ECG Synthesis via Diffusion-Based State Space Augmented Transformer
Abstract
:1. Introduction
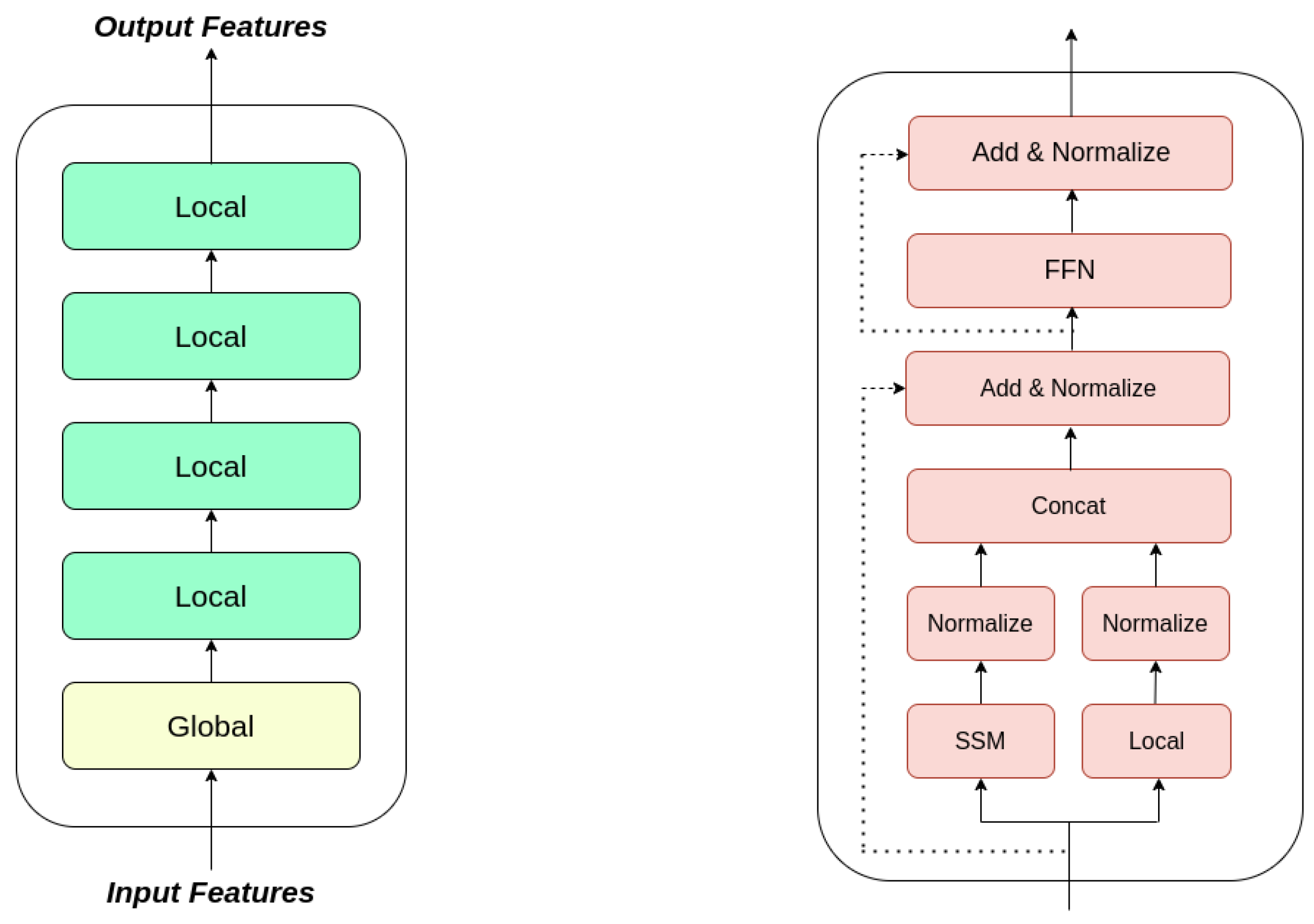
- Introduced a diffusion model for generating concise (10 s) 12-lead ECGs, incorporating a State Space Augmented Transformer [27] as a pivotal internal component.
- Enabled conditional generation of ECG samples, for different heart rhythms in a multilabel setting.
- Evaluated the quality of generated samples using metrics such as Dynamic Time Warping (DTW) and Maximum Mean Discrepancy (MMD). Additionally, to ensure authenticity, we evaluated how similar the performance of a pre-trained classifier is when applied to both generated and real ECG samples.
2. Related Work
3. Materials and Methods
3.1. Dataset
3.2. Diffusion Models
3.3. Structured State Space Models
3.4. State Space Augmented Transformer
3.5. Proposed Approach: DSAT-ECG
4. Results
4.1. Training Settings
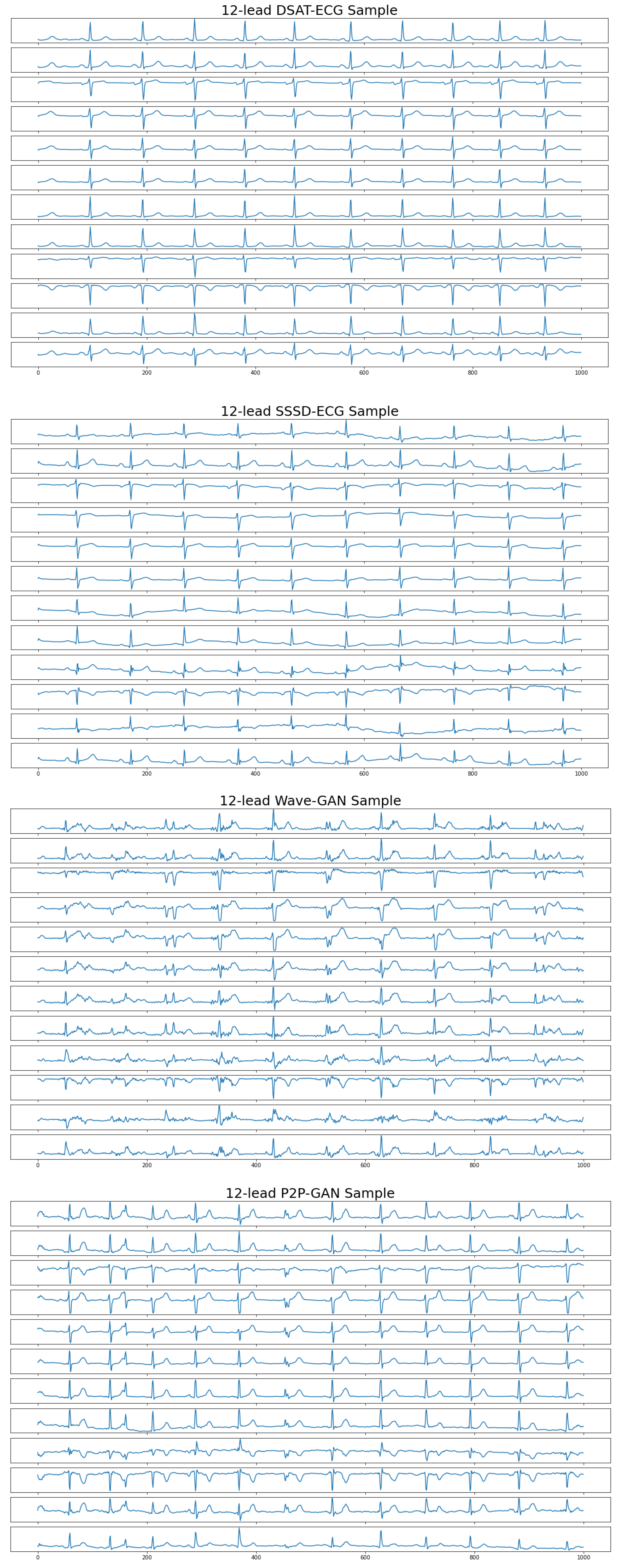
4.2. Quality Evaluation
4.3. Authenticity Evaluation
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hong, S.; Zhou, Y.; Shang, J.; Xiao, C.; Sun, J. Opportunities and challenges of deep learning methods for electrocardiogram data: A systematic review. Comp. Biol. Med. 2020, 122, 103801. [Google Scholar] [CrossRef] [PubMed]
- Petmezas, G.; Stefanopoulos, L.; Kilintzis, V.; Tzavelis, A.; Rogers, J.A.; Katsaggelos, A.K.; Maglaveras, N. State-of-the-Art Deep Learning Methods on Electrocardiogram Data: Systematic Review. JMIR Med. Inform. 2022, 10, e38454. [Google Scholar] [CrossRef]
- Wagner, P.; Strodthoff, N.; Bousseljot, R.D.; Samek, W.; Schaeffter, T. PTB-XL, a Large Publicly Available Electrocardiography Dataset. (Version 1.0.3). 2022. Available online: https://physionet.org/content/ptb-xl/1.0.3/ (accessed on 8 September 2023).
- Wagner, P.; Strodthoff, N.; Bousseljot, R.D.; Kreiseler, D.; Lunze, F.I.; Samek, W.; Schaeffter, T. PTB-XL, a large publicly available electrocardiography dataset. Sci. Data 2020, 7, 154. [Google Scholar] [CrossRef] [PubMed]
- Goldberger, A.L.; Amaral, L.A.N.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet. Circulation 2000, 101, E215–E220. [Google Scholar] [CrossRef] [PubMed]
- Mehari, T.; Strodthoff, N. Self-supervised representation learning from 12-lead ECG data. Comp. Biol. Med. 2021, 141, 105114. [Google Scholar] [CrossRef]
- Mehari, T.; Strodthoff, N. Advancing the State-of-the-Art for ECG Analysis through Structured State Space Models. arXiv 2022, arXiv:2211.07579. [Google Scholar]
- Xu, J.; Glicksberg, B.S.; Su, C.; Walker, P.; Bian, J.; Wang, F. Federated Learning for Healthcare Informatics. J. Healthcare Inform. Res. 2020, 5, 1–19. [Google Scholar] [CrossRef]
- Yin, H.; Mallya, A.; Vahdat, A.; Álvarez, J.M.; Kautz, J.; Molchanov, P. See through Gradients: Image Batch Recovery via GradInversion. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Nashville, TN, USA, 20–25 June 2021; pp. 16332–16341. [Google Scholar]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. arXiv 2014, arXiv:stat.ML/1406.2661. [Google Scholar] [CrossRef]
- Delaney, A.M.; Brophy, E.; Ward, T.E. Synthesis of Realistic ECG using Generative Adversarial Networks. arXiv 2019, arXiv:eess.SP/1909.09150. [Google Scholar]
- Zhu, F.; Fei, Y.; Fu, Y.; Liu, Q.; Shen, B. Electrocardiogram generation with a bidirectional LSTM-CNN generative adversarial network. Sci. Rep. 2019, 9, 6734. [Google Scholar] [CrossRef]
- Golany, T.; Lavee, G.; Yarden, S.; Radinsky, K. Improving ECG Classification Using Generative Adversarial Networks. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; Volume 34, pp. 13280–13285. [Google Scholar] [CrossRef]
- Rafi, T.H.; Woong Ko, Y. HeartNet: Self Multihead Attention Mechanism via Convolutional Network With Adversarial Data Synthesis for ECG-Based Arrhythmia Classification. IEEE Access 2022, 10, 100501–100512. [Google Scholar] [CrossRef]
- Li, X.; Metsis, V.; Wang, H.; Ngu, A. TTS-GAN: A Transformer-Based Time-Series Generative Adversarial Network; Springer: Berlin, Germnay, 2022; pp. 133–143. [Google Scholar] [CrossRef]
- Golany, T.; Freedman, D.; Radinsky, K. ECG ODE-GAN: Learning Ordinary Differential Equations of ECG Dynamics via Generative Adversarial Learning. In Proceedings of the AAAI Conference on Artificial Intelligence, Washington, DC, USA, 7–14 February 2021; Volume 35, pp. 134–141. [Google Scholar] [CrossRef]
- Chen, J.; Liao, K.; Wei, K.; Ying, H.; Chen, D.Z.; Wu, J. ME-GAN: Learning Panoptic Electrocardio Representations for Multi-view ECG Synthesis Conditioned on Heart Diseases. In Proceedings of the 39th International Conference on Machine Learning, Baltimore, MA, USA, 17–23 July 2022; Volume 162, pp. 3360–3370. [Google Scholar]
- Neifar, N.; Mdhaffar, A.; Ben-Hamadou, A.; Jmaiel, M.; Freisleben, B. Disentangling temporal and amplitude variations in ECG synthesis using anchored GANs. In Proceedings of the 37th ACM/SIGAPP Symposium on Applied Computing, Tallinn, Estonia, 27–31 March 2023; pp. 645–652. [Google Scholar] [CrossRef]
- Golany, T.; Radinsky, K.; Freedman, D. SimGANs: Simulator-Based Generative Adversarial Networks for ECG Synthesis to Improve Deep ECG Classification. In Proceedings of the 37th International Conference on Machine Learning, Virtual Event, 13–18 July 2020; pp. 3597–3606. [Google Scholar]
- Golany, T.; Radinsky, K. PGANs: Personalized Generative Adversarial Networks for ECG Synthesis to Improve Patient-Specific Deep ECG Classification. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 557–564. [Google Scholar] [CrossRef]
- Thambawita, V.; Isaksen, J.; Hicks, S.; Ghouse, J.; Ahlberg, G.; Linneberg, A.; Grarup, N.; Ellervik, C.; Olesen, M.; Hansen, T.; et al. DeepFake electrocardiograms using generative adversarial networks are the beginning of the end for privacy issues in medicine. Sci. Rep. 2021, 11, 21896. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Tang, Y.M.; Yu, K.M.; To, S. SLC-GAN: An automated myocardial infarction detection model based on generative adversarial networks and convolutional neural networks with single-lead electrocardiogram synthesis. Inform. Sci. 2022, 589, 738–750. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, Y.; Cai, X.; Yuan, X. E²GAN: End-to-End Generative Adversarial Network for Multivariate Time Series Imputation. In Proceedings of the Twenty-Eighth International Joint Conference on Artificial Intelligence, IJCAI-19, International Joint Conferences on Artificial Intelligence Organization, Macao, China, 10–16 August 2019; pp. 3094–3100. [Google Scholar] [CrossRef]
- Sohl-Dickstein, J.; Weiss, E.A.; Maheswaranathan, N.; Ganguli, S. Deep Unsupervised Learning using Nonequilibrium Thermodynamics. arXiv 2015, arXiv:cs.LG/1503.03585. [Google Scholar]
- Mukhopadhyay, S.; Gwilliam, M.; Agarwal, V.; Padmanabhan, N.; Swaminathan, A.; Hegde, S.; Zhou, T.; Shrivastava, A. Diffusion Models Beat GANs on Image Classification. arXiv 2023, arXiv:cs.CV/2307.08702. [Google Scholar]
- Ho, J.; Salimans, T.; Gritsenko, A.; Chan, W.; Norouzi, M.; Fleet, D.J. Video Diffusion Models. arXiv 2022, arXiv:cs.CV/2204.03458. [Google Scholar]
- Zuo, S.; Liu, X.; Jiao, J.; Charles, D.; Manavoglu, E.; Zhao, T.; Gao, J. Efficient Long Sequence Modeling via State Space Augmented Transformer. arXiv 2022, arXiv:cs.CL/2212.08136. [Google Scholar]
- Sang, Y.; Beetz, M.; Grau, V. Generation of 12-Lead Electrocardiogram with Subject-Specific, Image-Derived Characteristics Using a Conditional Variational Autoencoder. In Proceedings of the 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI), Kolkata, India, 28–31 March 2022; pp. 1–5. [Google Scholar] [CrossRef]
- Alcaraz, J.M.L.; Strodthoff, N. Diffusion-based conditional ECG generation with structured state space models. Comp. Biol. Med. 2023, 163, 107115. [Google Scholar] [CrossRef]
- Alcaraz, J.M.L.; Strodthoff, N. Diffusion-based Time Series Imputation and Forecasting with Structured State Space Models. arXiv 2022, arXiv:abs/2208.09399. [Google Scholar]
- Neifar, N.; Ben-Hamadou, A.; Mdhaffar, A.; Jmaiel, M. DiffECG: A Generalized Probabilistic Diffusion Model for ECG Signals Synthesis. arXiv 2023, arXiv:cs.CV/2306.01875. [Google Scholar]
- Adib, E.; Fernandez, A.S.; Afghah, F.; Prevost, J.J. Synthetic ECG Signal Generation Using Probabilistic Diffusion Models. IEEE Access 2023, 11, 75818–75828. [Google Scholar] [CrossRef]
- Ho, J.; Jain, A.; Abbeel, P. Denoising Diffusion Probabilistic Models. In Advances in Neural Information Processing Systems; Larochelle, H., Ranzato, M., Hadsell, R., Balcan, M., Lin, H., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2020; Volume 33, pp. 6840–6851. [Google Scholar]
- Gu, A.; Dao, T.; Ermon, S.; Rudra, A.; Ré, C. HiPPO: Recurrent Memory with Optimal Polynomial Projections. In Advances in Neural Information Processing Systems; Larochelle, H., Ranzato, M., Hadsell, R., Balcan, M., Lin, H., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2020; Volume 33, pp. 1474–1487. [Google Scholar]
- Gu, A.; Goel, K.; Ré, C. Efficiently Modeling Long Sequences with Structured State Spaces. arXiv 2022, arXiv:cs.LG/2111.00396. [Google Scholar]
- Ba, J.L.; Kiros, J.R.; Hinton, G.E. Layer Normalization. arXiv 2016, arXiv:stat.ML/1607.06450. [Google Scholar]
- Tay, Y.; Dehghani, M.; Abnar, S.; Shen, Y.; Bahri, D.; Pham, P.; Rao, J.; Yang, L.; Ruder, S.; Metzler, D. Long Range Arena: A Benchmark for Efficient Transformers. In Proceedings of the International Conference on Learning Representations, Virtual Event, 3–7 May 2021. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.u.; Polosukhin, I. Attention is All you Need. In Advances in Neural Information Processing Systems; Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Volume 30. [Google Scholar]
- Kong, Z.; Ping, W.; Huang, J.; Zhao, K.; Catanzaro, B. DiffWave: A Versatile Diffusion Model for Audio Synthesis. In Proceedings of the International Conference on Learning Representations, Virtual Event, 3–7 May 2021. [Google Scholar]
- Müller, M. Dynamic time warping. Inform. Retrieval Music Motion 2007, 2, 69–84. [Google Scholar] [CrossRef]
- Gretton, A.; Borgwardt, K.M.; Rasch, M.J.; Schölkopf, B.; Smola, A. A Kernel Two-Sample Test. J. Mach. Learn. Res. 2012, 13, 723–773. [Google Scholar]
- Hong, S.; Xu, Y.; Khare, A.; Priambada, S.; Maher, K.; Aljiffry, A.; Sun, J.; Tumanov, A. HOLMES: Health OnLine Model Ensemble Serving for Deep Learning Models in Intensive Care Units. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Virtual Event, 6–10 July 2020; pp. 1614–1624. [Google Scholar]

| Model | Avg DTW (Lower is Better) | Avg MMD (Lower is Better) |
|---|---|---|
| WaveGAN | 8.057 | 6.418 × |
| P2PGAN | 8.430 | 6.381 × |
| SSSD-ECG | 7.481 | 4.784 × |
| DSAT-ECG (Ours) | 7.174 | 4.603 × |
| Test Sample | Accuracy (Higher is Better) | AUROC (Higher is Better) |
|---|---|---|
| WaveGAN | 89.32 | 88.34 |
| P2PGAN | 89.67 | 89.58 |
| SSSD-ECG | 95.01 | 93.98 |
| DSAT-ECG (Ours) | 95.84 | 94.56 |
| Real | 96.38 | 94.67 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zama, M.H.; Schwenker, F. ECG Synthesis via Diffusion-Based State Space Augmented Transformer. Sensors 2023, 23, 8328. https://doi.org/10.3390/s23198328
Zama MH, Schwenker F. ECG Synthesis via Diffusion-Based State Space Augmented Transformer. Sensors. 2023; 23(19):8328. https://doi.org/10.3390/s23198328
Chicago/Turabian StyleZama, Md Haider, and Friedhelm Schwenker. 2023. "ECG Synthesis via Diffusion-Based State Space Augmented Transformer" Sensors 23, no. 19: 8328. https://doi.org/10.3390/s23198328
APA StyleZama, M. H., & Schwenker, F. (2023). ECG Synthesis via Diffusion-Based State Space Augmented Transformer. Sensors, 23(19), 8328. https://doi.org/10.3390/s23198328







