelectron. 2020, 163, 112290. [Google Scholar] [CrossRef]
- Kanwar, S.S.; Dunlay, C.J.; Simeone, D.M.; Nagrath, S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab A Chip 2014, 14, 1891–1900. [Google Scholar] [CrossRef]
- Zhang, P.; He, M.; Zeng, Y. Ultrasensitive microfluidic analysis of circulating exosomes using a nanostructured graphene oxide/polydopamine coating. Lab A Chip 2016, 16, 3033–3042. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, W.; Qiu, X.; Mei, Q.; Luo, Y.; Fu, W. Rapid and sensitive exosome detection with CRISPR/Cas12a. Anal. Bioanal. Chem. 2020, 412, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Sunkara, V.; Kim, C.-J.; Park, J.; Woo, H.-K.; Kim, D.; Ha, H.K.; Kim, M.-H.; Son, Y.; Kim, J.-R.; Cho, Y.-K. Fully automated, label-free isolation of extracellular vesicles from whole blood for cancer diagnosis and monitoring. Theranostics 2019, 9, 1851. [Google Scholar] [CrossRef]
- Li, Z.; Liu, C.; Cheng, Y.; Li, Y.; Deng, J.; Bai, L.; Qin, L.; Mei, H.; Zeng, M.; Tian, F. Cascaded microfluidic circuits for pulsatile filtration of extracellular vesicles from whole blood for early cancer diagnosis. Sci. Adv. 2023, 9, eade2819. [Google Scholar] [CrossRef]
- Liu, F.; Vermesh, O.; Mani, V.; Ge, T.J.; Madsen, S.J.; Sabour, A.; Hsu, E.-C.; Gowrishankar, G.; Kanada, M.; Jokerst, J.V. The exosome total isolation chip. ACS Nano 2017, 11, 10712–10723. [Google Scholar] [CrossRef]
- Zhou, Y.; Ma, Z.; Tayebi, M.; Ai, Y. Submicron particle focusing and exosome sorting by wavy microchannel structures within viscoelastic fluids. Anal. Chem. 2019, 91, 4577–4584. [Google Scholar] [CrossRef]
- Ayala-Mar, S.; Perez-Gonzalez, V.H.; Mata-Gómez, M.A.; Gallo-Villanueva, R.C.; González-Valdez, J. Electrokinetically driven exosome separation and concentration using dielectrophoretic-enhanced PDMS-based microfluidics. Anal. Chem. 2019, 91, 14975–14982. [Google Scholar] [CrossRef]
- Le, M.-C.N.; Fan, Z.H. Exosome isolation using nanostructures and microfluidic devices. Biomed. Mater. 2021, 16, 022005. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, H.-j.; Fine, D.; Schmulen, J.; Hu, Y.; Godin, B.; Zhang, J.X.; Liu, X. Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles. Lab A Chip 2013, 13, 2879–2882. [Google Scholar] [CrossRef]
- Yasui, T.; Yanagida, T.; Ito, S.; Konakade, Y.; Takeshita, D.; Naganawa, T.; Nagashima, K.; Shimada, T.; Kaji, N.; Nakamura, Y. Unveiling massive numbers of cancer-related urinary-microRNA candidates via nanowires. Sci. Adv. 2017, 3, e1701133. [Google Scholar] [CrossRef]
- Yeo, J.C.; Kenry, K.; Zhao, Z.; Zhang, P.; Wang, Z.; Lim, C.T. Label-free extraction of extracellular vesicles using centrifugal microfluidics. Biomicrofluidics 2018, 12, 024103. [Google Scholar] [CrossRef]
- Liu, C.; Guo, J.; Tian, F.; Yang, N.; Yan, F.; Ding, Y.; Wei, J.; Hu, G.; Nie, G.; Sun, J. Field-free isolation of exosomes from extracellular vesicles by microfluidic viscoelastic flows. ACS Nano 2017, 11, 6968–6976. [Google Scholar] [CrossRef]
- Wunsch, B.H.; Smith, J.T.; Gifford, S.M.; Wang, C.; Brink, M.; Bruce, R.L.; Austin, R.H.; Stolovitzky, G.; Astier, Y. Nanoscale lateral displacement arrays for the separation of exosomes and colloids down to 20 nm. Nat. Nanotechnol. 2016, 11, 936–940. [Google Scholar] [CrossRef] [PubMed]
- Santana, S.M.; Antonyak, M.A.; Cerione, R.A.; Kirby, B.J. Microfluidic isolation of cancer-cell-derived microvesicles from hetergeneous extracellular shed vesicle populations. Biomed. Microdevices 2014, 16, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.T.; Wunsch, B.H.; Dogra, N.; Ahsen, M.E.; Lee, K.; Yadav, K.K.; Weil, R.; Pereira, M.A.; Patel, J.V.; Duch, E.A. Integrated nanoscale deterministic lateral displacement arrays for separation of extracellular vesicles from clinically-relevant volumes of biological samples. Lab A Chip 2018, 18, 3913–3925. [Google Scholar] [CrossRef]
- Cho, S.; Jo, W.; Heo, Y.; Kang, J.Y.; Kwak, R.; Park, J. Isolation of extracellular vesicle from blood plasma using electrophoretic migration through porous membrane. Sens. Actuators B Chem. 2016, 233, 289–297. [Google Scholar] [CrossRef]
- Chen, H.; Yamakawa, T.; Inaba, M.; Nakano, M.; Suehiro, J. Characterization of extra-cellular vesicle dielectrophoresis and estimation of its electric properties. Sensors 2022, 22, 3279. [Google Scholar] [CrossRef]
- Zhang, H.; Chang, H.; Neuzil, P. DEP-on-a-chip: Dielectrophoresis applied to microfluidic platforms. Micromachines 2019, 10, 423. [Google Scholar] [CrossRef]
- Shi, L.; Rana, A.; Esfandiari, L. A low voltage nanopipette dielectrophoretic device for rapid entrapment of nanoparticles and exosomes extracted from plasma of healthy donors. Sci. Rep. 2018, 8, 6751. [Google Scholar] [CrossRef] [PubMed]
- Narji, N.F.N.M.; Ahmad, M.R. Dielectrophoresis-based microfluidic device for separation of potential cancer cells. Bull. Electr. Eng. Inform. 2020, 9, 2270–2277. [Google Scholar] [CrossRef]
- Ibsen, S.D.; Wright, J.; Lewis, J.M.; Kim, S.; Ko, S.-Y.; Ong, J.; Manouchehri, S.; Vyas, A.; Akers, J.; Chen, C.C. Rapid isolation and detection of exosomes and associated biomarkers from plasma. ACS Nano 2017, 11, 6641–6651. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Shao, H.; Weissleder, R.; Lee, H. Acoustic purification of extracellular microvesicles. ACS Nano 2015, 9, 2321–2327. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Ouyang, Y.; Wang, Z.; Zhang, R.; Huang, P.-H.; Chen, C.; Li, H.; Li, P.; Quinn, D.; Dao, M. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. USA 2017, 114, 10584–10589. [Google Scholar] [CrossRef] [PubMed]
- Evander, M.; Gidlöf, O.; Olde, B.; Erlinge, D.; Laurell, T. Non-contact acoustic capture of microparticles from small plasma volumes. Lab A Chip 2015, 15, 2588–2596. [Google Scholar] [CrossRef] [PubMed]
- Yeo, L.Y.; Friend, J.R. Surface acoustic wave microfluidics. Annu. Rev. Fluid Mech. 2014, 46, 379–406. [Google Scholar] [CrossRef]
- Destgeer, G.; Sung, H.J. Recent advances in microfluidic actuation and micro-object manipulation via surface acoustic waves. Lab A Chip 2015, 15, 2722–2738. [Google Scholar] [CrossRef]
- Wang, Z.; Li, F.; Rufo, J.; Chen, C.; Yang, S.; Li, L.; Zhang, J.; Cheng, J.; Kim, Y.; Wu, M. Acoustofluidic salivary exosome isolation: A liquid biopsy compatible approach for human papillomavirus–associated oropharyngeal cancer detection. J. Mol. Diagn. 2020, 22, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Schachermeyer, S.; Zhong, W. Flow Field-Flow Fractionation: Analysis of Biomolecules and Their Complexes; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Zhang, H.; Lyden, D. Asymmetric-flow field-flow fractionation technology for exomere and small extracellular vesicle separation and characterization. Nat. Protoc. 2019, 14, 1027–1053. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Shen, S.; Lee, S.; Dou, H. Field-flow fractionation: A gentle separation and characterization technique in biomedicine. TrAC Trends Anal. Chem. 2018, 108, 231–238. [Google Scholar] [CrossRef]
- Wu, B.; Chen, X.; Wang, J.; Qing, X.; Wang, Z.; Ding, X.; Xie, Z.; Niu, L.; Guo, X.; Cai, T. Separation and characterization of extracellular vesicles from human plasma by asymmetrical flow field-flow fractionation. Anal. Chim. Acta 2020, 1127, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Sitar, S.; Kejžar, A.; Pahovnik, D.; Kogej, K.; Tušek-Žnidarič, M.; Lenassi, M.; Žagar, E. Size characterization and quantification of exosomes by asymmetrical-flow field-flow fractionation. Anal. Chem. 2015, 87, 9225–9233. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.S.; Kim, J.Y.; Lee, J.C.; Moon, M.H. Investigation of lipidomic perturbations in oxidatively stressed subcellular organelles and exosomes by asymmetrical flow field–flow fractionation and nanoflow ultrahigh performance liquid chromatography–tandem mass spectrometry. Anal. Chim. Acta 2019, 1073, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.B.; Lee, G.B.; Moon, M.H. Size separation of exosomes and microvesicles using flow field-flow fractionation/multiangle light scattering and lipidomic comparison. Anal. Chem. 2022, 94, 8958–8965. [Google Scholar] [CrossRef]
- Kim, Y.B.; Yang, J.S.; Lee, G.B.; Moon, M.H. Evaluation of exosome separation from human serum by frit-inlet asymmetrical flow field-flow fractionation and multiangle light scattering. Anal. Chim. Acta 2020, 1124, 137–145. [Google Scholar] [CrossRef]
- Yang, J.S.; Lee, J.C.; Byeon, S.K.; Rha, K.H.; Moon, M.H. Size Dependent Lipidomic Analysis of Urinary Exosomes from Patients with Prostate Cancer by Flow Field-Flow Fractionation and Nanoflow Liquid Chromatography-Tandem Mass Spectrometry. Anal. Chem. 2017, 89, 2488–2496. [Google Scholar] [CrossRef]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef]
- Park, I.; Paeng, K.J.; Kang, D.; Moon, M.H. Performance of hollow-fiber flow field-flow fractionation in protein separation. J. Sep. Sci. 2005, 28, 2043–2049. [Google Scholar] [CrossRef]
- Zattoni, A.; Rambaldi, D.C.; Roda, B.; Parisi, D.; Roda, A.; Moon, M.H.; Reschiglian, P. Hollow-fiber flow field-flow fractionation of whole blood serum. J. Chromatogr. A 2008, 1183, 135–142. [Google Scholar] [CrossRef]
- Reschiglian, P.; Zattoni, A.; Roda, B.; Cinque, L.; Parisi, D.; Roda, A.; Dal Piaz, F.; Moon, M.H.; Min, B.R. On-line hollow-fiber flow field-flow fractionation-electrospray ionization/time-of-flight mass spectrometry of intact proteins. Anal. Chem. 2005, 77, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Zattoni, A.; Casolari, S.; Rambaldi, D.C.; Reschiglian, P. Hollow-fiber flow field-flow fractionation. Curr. Anal. Chem. 2007, 3, 310–323. [Google Scholar] [CrossRef]
- Marassi, V.; Roda, B.; Zattoni, A.; Tanase, M.; Reschiglian, P. Hollow fiber flow field-flow fractionation and size-exclusion chromatography with multi-angle light scattering detection: A complementary approach in biopharmaceutical industry. J. Chromatogr. A 2014, 1372, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Tanase, M.; Urbanska, A.M.; Zolla, V.; Clement, C.C.; Huang, L.; Morozova, K.; Follo, C.; Goldberg, M.; Roda, B.; Reschiglian, P. Role of carbonyl modifications on aging-associated protein aggregation. Sci. Rep. 2016, 6, 19311. [Google Scholar] [CrossRef] [PubMed]
- Marassi, V.; Beretti, F.; Roda, B.; Alessandrini, A.; Facci, P.; Maraldi, T.; Zattoni, A.; Reschiglian, P.; Portolani, M. A new approach for the separation, characterization and testing of potential prionoid protein aggregates through hollow-fiber flow field-flow fractionation and multi-angle light scattering. Anal. Chim. Acta 2019, 1087, 121–130. [Google Scholar] [CrossRef]
- Fukuda, J.; Iwura, T.; Yanagihara, S.; Kano, K. Separation and quantification of monoclonal-antibody aggregates by hollow-fiber-flow field-flow fractionation. Anal. Bioanal. Chem. 2014, 406, 6257–6264. [Google Scholar] [CrossRef] [PubMed]
- Marassi, V.; Maggio, S.; Battistelli, M.; Stocchi, V.; Zattoni, A.; Reschiglian, P.; Guescini, M.; Roda, B. An ultracentrifugation–hollow-fiber flow field-flow fractionation orthogonal approach for the purification and mapping of extracellular vesicle subtypes. J. Chromatogr. A 2021, 1638, 461861. [Google Scholar] [CrossRef]
- Kibria, G.; Ramos, E.K.; Lee, K.E.; Bedoyan, S.; Huang, S.; Samaeekia, R.; Athman, J.J.; Harding, C.V.; Lötvall, J.; Harris, L. A rapid, automated surface protein profiling of single circulating exosomes in human blood. Sci. Rep. 2016, 6, 36502. [Google Scholar] [CrossRef]
- Jia, Y.; Ni, Z.; Sun, H.; Wang, C. Microfluidic approaches toward the isolation and detection of exosome nanovesicles. Ieee Access 2019, 7, 45080–45098. [Google Scholar] [CrossRef]
- Zhou, Y.; Mahapatra, C.; Chen, H.; Peng, X.; Ramakrishna, S.; Nanda, H.S. Recent developments in fluorescent aptasensors for detection of antibiotics. Curr. Opin. Biomed. Eng. 2020, 13, 16–24. [Google Scholar] [CrossRef]
- Roda, A.; Mirasoli, M.; Michelini, E.; Di Fusco, M.; Zangheri, M.; Cevenini, L.; Roda, B.; Simoni, P. Progress in chemical luminescence-based biosensors: A critical review. Biosens. Bioelectron. 2016, 76, 164–179. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Han, H.; Naw, H.P.P.; Lammy, A.V.; Goh, C.H.; Boujday, S.; Steele, T.W. Real-time colorimetric hydration sensor for sport activities. Mater. Des. 2016, 90, 1181–1185. [Google Scholar] [CrossRef]
- Wang, Q.-L.; Huang, W.-X.; Zhang, P.-J.; Chen, L.; Lio, C.-K.; Zhou, H.; Qing, L.-S.; Luo, P. Colorimetric determination of the early biomarker hypoxia-inducible factor-1 alpha (HIF-1α) in circulating exosomes by using a gold seed-coated with aptamer-functionalized Au@ Au core-shell peroxidase mimic. Microchim. Acta 2020, 187, 61. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Liu, M.; Wang, L.; Yan, A.; He, W.; Chen, M.; Lan, J.; Xu, J.; Guan, L.; Chen, J. A visible and colorimetric aptasensor based on DNA-capped single-walled carbon nanotubes for detection of exosomes. Biosens. Bioelectron. 2017, 92, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Roda, A.; Guardigli, M. Analytical chemiluminescence and bioluminescence: Latest achievements and new horizons. Anal. Bioanal. Chem. 2012, 402, 69–76. [Google Scholar] [CrossRef]
- Arakawa, T.; Prestrelski, S.J.; Kenney, W.C.; Carpenter, J.F. Factors affecting short-term and long-term stabilities of proteins. Adv. Drug Deliv. Rev. 2001, 46, 307–326. [Google Scholar] [CrossRef]
- Babamiri, B.; Bahari, D.; Salimi, A. Highly sensitive bioaffinity electrochemiluminescence sensors: Recent advances and future directions. Biosens. Bioelectron. 2019, 142, 111530. [Google Scholar] [CrossRef]
- Cheng, N.; Du, D.; Wang, X.; Liu, D.; Xu, W.; Luo, Y.; Lin, Y. Recent advances in biosensors for detecting cancer-derived exosomes. Trends Biotechnol. 2019, 37, 1236–1254. [Google Scholar] [CrossRef]
- Singh, A.; Sharma, A.; Ahmed, A.; Sundramoorthy, A.K.; Furukawa, H.; Arya, S.; Khosla, A. Recent advances in electrochemical biosensors: Applications, challenges, and future scope. Biosensors 2021, 11, 336. [Google Scholar] [CrossRef] [PubMed]
- Kuntamung, K.; Jakmunee, J.; Ounnunkad, K. A label-free multiplex electrochemical biosensor for the detection of three breast cancer biomarker proteins employing dye/metal ion-loaded and antibody-conjugated polyethyleneimine-gold nanoparticles. J. Mater. Chem. B 2021, 9, 6576–6585. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Chen, H.; Zhou, Y.; Wu, J.; Ramakrishna, S.; Peng, X.; Nanda, H.S.; Zhou, Y. Recent advances in biosensors for detection of exosomes. Curr. Opin. Biomed. Eng. 2021, 18, 100280. [Google Scholar] [CrossRef]
- Umme, S.; Siciliano, G.; Primiceri, E.; Turco, A.; Tarantini, I.; Ferrara, F.; Chiriacò, M.S. Electrochemical Sensors for Liquid Biopsy and Their Integration into Lab-on-Chip Platforms: Revolutionizing the Approach to Diseases. Chemosensors 2023, 11, 517. [Google Scholar] [CrossRef]
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface plasmon resonance: A versatile technique for biosensor applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef]
- Bellassai, N.; D’Agata, R.; Jungbluth, V.; Spoto, G. Surface plasmon resonance for biomarker detection: Advances in non-invasive cancer diagnosis. Front. Chem. 2019, 7, 570. [Google Scholar] [CrossRef]
- Fleischmann, M.; Hendra, P.J.; McQuillan, A.J. Raman spectra of pyridine adsorbed at a silver electrode. Chem. Phys. Lett. 1974, 26, 163–166. [Google Scholar] [CrossRef]
- Szlag, V.M.; Rodriguez, R.S.; He, J.; Hudson-Smith, N.; Kang, H.; Le, N.; Reineke, T.M.; Haynes, C.L. Molecular affinity agents for intrinsic surface-enhanced Raman scattering (SERS) sensors. ACS Appl. Mater. Interfaces 2018, 10, 31825–31844. [Google Scholar] [CrossRef]
- Tahir, M.A.; Dina, N.E.; Cheng, H.; Valev, V.K.; Zhang, L. Surface-enhanced Raman spectroscopy for bioanalysis and diagnosis. Nanoscale 2021, 13, 11593–11634. [Google Scholar] [CrossRef]
- Yaraki, M.T.; Tukova, A.; Wang, Y. Emerging SERS biosensors for the analysis of cells and extracellular vesicles. Nanoscale 2022. [Google Scholar]
- Shin, Y.-H.; Gutierrez-Wing, M.T.; Choi, J.-W. Recent progress in portable fluorescence sensors. J. Electrochem. Soc. 2021, 168, 017502. [Google Scholar] [CrossRef]
- Esmaelpourfarkhani, M.; Ramezani, M.; Alibolandi, M.; Abnous, K.; Taghdisi, S.M. Time-resolved Fluorescence DNA-based Sensors for Reducing Background Fluorescence of Environment. J. Fluoresc. 2023, 33, 2145–2160. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.-X.; Xue, S.-F.; Chen, Z.-H.; Ma, S.-H.; Zhang, S.; Shi, G.; Zhang, M. Dual lanthanide-doped complexes: The development of a time-resolved ratiometric fluorescent probe for anthrax biomarker and a paper-based visual sensor. Biosens. Bioelectron. 2017, 94, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Meng, H.-M.; An, Y.; Geng, X.; Zhao, K.; Qu, L.; Li, Z. Structure-switching aptamer triggering hybridization displacement reaction for label-free detection of exosomes. Talanta 2020, 209, 120510. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Meng, L.; Ye, W.; Wang, Q.; Geng, S.; Sun, C. A sensitive detection assay based on signal amplification technology for Alzheimer’s disease’s early biomarker in exosome. Anal. Chim. Acta 2018, 1022, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Hummelen, J.C.; Luider, T.M.; Wynberg, H. [39] Stable 1, 2-dioxetanes as labels for thermochemiluminescent immunoassay. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1986; Volume 133, pp. 531–557. [Google Scholar]
- Di Fusco, M.; Quintavalla, A.; Lombardo, M.; Guardigli, M.; Mirasoli, M.; Trombini, C.; Roda, A. Organically modified silica nanoparticles doped with new acridine-1, 2-dioxetane analogues as thermochemiluminescence reagentless labels for ultrasensitive immunoassays. Anal. Bioanal. Chem. 2015, 407, 1567–1576. [Google Scholar] [CrossRef]
- Andronico, L.A.; Chen, L.; Mirasoli, M.; Guardigli, M.; Quintavalla, A.; Lombardo, M.; Trombini, C.; Chiu, D.T.; Roda, A. Thermochemiluminescent semiconducting polymer dots as sensitive nanoprobes for reagentless immunoassay. Nanoscale 2018, 10, 14012–14021. [Google Scholar] [CrossRef]
- Roda, A.; Zangheri, M.; Calabria, D.; Mirasoli, M.; Caliceti, C.; Quintavalla, A.; Lombardo, M.; Trombini, C.; Simoni, P. A simple smartphone-based thermochemiluminescent immunosensor for valproic acid detection using 1, 2-dioxetane analogue-doped nanoparticles as a label. Sens. Actuators B Chem. 2019, 279, 327–333. [Google Scholar] [CrossRef]
- Moroni, G.; Calabria, D.; Quintavalla, A.; Lombardo, M.; Mirasoli, M.; Roda, A.; Gioiello, A. Thermochemiluminescence-Based Sensitive Probes: Synthesis and Photophysical Characterization of Acridine-Containing 1, 2-Dioxetanes Focusing on Fluorophore Push-Pull Effects. ChemPhotoChem 2022, 6, e202100152. [Google Scholar] [CrossRef]
- Heyer, E.; Lory, P.; Leprince, J.; Moreau, M.; Romieu, A.; Guardigli, M.; Roda, A.; Ziessel, R. Highly Fluorescent and Water-Soluble Diketopyrrolopyrrole Dyes for Bioconjugation. Angew. Chem. 2015, 127, 3038–3042. [Google Scholar] [CrossRef]
- Coumans, F.A.; Brisson, A.R.; Buzas, E.I.; Dignat-George, F.; Drees, E.E.; El-Andaloussi, S.; Emanueli, C.; Gasecka, A.; Hendrix, A.; Hill, A.F. Methodological guidelines to study extracellular vesicles. Circ. Res. 2017, 120, 1632–1648. [Google Scholar] [CrossRef]
- Marassi, V.; Mattarozzi, M.; Toma, L.; Giordani, S.; Ronda, L.; Roda, B.; Zattoni, A.; Reschiglian, P.; Careri, M. FFF-based high-throughput sequence shortlisting to support the development of aptamer-based analytical strategies. Anal. Bioanal. Chem. 2022, 414, 5519–5527. [Google Scholar] [CrossRef]
- Marassi, V.; Giordani, S.; Reschiglian, P.; Roda, B.; Zattoni, A. Tracking heme-protein interactions in healthy and pathological human serum in native conditions by miniaturized FFF-multidetection. Appl. Sci. 2022, 12, 6762. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Patel, G.K.; Laurini, J.A.; Roveda, K.; Tan, M.C.; Patton, M.C.; Singh, S.; Taylor, W.; Singh, A.P. Exosomal Markers (CD63 and CD9) expression pattern using immunohistochemistry in resected malignant and non-malignant pancreatic specimens. Pancreas 2017, 46, 782. [Google Scholar]
- Rontogianni, S.; Synadaki, E.; Li, B.; Liefaard, M.C.; Lips, E.H.; Wesseling, J.; Wu, W.; Altelaar, M. Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun. Biol. 2019, 2, 325. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, M.; Djeghlaf, L.; Bataille, J.; Gamby, J.; Haghiri-Gosnet, A.M.; Pallandre, A. Logic digital fluidic in miniaturized functional devices: Perspective to the next generation of microfluidic lab-on-chips. Electrophoresis 2017, 38, 953–976. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Lan, Z.; Xu, L.; Zhu, Z.; Shu, X. A miniaturized apparatus based on a smartphone for microsecond-resolved luminescence lifetime imaging. Sens. Actuators B Chem. 2021, 343, 130086. [Google Scholar] [CrossRef]



| Biosensing Approach | Advantages | Limitations |
|---|---|---|
| Optical | Rapid response, easy to use, inexpensive, qualitative naked-eye detection, POC capability. | Low sensitivity, limited multiplexing capability. |
| EC | High sensitivity, rapid response, Inexpensive, multiplexing capability, reagent-less | Challenging surface functionalization, matrix effect, reproducibility problems |
| Fluorescence | High sensibility, rapid response, multiplex capability, reagent-less | Complex instrumentation required, high background for complex bio samples |
| Fluorescence (Time resolved) | High sensibility, rapid response, high selectivity, multiplex capability, reagent-less | Complex instrumentation required. |
| CL/BL | High sensibility, rapid response, POC capability | Reagent-dependent, |
| ECL | Higher s/n and specificity compared to CL | Challenging miniaturization, reagent-dependent |
| TCL | Reagent-less | Developmental stage technology |
| SPR | High sensitivity, real-time detection, label-free system | Non-specific absorption, proof of concept state, complex equipment required |
| SERS (Label-aided) | Superior sensitivity, multiplexing capability, simple manufacturing | Costly equipment, difficult data analysis |
| SERS (Label-free) | Provide additional structural information of the analyte | Very complex data analysis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marassi, V.; Giordani, S.; Placci, A.; Punzo, A.; Caliceti, C.; Zattoni, A.; Reschiglian, P.; Roda, B.; Roda, A. Emerging Microfluidic Tools for Simultaneous Exosomes and Cargo Biosensing in Liquid Biopsy: New Integrated Miniaturized FFF-Assisted Approach for Colon Cancer Diagnosis. Sensors 2023, 23, 9432. https://doi.org/10.3390/s23239432
Marassi V, Giordani S, Placci A, Punzo A, Caliceti C, Zattoni A, Reschiglian P, Roda B, Roda A. Emerging Microfluidic Tools for Simultaneous Exosomes and Cargo Biosensing in Liquid Biopsy: New Integrated Miniaturized FFF-Assisted Approach for Colon Cancer Diagnosis. Sensors. 2023; 23(23):9432. https://doi.org/10.3390/s23239432
Chicago/Turabian StyleMarassi, Valentina, Stefano Giordani, Anna Placci, Angela Punzo, Cristiana Caliceti, Andrea Zattoni, Pierluigi Reschiglian, Barbara Roda, and Aldo Roda. 2023. "Emerging Microfluidic Tools for Simultaneous Exosomes and Cargo Biosensing in Liquid Biopsy: New Integrated Miniaturized FFF-Assisted Approach for Colon Cancer Diagnosis" Sensors 23, no. 23: 9432. https://doi.org/10.3390/s23239432
APA StyleMarassi, V., Giordani, S., Placci, A., Punzo, A., Caliceti, C., Zattoni, A., Reschiglian, P., Roda, B., & Roda, A. (2023). Emerging Microfluidic Tools for Simultaneous Exosomes and Cargo Biosensing in Liquid Biopsy: New Integrated Miniaturized FFF-Assisted Approach for Colon Cancer Diagnosis. Sensors, 23(23), 9432. https://doi.org/10.3390/s23239432










