A New Method for Heart Disease Detection: Long Short-Term Feature Extraction from Heart Sound Data
Abstract
1. Introduction
- Heart sound classification remains a prominent topic of discussion, and the PhysioNet/CinC Challenge 2016 dataset is currently the most comprehensive and up-to-date collection of data available. With the dataset being made available to the public, numerous machine learning algorithms have been applied to the data, most of which divide signals into fragments and then analyze the features extracted from those fragments. This study stands out as the only one representing the features extracted from both small fragments and the entire heart sound signal.
- The main contribution of this work lies in its assertion that small fragments and the whole signal have distinct characteristics, and when used in combination, they increase classification accuracy. To validate this assertion, we employ various machine learning models utilizing a combination of features. The results indicate that the combined feature set boosts the classification accuracy on the publicly available portion of the PhysioNet dataset.
- Lastly, we propose a novel approach to eliminate extraneous peaks and determine the fundamental heart sounds.
2. Related Works
3. Materials and Methods
3.1. Dataset
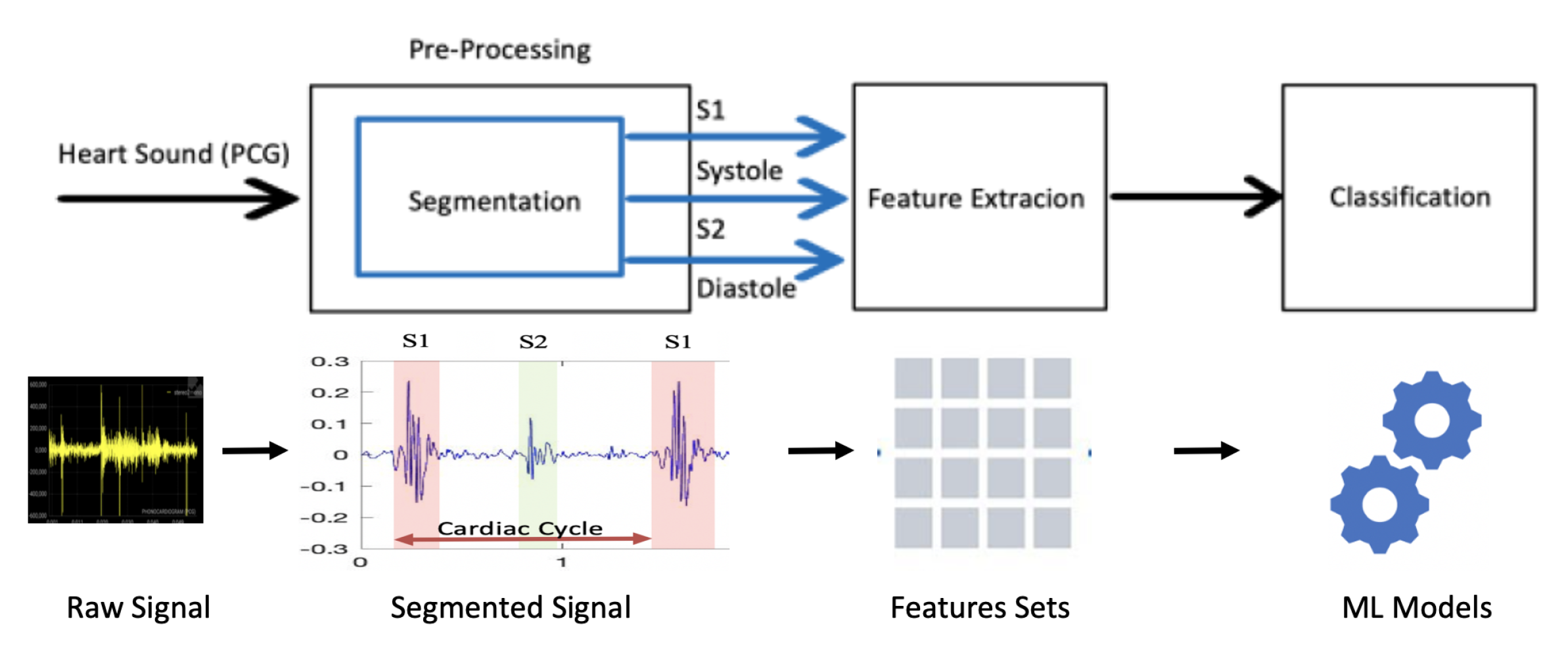
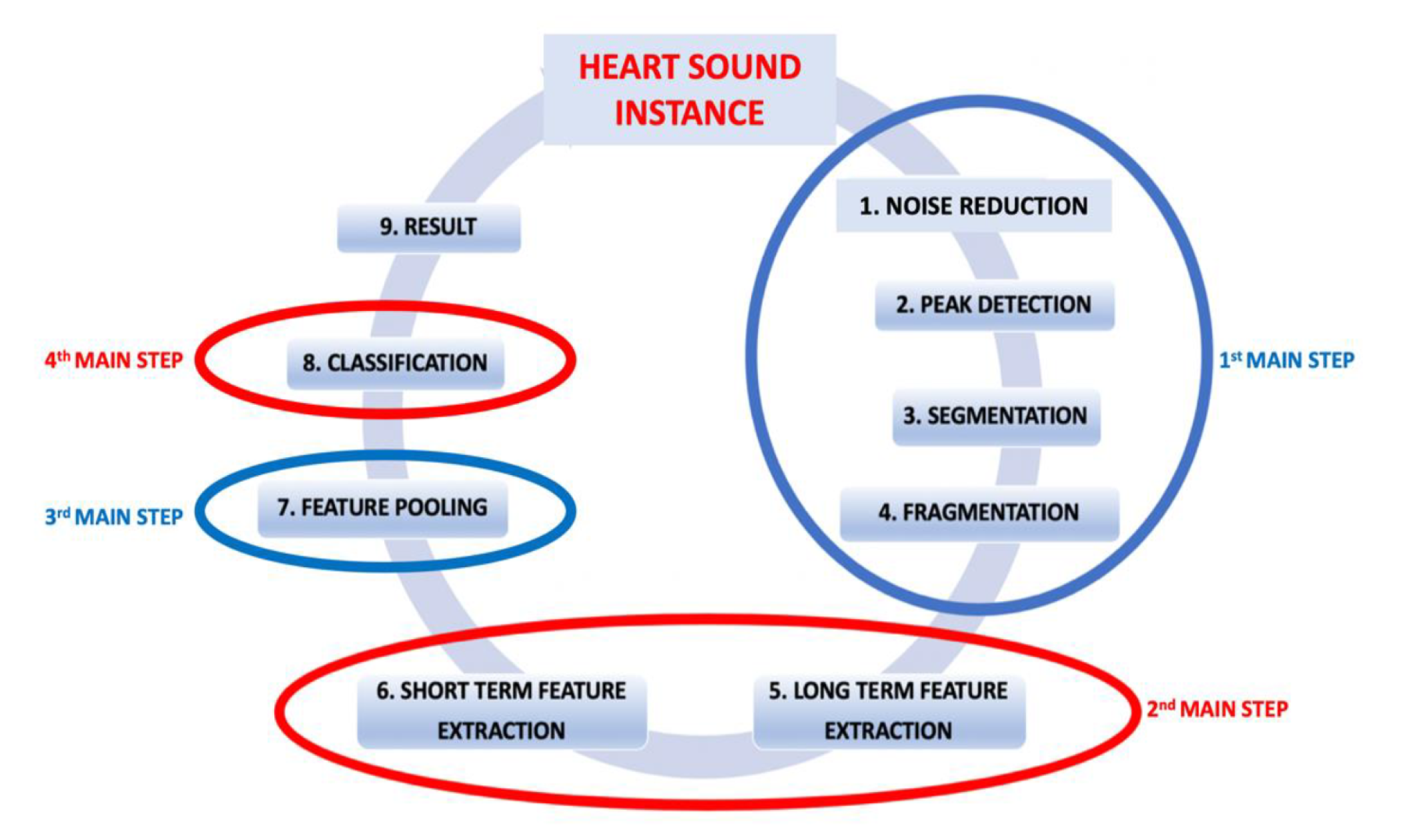
3.2. Proposed Method
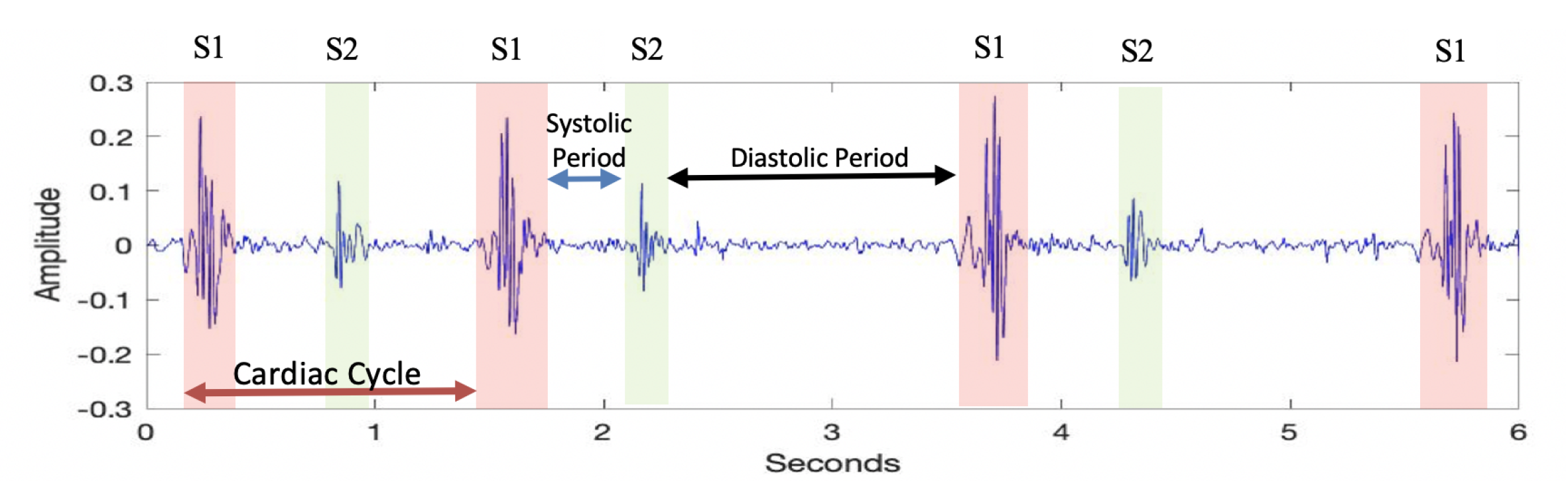
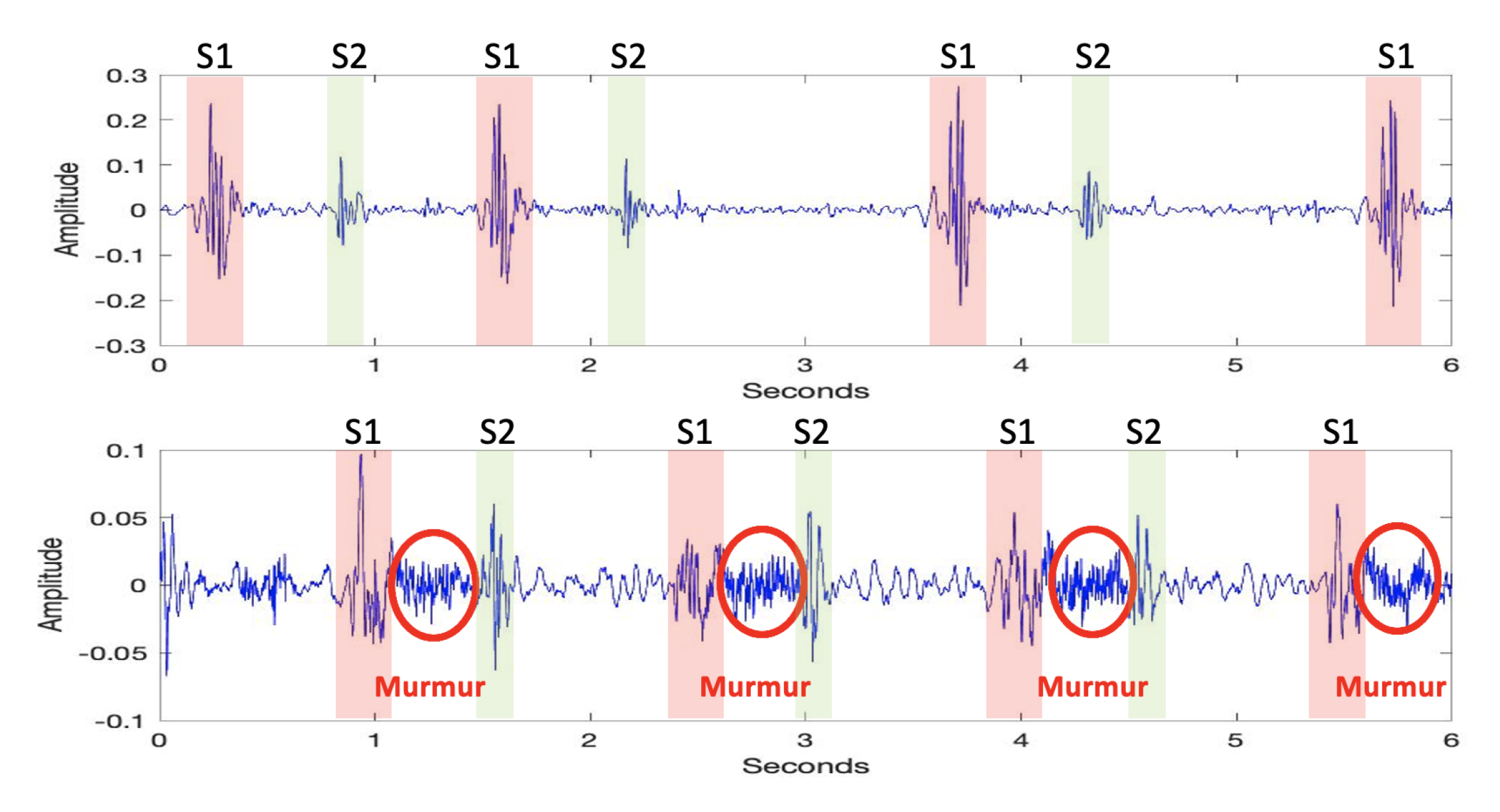
3.2.1. Pre-Processing
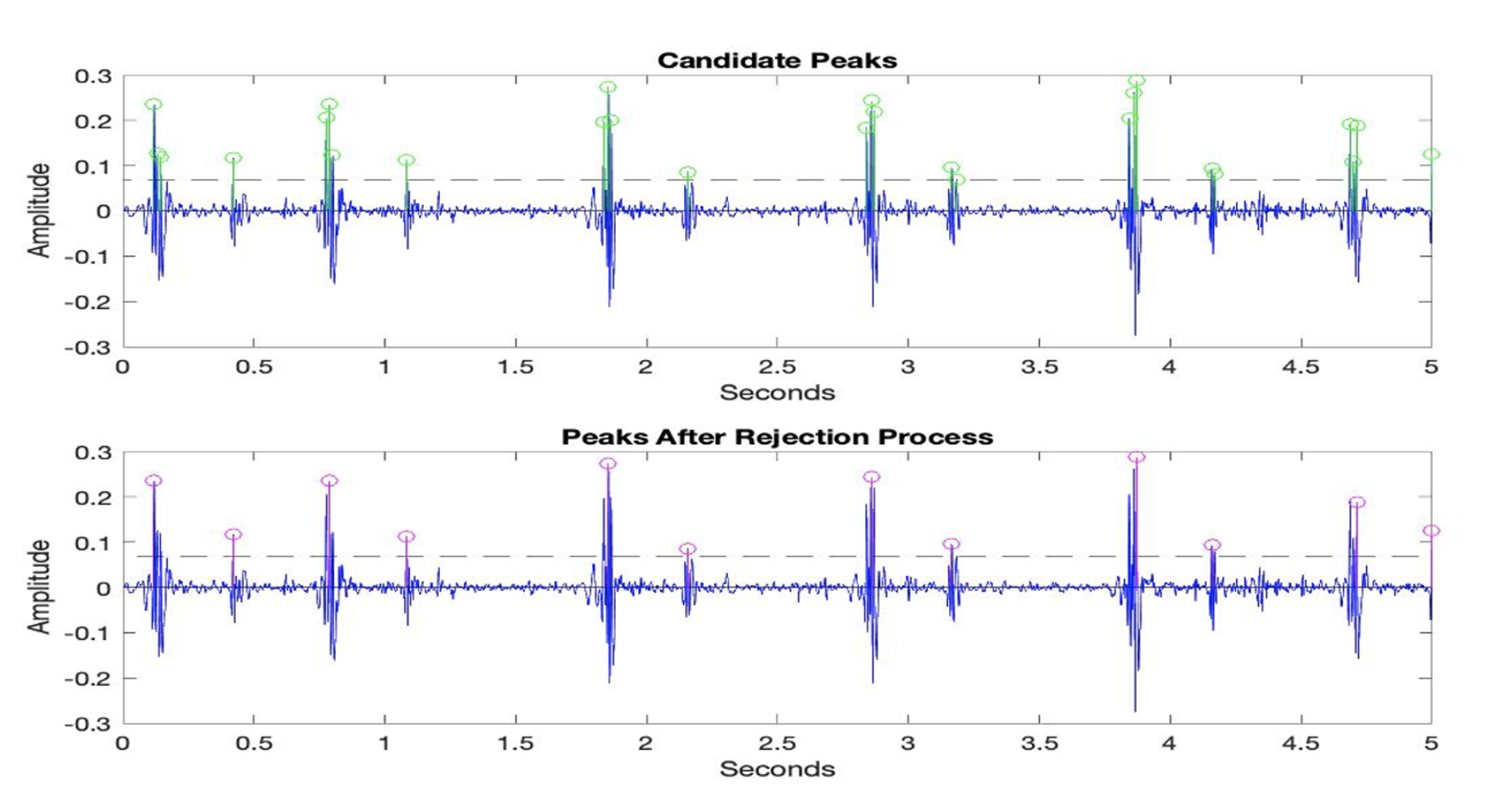
- If two neighboring peaks have a time interval of less than 50 ms, the peak with the smaller amplitude value is rejected, and the other peak is considered a prospective descriptor point.
- If two neighboring peaks have a time interval of less than 50 ms, the number of heartbeats in healthy individuals should be between 40 and 140 beats per min. Thus, the cardiac cycle, which consists of the systolic and diastolic periods, cannot be shorter than 400 ms or longer than 1500 ms.
- If two neighboring peaks have a time interval of less than 400 ms and more than 50 ms, the peak with the smaller amplitude value is rejected, and the other peak is considered a prospective descriptor point.
- If the time interval between the two peaks is more than 1500 ms, it indicates the presence of unidentified peaks. Thus, the threshold value is refined and the rejection steps are applied again.
3.2.2. Feature Extraction
3.2.3. Feature Pooling
3.2.4. Classification
4. Results and Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Algorithm | Metrics | Feature Sets | ||||
|---|---|---|---|---|---|---|
| STF | LTF | LSTF | SSTF | SLSTF | ||
| Fine Tree | Accuracy | 78.6% | 77.9% | 87.8% | 83.1% | 70.8% |
| Sensitivity | 84% | 92% | 86% | 91% | 78% | |
| Specificity | 93% | 53% | 94% | 69% | 93% | |
| Medium Tree | Accuracy | 77.9% | 76.6% | 84.9% | 84.4% | 71.4% |
| Sensitivity | 68% | 95% | 72% | 92% | 63% | |
| Specificity | 98% | 81% | 97% | 62% | 96% | |
| Coarse Tree | Accuracy | 72.2% | 75.7% | 83.5 | 80.2% | 56.2% |
| Sensitivity | 63% | 76% | 62% | 95% | 43% | |
| Specificity | 97% | 94% | 97% | 33% | 97% | |
| Algorithm | Metrics | Feature Sets | ||||
|---|---|---|---|---|---|---|
| STF | LTF | LSTF | SSTF | SLSTF | ||
| Gaussian Naive Bayes | Accuracy | 68% | 75.2% | 83.3% | 71.6% | 76% |
| Kernel Naive Bayes | Accuracy | 67% | 74.5% | 82.8% | 70.8% | 76% |
| Algorithm | Metrics | Feature Sets | ||||
|---|---|---|---|---|---|---|
| STF | LTF | LSTF | SSTF | SLSTF | ||
| Linear Support Vector Machine | Accuracy | 71.7% | 75.7% | 77% | 66.4% | 67.4% |
| Sensitivity | 64% | 76% | 65% | 57% | 58% | |
| Specificity | 96% | 94% | 97% | 97% | 97% | |
| Quadratic Support Vector Machine | Accuracy | 83.6% | 75.8% | 84.2% | 80.3% | 81.8% |
| Sensitivity | 80% | 99% | 80% | 75% | 77% | |
| Specificity | 96% | 97% | 96% | 98% | 98% | |
| Cubic Support Vector Machine | Accuracy | 88.3% | 74.4% | 89.7% | 86.3% | 89.3% |
| Sensitivity | 87% | 76% | 89% | 84% | 89% | |
| Specificity | 93% | 65% | 93% | 93% | 93% | |
| Fine Gaussian Support Vector Machine | Accuracy | 81.6% | 74.5% | 91.2% | 79.8% | 80.1% |
| Sensitivity | 96% | 84% | 97% | 96% | 97% | |
| Specificity | 71% | - | 73% | 62% | 73% | |
| Medium Support Vector Machine | Accuracy | 84.1% | 77.9% | 87.4% | 83.5% | 83.9% |
| Sensitivity | 79% | 78% | 80% | 79% | 79% | |
| Specificity | 98% | - | 98% | 98% | 98% | |
| Coarse Support Vector Machine | Accuracy | 72.1% | 76.7% | 77.5% | 69.7% | 69.7% |
| Sensitivity | 64% | 77% | 66% | 61% | 61% | |
| Specificity | 98% | - | 98% | 98% | 98% | |
| Algorithm | Metrics | Feature Sets | ||||
|---|---|---|---|---|---|---|
| STF | LTF | LSTF | SSTF | SLSTF | ||
| Fine K-Nearest Neighbor | Accuracy | 84.1% | 85.4% | 86.2% | 83.1% | 85.9% |
| Sensitivity | 97% | 94% | 97% | 96% | 97% | |
| Specificity | 88% | 99% | 90% | 85% | 91% | |
| Medium K-Nearest Neighbor | Accuracy | 84.4% | 80.5% | 84.6% | 84.1% | 84.6% |
| Sensitivity | 80% | 88% | 80% | 79% | 80% | |
| Specificity | 99% | - | 99% | 99% | 99% | |
| Coarse K-Nearest Neighbor | Accuracy | 73.2% | 76.2% | 74.5% | 73.6% | 73.8% |
| Sensitivity | 65% | 96% | 64% | 66% | 66% | |
| Specificity | 98% | 84% | 98% | 98% | 98% | |
| Cosine K-Nearest Neighbor | Accuracy | 84.6% | 81% | 85.1% | 83.2% | 84.7% |
| Sensitivity | 80% | 87% | 81% | 78% | 80% | |
| Specificity | 99% | 39% | 99% | 98% | 99% | |
| Cubic K-Nearest Neighbor | Accuracy | 83.7% | 81.2% | 84.7% | 83.5% | 84.1% |
| Sensitivity | 79% | 87% | 79% | 79% | 79% | |
| Specificity | 99% | 37% | 99% | 99% | 99% | |
| Weighted K-Nearest Neighbor | Accuracy | 87.3% | 86% | 89.9% | 86.9% | 88% |
| Sensitivity | 84% | 97% | 85% | 83% | 85% | |
| Specificity | 98% | 95% | 98% | 98% | 99% | |
| Algorithm | Metrics | Feature Sets | ||||
|---|---|---|---|---|---|---|
| STF | LTF | LSTF | SSTF | SLSTF | ||
| Boosted Tree | Accuracy | 82.6% | 77.3% | 84.1% | 76.7% | 78.3% |
| Sensitivity | 78% | 98% | 80% | 70% | 72% | |
| Specificity | 99% | 88% | 99% | 96% | 97% | |
| Bagged Tree | Accuracy | 89.1% | 91% | 91.3% | 86.8% | 90% |
| Sensitivity | 88% | 97% | 90% | 86% | 88% | |
| Specificity | 93% | 95% | 95% | 90% | 95% | |
| Subspace Discriminant | Accuracy | 62.3% | 76% | 73.8% | 50.9% | 52.5% |
| Sensitivity | 51% | 99% | 53% | 36% | 38% | |
| Specificity | 98% | 96% | 97% | 98% | 98% | |
| Subspace K-Nearest Neighbor | Accuracy | 90.3% | 89% | 92.7% | 90.5% | 90.1% |
| Sensitivity | 96% | 97% | 96% | 96% | 96% | |
| Specificity | 82% | 94% | 82% | 82% | 83% | |
| RUSBoosted Trees | Accuracy | 87% | 71% | 88.1% | 83% | 85.2% |
| Sensitivity | 87% | 70% | 87% | 82% | 85% | |
| Specificity | 89% | 72% | 91% | 85% | 87% | |
References
- WHO. World Statistics on Cardiovascular Disease. Available online: https://www.who.int/en/news-room/fact-sheets/detail/cardiovascular-diseases-(CVDs) (accessed on 10 June 2023).
- Thiyagaraja, S.R.; Dantu, R.; Shrestha, P.L.; Chitnis, A.; Thompson, M.A.; Anumandla, P.T.; Sarma, T.; Dantu, S. A novel heart-mobile interface for detection and classification of the heart sound. Biomed. Signal Process. Control 2018, 45, 313–324. [Google Scholar] [CrossRef]
- Cardiosleeve, the World’s First Stethoscope Attachment to Provide ECG, Digital Auscultation and Instant Analysis, Rijuven Corporation. Available online: https://cardiosleeve.myshopify.com/ (accessed on 10 June 2023).
- Sensicardiac, A Way to Provide Quantitative Heart Screenings. Available online: https://sensicardiac.com/ (accessed on 10 June 2023).
- Eko Devices, Eko Core Digital Stethoscope. Available online: https://ekodevices.com/ (accessed on 10 June 2023).
- Liu, C.; Springer, D.; Li, Q.; Moody, B.; Juan, R.A.; Chorro, F.J.; Castells, F.; Roig, J.M.; Silva, I.; Johnson, A.E.; et al. An open-access database for the evaluation of heart sound algorithms. Physiol. Meas. 2016, 37, 2181. [Google Scholar] [CrossRef]
- Han, W.; Yang, Z.; Lu, J.; Xie, S. Supervised threshold-based heart sound classification algorithm. Physiol. Meas. 2018, 39, 115011. [Google Scholar] [CrossRef] [PubMed]
- Latif, S.; Usman, M.; Rana, R.; Quadir, J. Phonocardiographic Sensing using Deep Learning for Abnormal Heartbeat Detection. IEEE Sens. J. 2018, 18, 9393–9400. [Google Scholar] [CrossRef]
- Schmidt, S.E.; Holst-Hansen, C.; Graff, C.; Toft, E.; Struijk, J. Segmentation of heart sound recordings by a duration-dependent hidden Markov model. Physiol. Meas. 2010, 31, 513–529. [Google Scholar] [CrossRef]
- Ari, S.; Hembram, K.; Saha, G. Detection of cardiac abnormality from PCG signal using LMS based least square SVM classier. Expert Syst. Appl. 2010, 37, 8019–8026. [Google Scholar] [CrossRef]
- Avendano-Valencia, L.D.; Godino-Llorente, J.I.; Blanco-Velasco, M.; Castellanos-Dominguez, G. Feature extraction from parametric time-frequency representations for heart murmur detection. Ann. Biomed. Eng. 2010, 38, 2716–2732. [Google Scholar] [CrossRef]
- Bentley, P.; Nordehn, G.; Coimbra, M.; Mannor, S.; Getz, R. The PASCAL Classifying Heart Sounds Challenge. 2011. Available online: www.peterjbentley.com/heartchallenge/index.html (accessed on 10 June 2023).
- Gharehbaghi, A.; Dutoir, T.; Sepehri, A.; Hult, P.; Ask, P. An Automatic Tool for Pediatric Heart Sounds Segmentation Computing in Cardiology. In Proceedings of the 2011 Computing in Cardiology, Hangzhou, China, 18–21 September 2011; pp. 37–40. [Google Scholar]
- Li, T.; Tang, H.; Qiu, T.S.; Park, Y. Best subsequence selection of heart sound recording based on degree of sound periodicity. Electron. Lett. 2011, 47, 841–843. [Google Scholar] [CrossRef]
- Moukadem, A.; Dieterlen, A.; Hueber, N.; Brandt, C. Localization of Heart Sounds Based on S Transform and Radial Basis Function. In Proceedings of the Neural Network 15th Nordic-Baltic Conference on Biomedical Engineering and Medical Physics, Aalborg, Denmark, 14–17 June 2011; pp. 168–171. [Google Scholar]
- Tang, H.; Li, T.; Qiu, T.S.; Park, Y. Segmentation of heart sounds based on dynamic clustering. Biomedical Signal Process. Control 2012, 7, 509–516. [Google Scholar]
- Uguz, H. Adaptive neuro-fuzzy inference system for diagnosis of the heart valve diseases using wavelet transform with entropy. Neural Comput. 2012, 21, 1617–1628. [Google Scholar] [CrossRef]
- Moukadem, A.; Dieterlena, A.; Hueberb, N.; Brandtc, C. A robust heart sounds segmentation module based on Stransform. Biomed. Signal Process. Control 2013, 8, 273–281. [Google Scholar] [CrossRef]
- Naseri, H.; Homaeinezhad, M.R. Detection and boundary identification of phonocardiogram sounds using an expert frequency-energy based metric. Ann. Biomed. Eng. 2013, 41, 279–292. [Google Scholar] [CrossRef]
- Castro, A.; Vinhoza, T.T.; Mattos, S.S.; Coimbra, M.T. Heart Sound Segmentation of Pediatric Auscultations Using Wavelet Analysis. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Osaka, Japan, 3–7 July 2013; pp. 3909–3912. [Google Scholar]
- Sun, S.; Jiang, Z.; Wang, H.; Fang, Y. Automatic moment segmentation and peak detection analysis of heart sound pattern via short-time modified Hilbert transform Comput. Methods Programs Biomed. 2014, 114, 219–230. [Google Scholar] [CrossRef]
- Varghees, V.N.; Ramachandran, K. A novel heart sound activity detection framework for automated heart sound analysis. Biomed. Signal Process. Control 2014, 13, 174–188. [Google Scholar] [CrossRef]
- Pedrosa, J.; Castro, A.; Vinhoza, T.T.V. Automatic Heart Sound Segmentation and Murmur Detection in Pediatric Phonocardiograms. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; pp. 2294–2297. [Google Scholar]
- Papadaniil, C.D.; Hadjileontiadis, L.J. Efficient heart sound segmentation and extraction using ensemble empirical mode decomposition and kurtosis features. IEEE J. Biomed. Health Inform. 2014, 18, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.N.; Guo, X.M.; Ding, X.R. A novel hybrid energy fraction and entropy-based approach for systolic heart murmur identification. Expert Syst. Appl. 2015, 42, 2710–2721. [Google Scholar] [CrossRef]
- Patidar, S.; Pachori, R.B.; Garg, N. Automatic diagnosis of septal defects based on tunable-Q wavelet transform of cardiac sound signals. Expert Syst. Appl. 2015, 42, 3315–3326. [Google Scholar] [CrossRef]
- Gharehbaghi, A.; Ekman, I.; Ask, P. Nylander E and Janerot-Sjoberg B. Assessment of aortic valve stenosis severity using intelligent phonocardiography. Int. J. Cardiol. 2015, 198, 58–60. [Google Scholar] [CrossRef]
- Springer, D.B.; Tarassenko, L.; Clifford G, D. Logistic regression-HSMM-based heart sound segmentation. IEEE Trans. Biomed. Eng. 2016, 63, 822–832. [Google Scholar] [CrossRef]
- Tsao, Y.; Lin, T.H.; Chen, F.; Chang, Y.F.; Cheng, C.H.; Tsai, K.H. Robust S1 and S2 heart sound recognition based on spectral restoration and multi-style training. Biomed. Signal Process. Control 2019, 49, 173–180. [Google Scholar] [CrossRef]
- Dominguez-Morales, J.P.; Jimenez-Fernandez, A.F.; Dominguez-Morales, M.J.; Jimenez-Moreno, G. Deep neural networks for the recognition and classification of heart murmurs using neuromorphic auditory sensors. IEEE Trans. Biomed. Circuits Syst. 2017, 12, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.; Mermelstein, P. Comparison of parametric representations for monosyllabic word recognition in continuously spoken sentences. IEEE Trans. Acoust. Speech Signal Process. 1980, 28, 357–366. [Google Scholar] [CrossRef]
- Zewoudie, A.W.; Luque, J.; Hernando, J. The use of long-term features for GMM-and i-vector-based speaker diarization systems. EURASIP J. Audio Speech Music. Process. 2018, 2018, 14. [Google Scholar] [CrossRef]
- Kay, Y.M.; Akay, M.; Welkowitz, W.; Kostis, J. Noninvasive detection of coronary artery disease. IEEE Eng. Med. Biol. 1994, 13, 761–764. [Google Scholar] [CrossRef]
- Bentley, P.M.; Grant, P.M.; McDonnell, J.T.E. Time-frequency and time-scale techniques for the classification of native and bioprosthetic heart valve sounds. IEEE Trans Biomed Eng. 1998, 45, 125–128. [Google Scholar] [CrossRef]
- Saracoglu, R. Hidden Markov model-based classification of heart valve disease with PCA for dimension reduction. Eng. Appl. Artif. Intell. 2012, 25, 1523–1528. [Google Scholar] [CrossRef]
- Heart Sound Classifier Example. Available online: https://www.mathworks.com/matlabcentral/fileexchange/65286-heart-sound-classifier (accessed on 10 June 2023).
- Sujadevi, V.G.; Mohan, N. A hybrid method for fundamental heart sound segmentation using group sparsity denoising and variational mode decomposition. Biomed. Eng. Lett. 2019, 9, 413–424. [Google Scholar] [CrossRef]
- Rouis, M.; Ouafi, A.; Sbaa, S. Optimal level and order detection in wavelet decomposition for PCG signal denoising. Biomed. Eng. Biomed. Tech. 2019, 64, 163–176. [Google Scholar] [CrossRef]
- Chowdhury, T.H.; Poudel, K.N.; Hu, Y. Time-Frequency Analysis, Denoising, Compression, Segmentation, and Classification of PCG Signals. IEEE Access 2017, 8, 160882–160890. [Google Scholar] [CrossRef]
| Feature | Mathematical Description | Explanation |
|---|---|---|
| 1 | The mean value of the current signal. | |
| 2 | The median value of the current signal. | |
| 3 | The standard deviation of the current signal. | |
| 4 | The mean absolute deviation. | |
| 5 | th term | first quartile of the current signal (Q1). |
| 6 | th term | third quartile of the current signal (Q3). |
| 7 | Q3–Q1 | The interquartile range, which is the difference between the first and third quartiles. |
| 8 | The skewness of the current signal. is an individual score; “m” is the population mean; “s” is the population standard deviation; N is the population size. | |
| 9 | Kurtosis of the current signal.
is an individual score; “m” is the population mean; “s” is the population standard deviation; N is the population size. | |
| 10 | Shannon entropy value of the current signal. | |
| 11 | After applying the fast Fourier transform, the Shannon energy is computed (spectral entropy of the current signal). Note: denotes the Fourier transform of the current signal. | |
| 12 | The maximum frequency (Hz) after applying the fast Fourier transform. | |
| 13 | The maximum frequency spectrum value after applying the fast Fourier transform. | |
| 14 | The ratio of the energy of the maximum frequency to the total energy. | |
| 5th to 27th | 13 coefficients from the MFFCs | Mel Frequency Cepstral Coefficients |
| Feature | Mathematical Description | Explanation |
|---|---|---|
| 1 | This feature is used to represent the mean value of the systolic intervals. | |
| 2 | This feature is used to represent the mean value of the diastolic intervals. | |
| 3 | This feature is used to represent the standard deviation of the systolic intervals. | |
| 4 | This feature is used to represent the standard deviation of the systolic intervals. | |
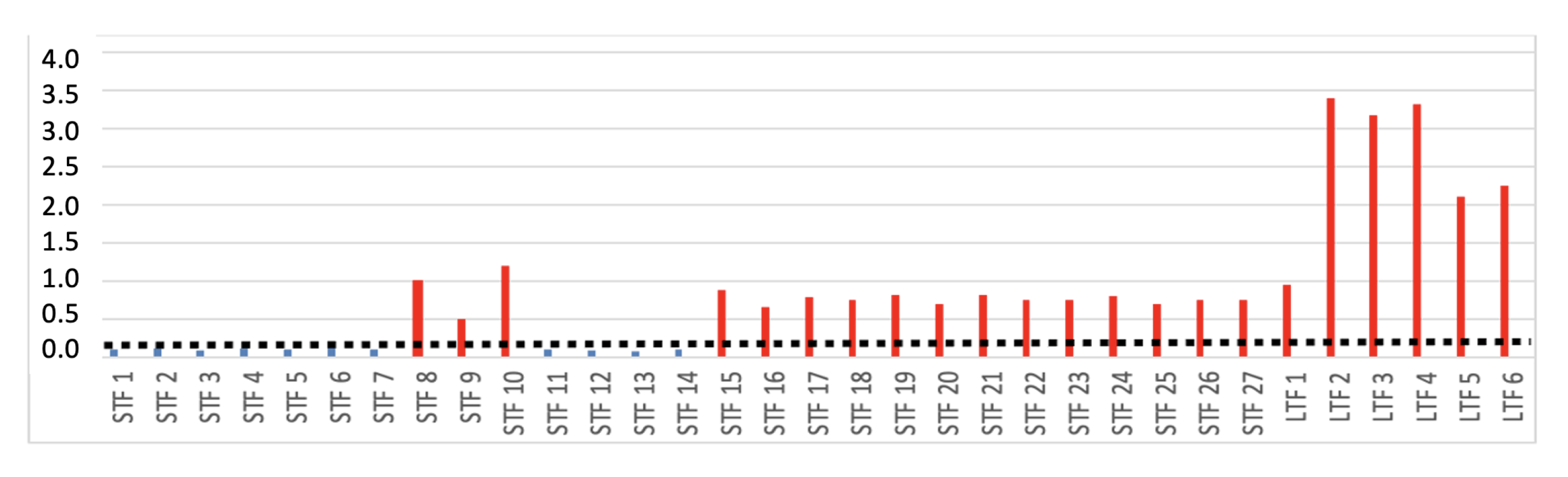
| 5 | Ratio of rejected peaks to total peaks. | |
| 6 | Amplitude value of rejected peaks to all. |
| Abbreviation | Number of Features | Explanation |
|---|---|---|
| STF | 27 | Short-term features. |
| LST | 6 | Long-term features. |
| LSTF | 33 | Short-term features + long-term features. |
| SSTF | 16 | Short-term features after feature reduction. |
| SLSTF | 22 | Short-term features after feature reduction + long-term features. |
| Technique | Algorithms | |||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| Decision Trees | Fine Tree | Medium Tree | Coarse Tree | |||
| Naive Bayes (NB) | Gaussian NB | Kernel NB | ||||
| Support Vector Machines (SVMs) | Linear SVM | Quadratic SVM | Cubic SVM | Fine SVM | Medium SVM | Coarse SVM |
| K-Nearest Neighbors (KNNs) | Fine KNN | Medium KNN | Coarse KNN | Cosine KNN | Cubic KNN | Weighted KNN |
| Ensemble Methods | Boosted Tree | Bagged Tree | Subspace Discriminant | Subspace KNN | RUSBoosted Tree | |
| Technique | Feature Sets | ||||
|---|---|---|---|---|---|
| STF | LTF | LSTF | SSTF | SLSTF | |
| Decision Trees | 78.6% | 77.9% | 87.8% | 83% | 70% |
| Naive Bayes | 68% | 75.2% | 83.3% | 71.6% | 76% |
| Support Vector Machines | 81.6% | 74.5% | 91.2% | 79% | 80.5% |
| K-Nearest Neighbors | 87.3% | 86% | 89.9% | 86.9% | 88% |
| Ensemble Methods | 90.3% | 89% | 92.7% | 90.5% | 90.1% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guven, M.; Uysal, F. A New Method for Heart Disease Detection: Long Short-Term Feature Extraction from Heart Sound Data. Sensors 2023, 23, 5835. https://doi.org/10.3390/s23135835
Guven M, Uysal F. A New Method for Heart Disease Detection: Long Short-Term Feature Extraction from Heart Sound Data. Sensors. 2023; 23(13):5835. https://doi.org/10.3390/s23135835
Chicago/Turabian StyleGuven, Mesut, and Fatih Uysal. 2023. "A New Method for Heart Disease Detection: Long Short-Term Feature Extraction from Heart Sound Data" Sensors 23, no. 13: 5835. https://doi.org/10.3390/s23135835
APA StyleGuven, M., & Uysal, F. (2023). A New Method for Heart Disease Detection: Long Short-Term Feature Extraction from Heart Sound Data. Sensors, 23(13), 5835. https://doi.org/10.3390/s23135835







