An Explainable EEG-Based Human Activity Recognition Model Using Machine-Learning Approach and LIME
Abstract
:1. Introduction
- Development of machine-learning classification models capable of recognizing resting, motor, and cognitive activities using EEG spectral features.
- Utilization of the LIME method to interpret the ML activity classification models and provide visual representations of the contributions made by EEG features for clinical reasoning in the context of human activity recognition (HAR).
2. Materials and Methods
2.1. Study Design
2.2. EEG Data Acquisition
2.3. Pre-Processing
2.4. EEG Feature Extraction
2.5. Feature Selection
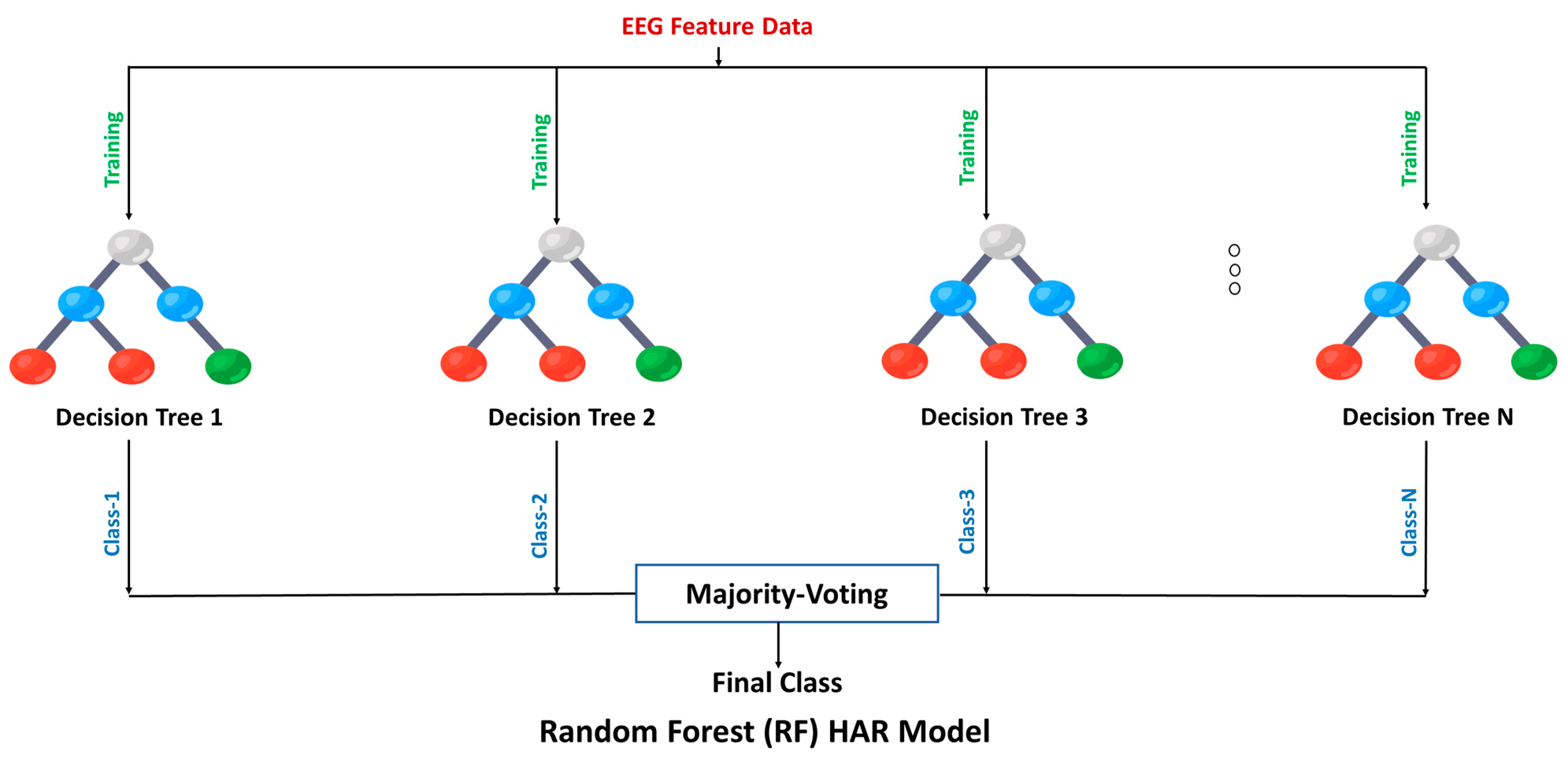
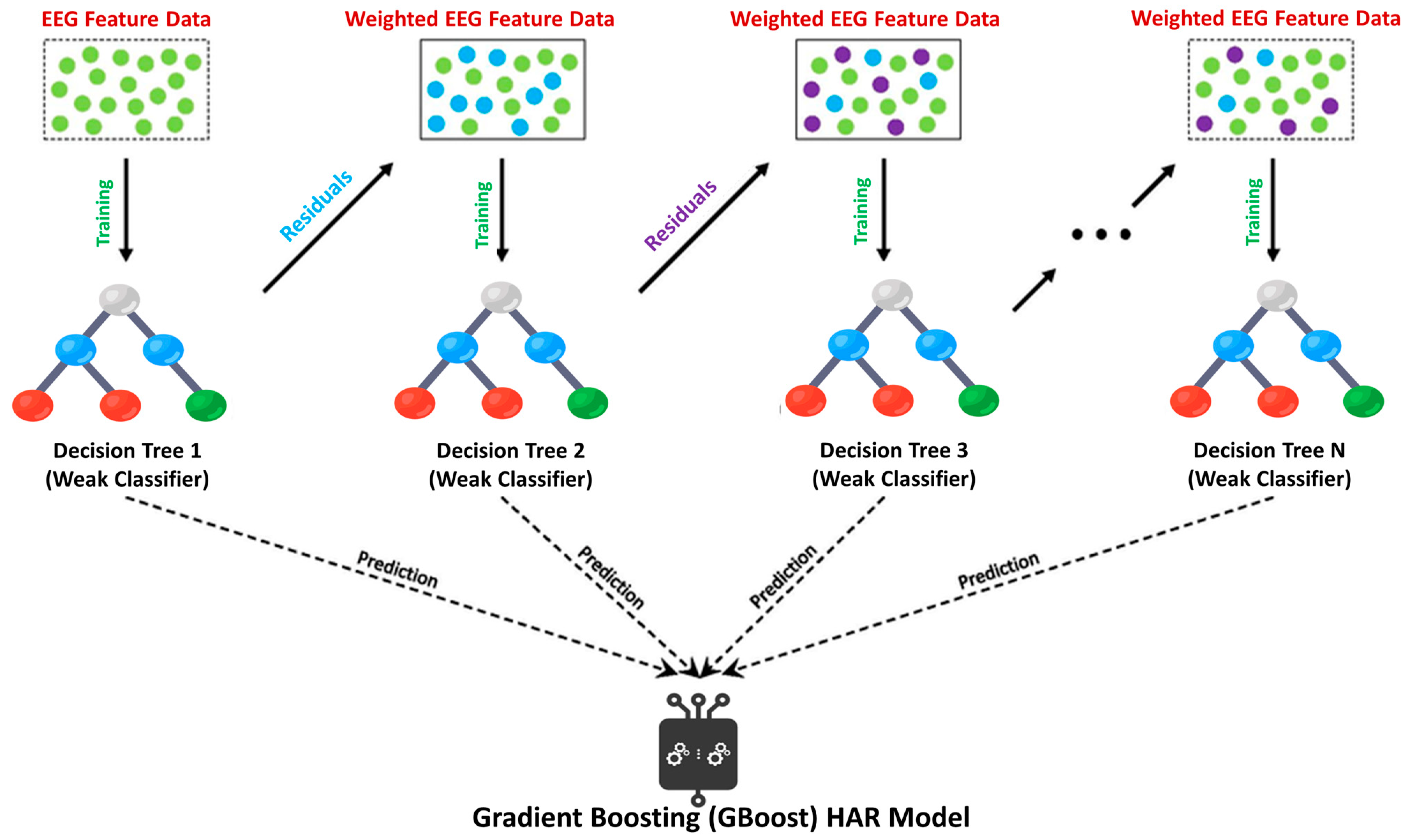
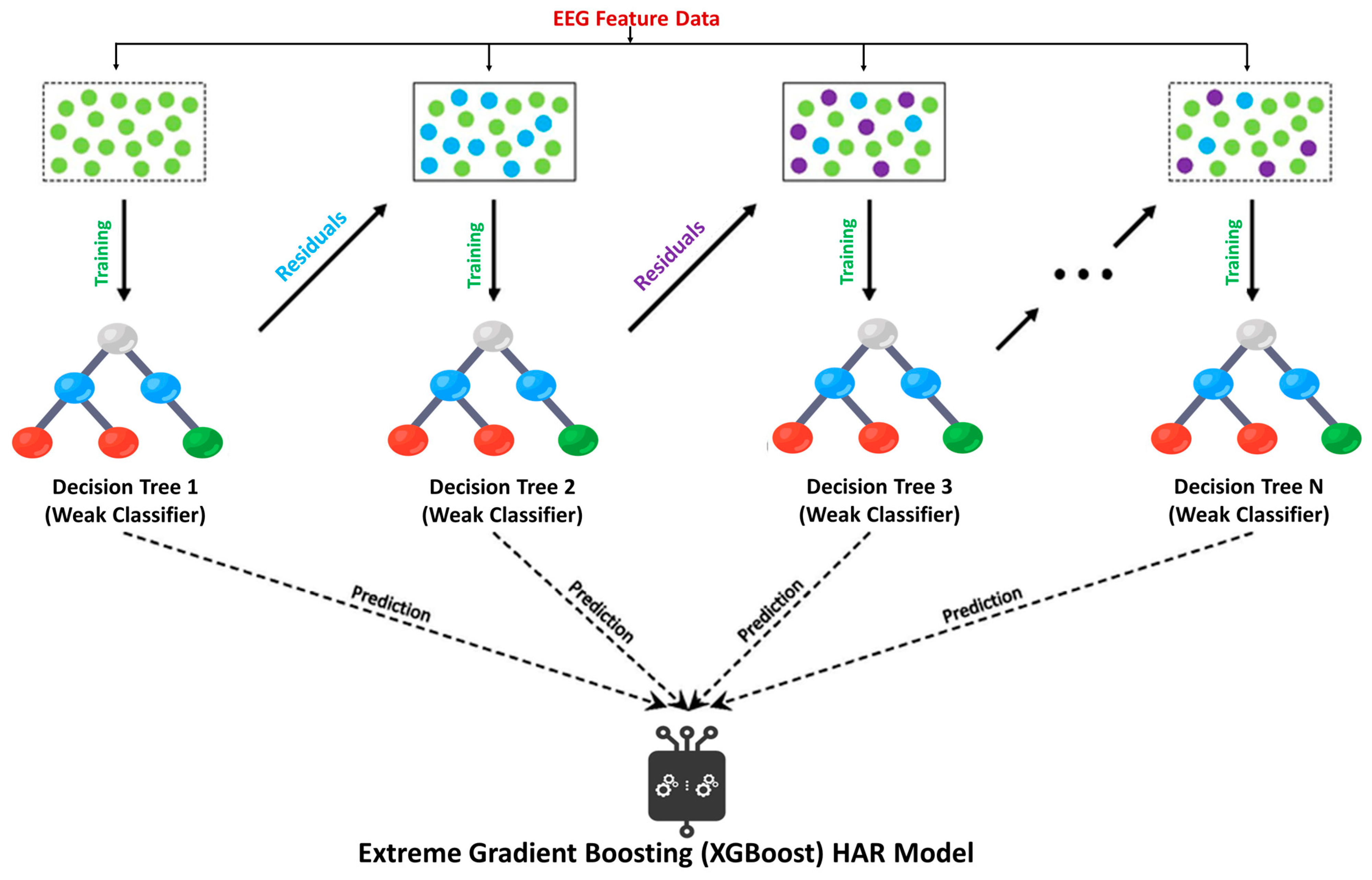
2.6. Machine Learning Classification
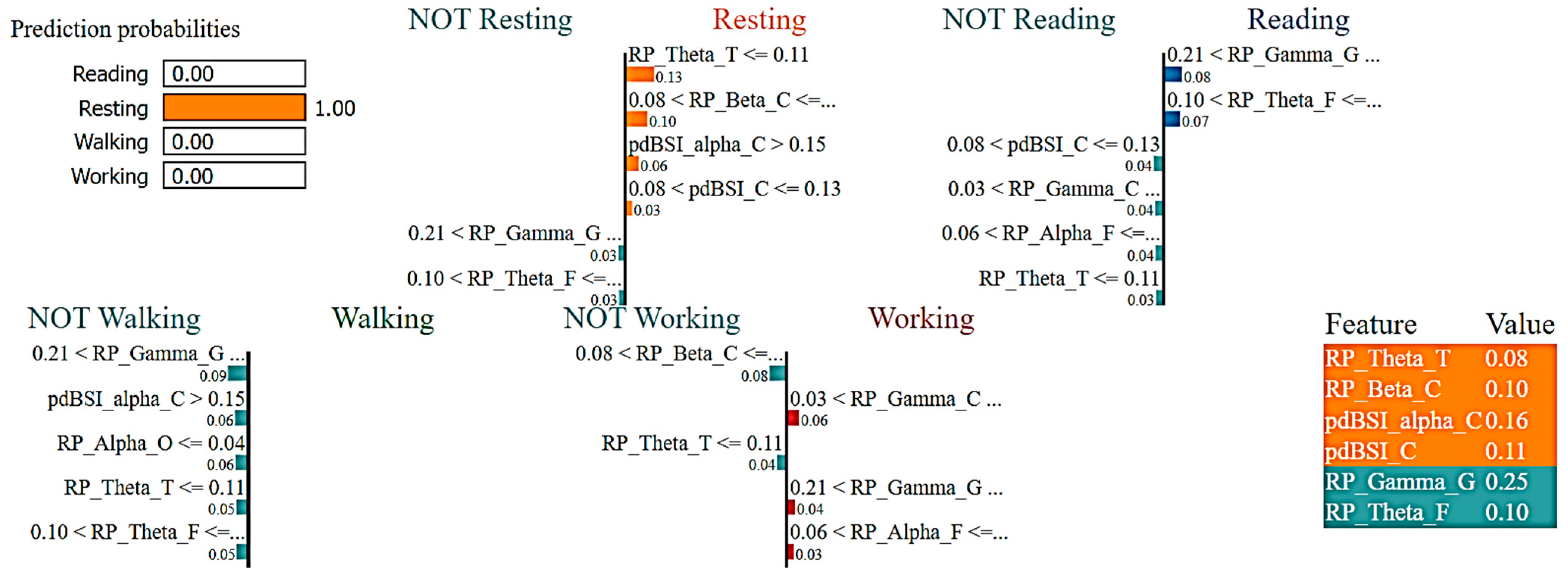
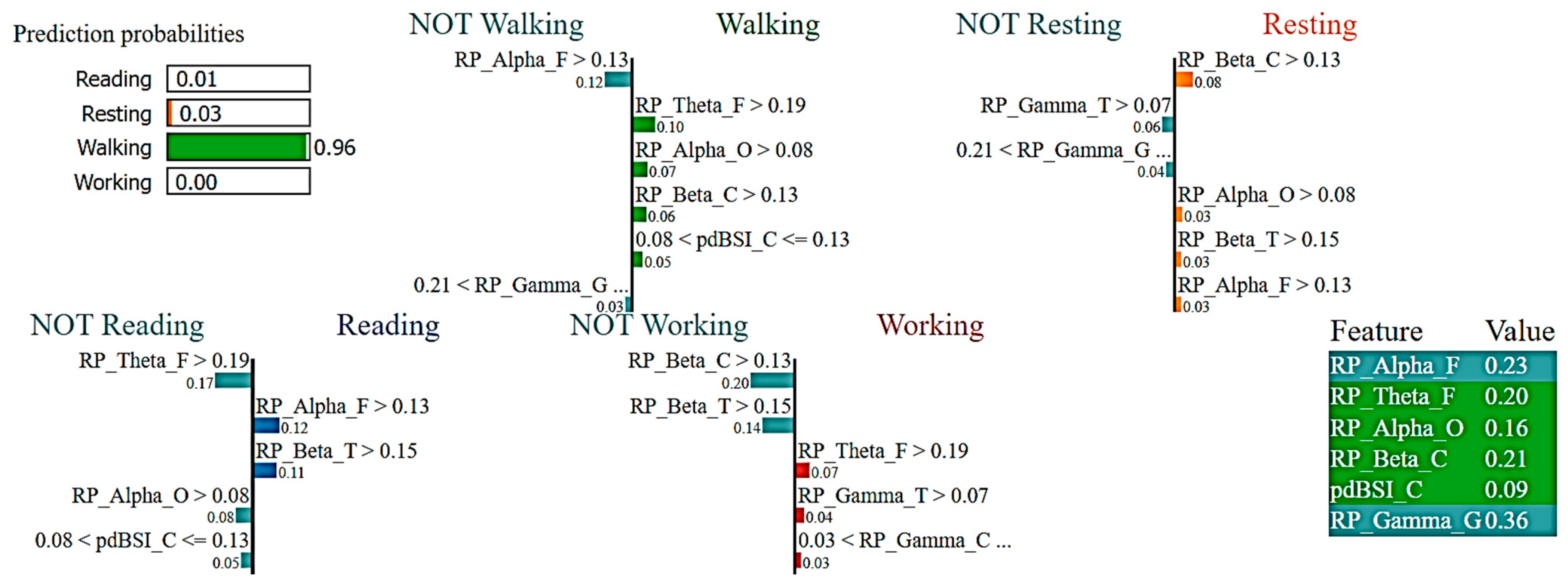
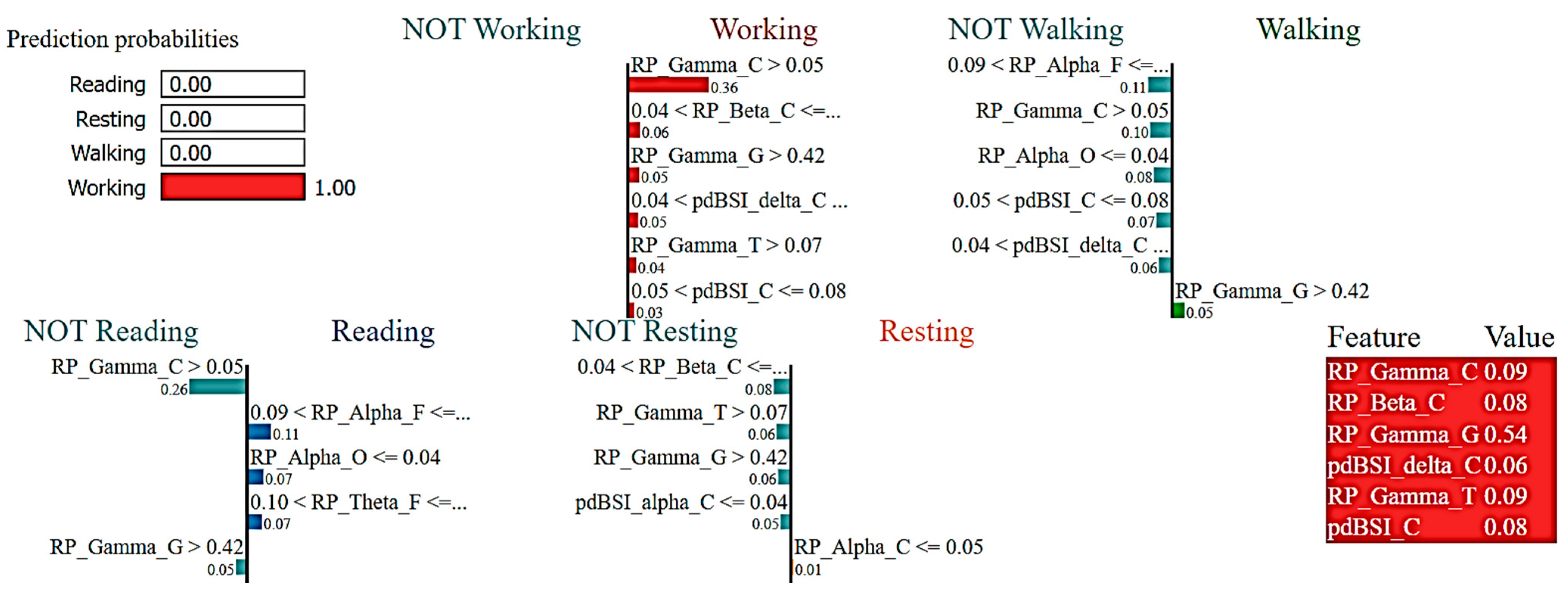
2.7. eXplainable Artificial Intelligence (XAI)
3. Results
3.1. Activity Recognition Model Using Machine Learning Approach
3.1.1. Hyperparameter Tuning
3.1.2. ML Classification Results
3.2. Interpretations of ML Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kwapisz, J.R.; Weiss, G.M.; Moore, S.A. Activity recognition using cell phone accelerometers. ACM SigKDD Explor. Newsl. 2011, 12, 74–82. [Google Scholar] [CrossRef]
- Cham, T.-J.; Rehg, J.M. A multiple hypothesis approach to figure tracking. In Proceedings of the 1999 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (Cat. No PR00149), Fort Collins, CO, USA, 23–25 June 1999; pp. 239–244. [Google Scholar]
- Khan, A.M.; Lee, Y.K.; Lee, S.Y.; Kim, T.S. A triaxial accelerometer-based physical-activity recognition via augmented-signal features and a hierarchical recognizer. IEEE Trans. Inf. Technol. Biomed. 2010, 14, 1166–1172. [Google Scholar] [CrossRef]
- Hussain, I.; Park, S.J. Big-ecg: Cardiographic predictive cyber-physical system for stroke management. IEEE Access 2021, 9, 123146–123164. [Google Scholar] [CrossRef]
- Hussain, I.; Park, S.-J. Prediction of myoelectric biomarkers in post-stroke gait. Sensors 2021, 21, 5334. [Google Scholar] [CrossRef]
- Park, S.J.; Hussain, I.; Hong, S.; Kim, D.; Park, H.; Benjamin, H.C.M. Real-time gait monitoring system for consumer stroke prediction service. In Proceedings of the IEEE International Conference on Consumer Electronics (ICCE), Las Vegas, NV, USA, 4–6 January 2020; pp. 1–4. [Google Scholar] [CrossRef]
- Hussain, I.; Park, S.-J. Quantitative evaluation of task-induced neurological outcome after stroke. Brain Sci. 2021, 11, 900. [Google Scholar] [CrossRef]
- Salehzadeh, A.; Calitz, A.P.; Greyling, J. Human activity recognition using deep electroencephalography learning. Biomed. Signal Process. Control 2020, 62, 102094. [Google Scholar] [CrossRef]
- Huynh-The, T.; Hua, C.-H.; Tu, N.A.; Kim, D.-S. Physical activity recognition with statistical-deep fusion model using multiple sensory data for smart health. IEEE Internet Things J. 2020, 8, 1533–1543. [Google Scholar] [CrossRef]
- Qiu, S.; Zhao, H.; Jiang, N.; Wang, Z.; Liu, L.; An, Y.; Zhao, H.; Miao, X.; Liu, R.; Fortino, G. Multi-sensor information fusion based on machine learning for real applications in human activity recognition: State-of-the-art and research challenges. Inf. Fusion 2022, 80, 241–265. [Google Scholar] [CrossRef]
- Hussain, I.; Park, S.J.; Hossain, M.A. Cloud-based clinical physiological monitoring system for disease prediction. In Proceedings of the 2nd International Conference on Smart Computing and Cyber Security; Springer Nature: Singapore, 2022; pp. 268–273. [Google Scholar] [CrossRef]
- Hussain, I.; Hossain, M.A.; Park, S.-J. A healthcare digital twin for diagnosis of stroke. In Proceedings of the 2021 IEEE International Conference on Biomedical Engineering, Computer and Information Technology for Health (BECITHCON), Dhaka, Bangladesh, 4–5 December 2021. [Google Scholar]
- Park, S.J.; Hong, S.; Kim, D.; Hussain, I.; Seo, Y. Intelligent in-car health monitoring system for elderly drivers in connected car. In Proceedings of the 20th Congress of the International Ergonomics Association (IEA 2018), Florence, Italy, 26–30 August 2018; Bagnara, S., Tartaglia, R., Albolino, S., Alexander, T., Fujita, Y., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 40–44. [Google Scholar] [CrossRef]
- Park, S.J.; Hong, S.; Kim, D.; Seo, Y.; Hussain, I. Knowledge based health monitoring during driving. In Proceedings of the HCI International 2018—Posters’ Extended Abstracts, Las Vegas, NV, USA, 15–20 July 2018; Stephanidis, C., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 387–392. [Google Scholar]
- Park, S.; Hong, S.; Kim, D.; Yu, J.; Hussain, I.; Park, H.; Benjamin, H. Development of intelligent stroke monitoring system for the elderly during sleeping. In Proceedings of the 15th World Sleep Congress, Vancouver, Canada, 20–25 September 2019; Elsevier: Amsterdam, The Netherlands, 2019; Volume 64, p. S294. [Google Scholar]
- Park, H.; Hong, S.; Hussain, I.; Kim, D.; Seo, Y.; Park, S.J. Gait monitoring system for stroke prediction of aging adults. In Proceedings of the International Conference on Applied Human Factors and Ergonomics, Washington, DC, USA, 24–28 July 2019; Advances in Human Factors in Wearable Technologies and Game Design, Ahram, T., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 93–97. [Google Scholar] [CrossRef]
- Hussain, I.; Young, S.; Kim, C.H.; Benjamin, H.C.M.; Park, S.J. Quantifying physiological biomarkers of a microwave brain stimulation device. Sensors 2021, 21, 1896. [Google Scholar] [CrossRef]
- Hussain, I.; Park, S.-J. Healthsos: Real-time health monitoring system for stroke prognostics. IEEE Access 2020, 8, 213574–213586. [Google Scholar] [CrossRef]
- Hong, S.; Kim, D.; Park, H.; Seo, Y.; Iqram, H.; Park, S. Gait Feature Vectors for Post-stroke Prediction using Wearable Sensor. Korean Soc. Emot. Sensib. 2019, 22, 55–64. [Google Scholar] [CrossRef]
- Salem, H.; El-Hasnony, I.M.; Kabeel, A.E.; El-Said, E.M.S.; Elzeki, O.M. Deep learning model and classification explainability of renewable energy-driven membrane desalination system using evaporative cooler. Alex. Eng. J. 2022, 61, 10007–10024. [Google Scholar] [CrossRef]
- Islam, M.S.; Hussain, I.; Rahman, M.M.; Park, S.J.; Hossain, M.A. Explainable artificial intelligence model for stroke prediction using eeg signal. Sensors 2022, 22, 9859. [Google Scholar] [CrossRef]
- Hussain, I.; Young, S.; Park, S.-J. Driving-Induced Neurological Biomarkers in an Advanced Driver-Assistance System. Sensors 2021, 21, 6985. [Google Scholar] [CrossRef]
- Hyvarinen, A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans. Neural Netw. 1999, 10, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Goldenholz, D.M.; Ahlfors, S.P.; Hämäläinen, M.S.; Sharon, D.; Ishitobi, M.; Vaina, L.M.; Stufflebeam, S.M. Mapping the signal-to-noise-ratios of cortical sources in magnetoencephalography and electroencephalography. Hum. Brain Mapp. 2009, 30, 1077–1086. [Google Scholar] [CrossRef] [PubMed]
- Gramfort, A.; Luessi, M.; Larson, E.; Engemann, D.A.; Strohmeier, D.; Brodbeck, C.; Goj, R.; Jas, M.; Brooks, T.; Parkkonen, L.; et al. Meg and eeg data analysis with mne-python. Front. Neurosci. 2013, 7, 267. [Google Scholar] [CrossRef] [PubMed]
- Welch, P. The use of fast fourier transform for the estimation of power spectra: A method based on time averaging over short, modified periodograms. IEEE Trans. Audio Electroacoust. 1967, 15, 70–73. [Google Scholar] [CrossRef]
- Sheorajpanday, R.V.A.; Nagels, G.; Weeren, A.J.T.M.; van Putten, M.J.A.M.; De Deyn, P.P. Reproducibility and clinical relevance of quantitative eeg parameters in cerebral ischemia: A basic approach. Clin. Neurophysiol. 2009, 120, 845–855. [Google Scholar] [CrossRef]
- Van Putten, M.J.A.M. The revised brain symmetry index. Clin. Neurophysiol. 2007, 118, 2362–2367. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Freund, Y.; Schapire, R.; Abe, N. A short introduction to boosting. J. Jpn. Soc. Artif. Intell. 1999, 14, 1612. [Google Scholar]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Stone, M. Cross-validatory choice and assessment of statistical predictions. J. R. Stat. Soc. Ser. B 1974, 36, 111–133. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2d graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M.L. Seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. Why should i trust you? Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Zhang, J.; Yin, Z.; Chen, P.; Nichele, S. Emotion recognition using multi-modal data and machine learning techniques: A tutorial and review. Inf. Fusion 2020, 59, 103–126. [Google Scholar] [CrossRef]
- Herbert, A.; Esparham, A. Mind–body therapy for children with attention-deficit/hyperactivity disorder. Children 2017, 4, 31. [Google Scholar] [CrossRef]
- Rapin, I.; Tuchman, R.F. Autism: Definition, neurobiology, screening, diagnosis. Pediatr. Clin. N. Am. 2008, 55, 1129–1146. [Google Scholar] [CrossRef] [PubMed]
- Price, C.J. A review and synthesis of the first 20 years of pet and fmri studies of heard speech, spoken language and reading. Neuroimage 2012, 62, 816–847. [Google Scholar] [CrossRef] [PubMed]
- Humphries, C.; Binder, J.R.; Medler, D.A.; Liebenthal, E. Syntactic and semantic modulation of neural activity during auditory sentence comprehension. J. Cogn. Neurosci. 2006, 18, 665–679. [Google Scholar] [CrossRef]
- Lam, N.H.; Schoffelen, J.-M.; Uddén, J.; Hultén, A.; Hagoort, P. Neural activity during sentence processing as reflected in theta, alpha, beta, and gamma oscillations. Neuroimage 2016, 142, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Tyng, C.M.; Amin, H.U.; Saad, M.N.; Malik, A.S. The influences of emotion on learning and memory. Front. Psychol. 2017, 8, 1454. [Google Scholar] [CrossRef] [PubMed]
- Bimbi, M.; Festante, F.; Coudé, G.; Vanderwert, R.E.; Fox, N.A.; Ferrari, P.F. Simultaneous scalp recorded eeg and local field potentials from monkey ventral premotor cortex during action observation and execution reveals the contribution of mirror and motor neurons to the mu-rhythm. Neuroimage 2018, 175, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Senkowski, D.; Herrmann, C.S. Effects of task difficulty on evoked gamma activity and erps in a visual discrimination task. Clin. Neurophysiol. 2002, 113, 1742–1753. [Google Scholar] [CrossRef]
- Wagner, J.; Solis-Escalante, T.; Scherer, R.; Neuper, C.; Müller-Putz, G. It’s how you get there: Walking down a virtual alley activates premotor and parietal areas. Front. Hum. Neurosci. 2014, 8, 93. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, H.; Kaneko, N.; Masugi, Y.; Ogawa, T.; Watanabe, K.; Nakazawa, K. Gait-phase-dependent and gait-phase-independent cortical activity across multiple regions involved in voluntary gait modifications in humans. Eur. J. Neurosci. 2021, 54, 8092–8105. [Google Scholar] [CrossRef]



| Random Forest Classifier (Accuracy = 80.33%) | |||
|---|---|---|---|
| Activity Class | Precision | Recall | F1-Score |
| Reading | 0.77 | 0.71 | 0.74 |
| Resting | 0.78 | 0.88 | 0.82 |
| Walking | 0.78 | 0.82 | 0.80 |
| Working | 0.89 | 0.82 | 0.86 |
| Weighted Average | 0.81 | 0.80 | 0.80 |
| XGB Classifier (Accuracy = 79.70%) | |||
|---|---|---|---|
| Activity Class | Precision | Recall | F1-Score |
| Reading | 0.78 | 0.7 | 0.74 |
| Resting | 0.78 | 0.89 | 0.83 |
| Walking | 0.76 | 0.77 | 0.76 |
| Working | 0.88 | 0.85 | 0.86 |
| Weighted Average | 0.80 | 0.80 | 0.80 |
| Gradient Boosting Classifier (Accuracy = 78.94%) | |||
|---|---|---|---|
| Activity Class | Precision | Recall | F1-Score |
| Reading | 0.75 | 0.7 | 0.73 |
| Resting | 0.76 | 0.87 | 0.81 |
| Walking | 0.75 | 0.79 | 0.77 |
| Working | 0.9 | 0.81 | 0.86 |
| Weighted Average | 0.79 | 0.79 | 0.79 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussain, I.; Jany, R.; Boyer, R.; Azad, A.; Alyami, S.A.; Park, S.J.; Hasan, M.M.; Hossain, M.A. An Explainable EEG-Based Human Activity Recognition Model Using Machine-Learning Approach and LIME. Sensors 2023, 23, 7452. https://doi.org/10.3390/s23177452
Hussain I, Jany R, Boyer R, Azad A, Alyami SA, Park SJ, Hasan MM, Hossain MA. An Explainable EEG-Based Human Activity Recognition Model Using Machine-Learning Approach and LIME. Sensors. 2023; 23(17):7452. https://doi.org/10.3390/s23177452
Chicago/Turabian StyleHussain, Iqram, Rafsan Jany, Richard Boyer, AKM Azad, Salem A. Alyami, Se Jin Park, Md Mehedi Hasan, and Md Azam Hossain. 2023. "An Explainable EEG-Based Human Activity Recognition Model Using Machine-Learning Approach and LIME" Sensors 23, no. 17: 7452. https://doi.org/10.3390/s23177452
APA StyleHussain, I., Jany, R., Boyer, R., Azad, A., Alyami, S. A., Park, S. J., Hasan, M. M., & Hossain, M. A. (2023). An Explainable EEG-Based Human Activity Recognition Model Using Machine-Learning Approach and LIME. Sensors, 23(17), 7452. https://doi.org/10.3390/s23177452










