Abstract
Quick, efficient, fully automated open-source programs to segment muscle and adipose tissues from computed tomography (CT) images would be a great contribution to body composition research. This study examined the concordance of cross-sectional areas (CSA) and densities for muscle, visceral adipose tissue (VAT), subcutaneous adipose tissue (SAT), and intramuscular adipose tissue (IMAT) from CT images at the third lumbar (L3) between an automated neural network (test method) and a semi-automatic human-based program (reference method). Concordance was further evaluated by disease status, sex, race/ethnicity, BMI categories. Agreement statistics applied included Lin’s Concordance (CCC), Spearman correlation coefficient (SCC), Sorensen dice-similarity coefficient (DSC), and Bland–Altman plots with limits of agreement (LOA) within 1.96 standard deviation. A total of 420 images from a diverse cohort of patients (60.35 ± 10.92 years; body mass index (BMI) of 28.77 ± 7.04 kg/m2; 55% female; 53% Black) were included in this study. About 30% of patients were healthy (i.e., received a CT scan for acute illness or pre-surgical donor work-up), while another 30% had a diagnosis of colorectal cancer. The CCC, SCC, and DSC estimates for muscle, VAT, SAT were all greater than 0.80 (>0.80 indicates good performance). Agreement analysis by diagnosis showed good performance for the test method except for critical illness (DSC 0.65–0.87). Bland–Altman plots revealed narrow LOA suggestive of good agreement despite minimal proportional bias around the zero-bias line for muscle, SAT, and IMAT CSA. The test method shows good performance and almost perfect concordance for L3 muscle, VAT, SAT, and IMAT per DSC estimates, and Bland–Altman plots even after stratification by sex, race/ethnicity, and BMI categories. Care must be taken to assess the density of the CT images from critically ill patients before applying the automated neural network (test method).
1. Introduction
Computed tomography (CT) images have been used for assessment of regional body composition in many clinical populations including cancer patients [1,2,3,4,5,6,7]. However, the technique for analyzing CT images is laborious, time-consuming, costly, and requires intensive training [8,9]. A commonly applied reference technique is a semi-automated method with human-analyzed correction of segmentation on CT imaging using the medical imaging software, SliceOmatic plus ABACS (TomoVision and Voronoi Health Analytics, Montreal, QC, Canada). The manual correction process takes approximately 15–20 min, which translates to approximately 1500 min or 60 h of work to analyze 100 CT images. Thus, the biggest limitation to CT body composition research is the lack of relatively easy and inexpensive programs or tools to detect and quantify muscle and adipose tissues at single or multiple spinal landmarks quickly and accurately. Various automated programs or neural networks have already been developed and tested (Paris, 2020, Automated body composition analysis of clinically acquired computed tomography scans using neural networks). A comprehensive list of neural networks with DSC estimates for various landmarks was recently published; however, most of these are not open-source programs. Paris et al. recently developed and validated a completely automated, neural-network, open-source framework called AutoMATiCA for the expeditious analysis of large repositories of CT images for cross-sectional areas (CSA) of various abdominal tissues (https://gitlab.com/Michael_Paris/AutoMATiCA accessed on 22 June 2021) [9]. In comparison to the human-analyzed technique, the AutoMATiCA program analyzes an image for various abdominal tissues in approximately 350 milliseconds per CT image, which translates to 35 min for complete analysis of 100 images.
In their paper, Paris et al. confirmed a high degree of agreement between this new program, AutoMATiCA (automated neural network), and a reference human-analyzed technique for CT images using the SliceOmatic software [9]. Following validation, Bland–Altman plots comparing neural-network analysis to human-analyzed CSA for different body composition parameters including muscle and various adipose tissues (intermuscular, visceral, subcutaneous) showed no biases (within limits of agreement). In addition, the authors reported strong agreement between the two methods using the Sorenson dice-similarity coefficient (DSC) of >0.80 [9,10]. For muscle, the DSC was 0.983 ± 0.013l between the human-analyzed technique and the network-predicted segmentation, for intermuscular adipose tissue (IMAT) it was 0.900 ± 0.034, for visceral adipose tissue (VAT), the DSC was 0.979 ± 0.019, and for subcutaneous adipose tissue (SAT), it was 0.986 ± 0.016 [9].
Given the need for validation among diverse patient populations and confirmation of the findings as reported by Paris et al., we tested the fully automatic program (automated neural network—AutoMATiCA) against SliceOmatic plus ABACS (reference method) with the human-based manual correction for analyzing CT images [9]. Additionally, the level of agreement using DSC and Bland–Altman plots as identified by Paris et al. needs to be further examined in subgroups of diverse communities: by sex, race/ethnicity, body mass index (BMI), and disease status. Thus, the purpose of this study was twofold: first, to confirm the concordance of a newly developed automated neural network (AutoMATiCA) for analyzing CT images for body composition at third lumbar (L3) vertebra with a reference semi-automatic human–based manual technique using the SliceOmatic plus ABACS Software; and second, to further examine the concordance between the two methods by disease status, sex, race/ethnicity, and BMI categories.
2. Materials and Methods
2.1. Study Design and Sample Population
A retrospective review was conducted on various adults at a major tertiary medical center who underwent CT abdominal imaging. Clinically acquired CT images obtained previously for research studies using electronic medical records of various adult medical and surgical patient populations (>18 years of age) were included. These images were previously analyzed with the reference software (SliceOmatic plus ABACS) using the semi-automatic plus human-based manual analysis technique as part of past research collaborations. CT images were obtained from retrospective cohort studies in various diverse clinical populations (breast cancer, 3%; COVID-19; 5%; colorectal cancer, 30%; critical illness, 9%; healthy controls, 31%; and metastatic breast cancer, 22%) at Rush University Medical Center (RUMC), University of Illinois at Chicago, and Loyola University Medical Center. CT images were previously anonymized. CT images included in this study were analyzed centrally by trained experts at RUMC Department of Clinical Nutrition. CT images were excluded if the images were poor quality or unevaluable (i.e., anasarca, poor positioning, excessive cutoffs, grainy, etc.). A total of 418 unique patients contributed 420 images for this study. Two patients provided two images to the study. The research studies providing anonymized CT images were previously approved according to institutional research review boards. Additionally, this study was reviewed and approved by the RUMC Institutional Review Board in accordance with protocols for human subject research.
The demographic and clinical variables collected for each adult patient were age (years), sex (male, female), race/ethnicity (White, Black, other, refused or unknown), clinical diagnosis, height (cm), weight (kg), and body mass index (BMI, kg/m2).
2.2. SliceOmatic Plus ABACS Analysis (Reference Method)
A total of 420 CT images at the third lumbar (L3) landmark previously analyzed for CSA and density of adipose tissues and muscle from clinically indicated CT scans were included in this study. Using standard protocols for abdominal body composition analysis, trained experts previously analyzed each single image to determine CSA (cm2) for SAT, VAT, IMAT, and muscle using medical imaging applications SliceOmatic plus ABACS v4.3 (TomoVision and Voronoi Health Analytics, Montreal, QC, Canada) [2,11]. The medical imaging software package permits segmental demarcation of each tissue compartment according to specific Hounsfield unit (HU). The HU scale from −1000 (representing air) to +2000 (representing bone) was developed by Sir Godfrey Hounsfield, inventor of the CT scanner, to measure the radiographic attenuation of images taken during a CT scan. The HU tissue specific threshold for VAT is −150 to −50 HU, for SAT and IMAT is −190 to −30 HU, and for muscle is −29 to 150 HU [2]. The tissue specific thresholds are preprogrammed. Density (mean HU) was also determined by the software for each adipose and skeletal tissue. Tissue boundaries were corrected manually as needed. The complete analysis of a single CT image took approximately 15–20 min. Intra-class coefficient of variations for body composition analysis for trained experts were previously examined and recorded to be less than 2%.
2.3. AutoMATiCA Analysis (Test Method)
AutoMATiCA conducts an automatic segmental analysis that yields estimates of cross-sectional area (CSA) and density (mean Hounsfield (HU) unit) of each adipose and muscle for each CT image at the L3 landmark [2]. To this end, the same 420 CT images analyzed segmentally with the reference method were processed for CSA (cm2) and tissue density using the test method, AutoMATiCA. Briefly, CT images were loaded into the program, a location to save the images and results was identified, and automated segmentation was performed the default segmentation determines CSA and density estimates for all the tissues at the L3 landmark (i.e., muscle, VAT, SAT, IMAT). Default HU ranges (HU thresholds of −150 to −50 HU for VAT, −190 to −30 HU for SAT and IMAT, and −29 to 150 HU for muscle) were maintained for body composition analysis. The option to save pictures in jpeg format of each image after segmentation was also selected. Segmental analysis for CSA and density estimates of each tissue using the test method took approximately 350 milliseconds per CT image. After the analysis, the segmentation of CSA for muscle, VAT, SAT, and IMAT was visually inspected using the picture files produced by the test method to evaluate if any of the images were incompletely segmented (defined as absence of color tagging or erroneous color tagging). The program neatly provides pictures (i.e., jpeg files) of all CT images in one folder, which enables the visual inspection of images at one time using Windows Explorer. Visual inspection took approximately 10 min to complete for the 420 images.
2.4. Statistical Analyses
Categorical variables were presented as percentage frequencies, with continuous variables presented as means ± standard deviations. Correlation between the two methods was evaluated using Lin’s concordance correlation coefficient (CCC) [12], intraclass correlation coefficient (ICC), and Spearman correlation coefficient. Agreement between the AutoMATiCA and human-based manual analysis technique were evaluated using: Dice similarity coefficient (DSC) and Bland–Altman plots [13]. The DSC values were compared across disease states, sex, and race/ethnicity using Mann–Whitney U test or Kruskal–Wallis Test. A score of ‘1’ for DSC implies perfect agreement, whereas a score of ‘0’ indicates no overlap or agreement. Bland–Altman plots were used to evaluate agreement between the two methods and to also determine limits of agreement (LOA) within 1.96 standard deviations. Statistical significance was defined as p < 0.05. Analyses were performed with SAS v9.4 (SAS Institute, Cary, NC, USA).
3. Results
3.1. Patient Characteristics
The sample consisted of 231 female patients (55%) and 187 male patients with various diagnoses including patients designated as healthy adults (n = 128) with acute illness (i.e., abdominal pain, hernia work-up) or receiving pre-surgical evaluations (i.e., organ donor, elective surgery work-up), patients with colorectal cancer (n = 127), and patients with metastatic breast cancer (n = 92), as shown in Table 1. Most patients were between 40–65 years of age (62%) and identified as either non-Hispanic Black (53%) or White (40%). The sample was mostly overweight (35%), followed by obese (34%) and low-normal (30%) BMI.

Table 1.
Patient Characteristics.
3.2. Body Composition Comparisons
The muscle, VAT, SAT, and IMAT CSA and tissue densities for the test and reference methods are shown in Table 2. Briefly, the muscle CSA for the test method versus the reference method was 143.19 ± 36 cm2 vs. 139.34 ± 37.91 cm2, respectively (Table 2). The VAT CSA for the test method was 122.08 ± 95.75 cm2 vs. 117.82 ± 93.56 cm2. The SAT CSA for the test method was 226.32 ± 142.55 cm2 vs. 215.41 ± 140.76 cm2 for the reference method. The muscle density for the test method was 35.52 ± 10.83 HU compared to 37.43 ± 16.34 HU for the reference method and for VAT HU it was −86.95 ± 9.83 HU vs. −87.10 ± 13.18 HU, respectively.

Table 2.
Summary of body composition parameters following segmental tissue analysis using test and reference methods.
3.3. Correlation and Agreement Comparisons
Table 3 highlights the correlation and agreement between the test reference method using agreement statistics. The CCC, ICC, and Spearman’s correlation for muscle, VAT, SAT were all greater than 0.80, indicating strong positive correlations. The DSC estimates reflecting the overall accuracy of the test method compared to the reference for CSA and densities was lowest for IMAT CSA (0.83 ± 0.15) and highest for VAT density (0.99 ± 0.07) demonstrating good to almost perfect agreement.

Table 3.
Summary of correlation and agreement statistics for patient cohort measured by Lin’s concordance correlation coefficient, intra-class correlation, Spearman correlation coefficients, and Bland-Altman summary statistics including assessment of proportional bias using Pearson correlation coefficients.
3.4. Agreement Comparisons by Subgroups
The lowest DSC estimates were observed for patients with critical illness (n = 37) as shown in Table 4. The lowest DSC value observed in these patients was 0.65 for VAT CSA and highest for muscle CSA (0.87). In comparison, DSC estimates for all other diagnoses including metastatic breast cancer ranged between 0.91 (good agreement) to 0.99 (perfect agreement) excluding critical illness. Small (DSC ≤ 0.15) yet statistically significant differences in DSC scores were observed across diagnosis categories with critical illness having the largest difference for VAT CSA (DSC = 0.33). Per visual inspection of analyzed images by test method, a total of 12 of 28 images were incompletely segmented with the test method (Figure 1).

Table 4.
Dice similarity coefficient estimates across diagnosis.
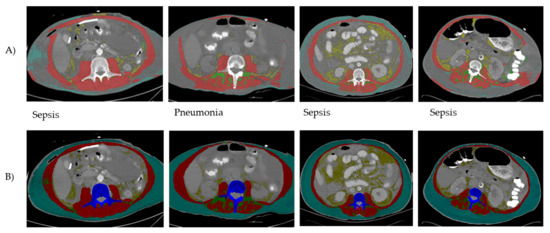
Figure 1.
Examples of CT images following AutoMATiCA (test method) analysis (A) compared to semi-automated SliceOmatic (reference method) analysis (B). Discrepancies in tagging (i.e., coloring of different tissues) by the test method were noticed for all the abdominal tissues at this landmark (skeletal muscle and adipose tissues). The inconsistent performance of the test method was noted mainly in patients with a critical illness diagnosis.
DSC estimates for muscle CSA and tissue densities stratified by sex and race/ethnicity remained in almost perfect agreement (DSC ≥ 0.97) with the exception of IMAT CSA (Table 5 and Supplemental Table S1 and Table S2). The IMAT CSA had the lowest DSC estimates for sex (females: 0.85 ± 0.15; males 0.80 ± 0.13), race/ethnicity (Blacks, n = 223: 0.82 ± 0.14; Whites, n = 169: 0.85 ± 0.15; Other, n = 28: 0.85 ± 0.18), and sex and race/ethnic categories as observed for the full sample. For BMI categories, the lowest DSC estimates were shown for IMAT CSA (DSC 0.82–0.84) compared to near perfect agreement for other body composition parameters (DSC > 0.90), as shown in Supplemental Table S3 (Supplemental Materials). The DSC for other parameters were consistent with strong to perfect agreement. Additionally, small (DSC ≤ 0.21) but statistically significant differences in DSC scores were observed by sex, race, sex and race/ethnic categories, and by BMI. As before, DSC estimates for remaining body composition by subgrouping consistent with good (>0.80) for IMAT CSA to near perfect agreement for other CSAs and densities (DSC 0.91–0.99).

Table 5.
DSC by Sex and Race/ethnic categories.
Bland–Altman plots of CSA and densities for muscle, VAT, SAT, and IMAT showed narrow LOA of 3.85 [−29.99 and 37.70] cm2, 4.26 [−84.45 and 92.97] cm2, 10.91 [−97.98 and 119.79] cm2, −3.65 [7.30 and −17.95] cm2, −0.63 [−25.71 and 24.45] HU, −0.53 [−16.50 and 15.45] HU, 0.45 [−13.23 and 14.13] HU, 0.26 [−11.19 and 11.72] HU, respectively, supportive of good to excellent agreement with minimal proportional bias between the test and reference method for the full sample (Table 3, Figure 2 and Figure 3). Specific analysis of proportional bias revealed statistically significant bias for muscle CSA and muscle, VAT, SAT, and IMAT densities for the full sample (Table 3). The average bias line for IMAT CSA density was below the zero bias line, suggesting that, on average, the test method measurements are lower than the measurements estimated by the reference method.
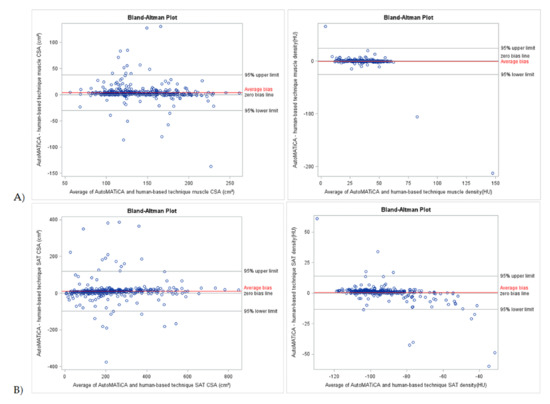
Figure 2.
Bland-Altman plots of body composition parameters between test (AutoMATica) and reference (SliceOmatic plus ABACS + manual correction) method for entire sample. Plots show cross-sectional area (CSA) and density for muscle (A) and subcutaneous adipose tissue (SAT, B). Limits of agreement within 1.96 standard deviations are shown with average bias (red line) for each plot.
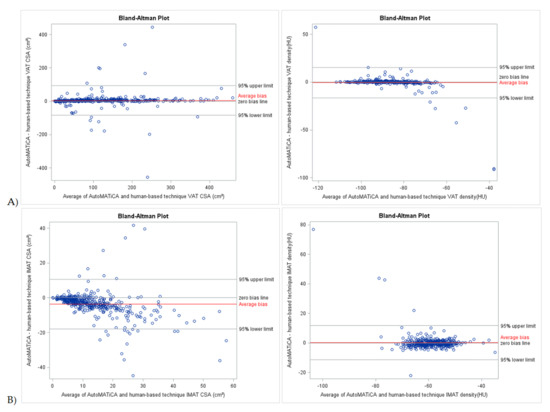
Figure 3.
Bland-Altman plots of body composition parameters between test (AutoMATica) and reference (SliceOmatic plus ABACS + manual correction) method for entire sample. Plots show cross-sectional area (CSA) and density visceral adipose tissue (VAT, A), and intermuscular adipose tissue (IMAT, B). Limits of agreement within 1.96 standard deviations are shown with average bias (red line) for each plot.
Additional Bland–Altman and proportional bias analyses for sex, race, sex and race/ethnic categories and BMI are available in the Supplementary Materials. Proportional biases observed for the full sample remained for the same body composition parameters when explored by sex and race/ethnic categories with these exceptions: (1) proportional bias was not observed for muscle CSA and IMAT density in the Black subjects, and (2) proportional bias was not observed for muscle and IMAT CSA in the others category ( Supplemental Tables S4–S7).
4. Discussion
The validity of the test method (AutoMATiCA) was supported by the DSC scores for muscle, VAT, and SAT CSA at the L3 landmark, as observed in this study, which are consistent with estimates reported by others using automated segmentation neural networks [9,14,15]. These findings were consistent with Paris et al. (AutoMATiCA developer), who reported DCS estimates > 0.97 for muscle, VAT, and SAT and good accuracy for IMAT CSA (DSC > 0.88) [9]. Corresponding Bland–Altman plots of these body composition parameters revealed narrow LOA for the diverse cohort of patients evaluated in this study. Additionally, the lower DSC estimates for VAT, SAT, and IMAT CSA in this study were observed for the small cohort of critically ill patients (DSC > 0.65) but higher for muscle CSA (DSC > 0.87), suggesting better estimation of muscle CSA and overall good concordance between test and reference methods specific for this tissue. However, the DSC scores for muscle (0.87), IMAT (0.69), VAT (0.65), and SAT (0.69) for critically ill patients (n= 37) in the intensive care unit (ICU) in this study were much lower compared to scores reported for ICU patients (n = 30) by Paris et al., (IMAT DSC = 0.88; VAT DSC = 0.98; SAT DSC = 0.98) [9]. This difference in scores for IMAT, VAT, and SAT between the reference and test method suggests that CT images of critically ill patients analyzed using the test method (AutoMATiCA) should be reviewed post-analysis to determine which images may need to be reanalyzed. Critically ill patients often have fluid retention and anasarca, which can impact body composition analysis, which has been similarly suggested in patients with severe COVID-19 illness requiring hospitalization [16]. Thus, the presence of fluid retention and anasarca in critically ill patients may have limited the ability of the automated neural network, AutoMATiCA (test method), to differentiate tissue voxels in muscle and other tissues. Additionally, these data showed good to near perfect agreement (DSC > 0.80–0.99) between the two methods for most tissues including IMAT CSA following stratification by sex, BMI categories, race/ethnicity, and sex and race/ethnicity categories. Although small statistically significant differences in DSC scores were observed when the CT data were stratified by sex, race, and BMI, these estimates are likely clinically irrelevant. Such evidence further validates the utility of the test method for quick, efficient, accurate measurements of CSA and densities of abdominal tissues regardless of demographic characteristics.
Two recently published research studies also examine newer prototypes of automated artificial intelligence (AI) neural networks compared to human-based segmentation protocols for muscle, VAT, SAT, and IMAT CSA. Borrelli et al. examined the accuracy of an AI method to analyze CT images for muscle and SAT volume compared to a single CT image at the L3 landmark segmentation analysis (reference method) using a training group of 50 patients and a test group of 74 cancer patients providing two images each [14]. Their estimates of DSC for SAT (0.96, range 0.82–0.97) and muscle (0.94, range 0.82–0.97) volumes were similar to estimates for SAT CSA (0.93) and muscle CSA (0.97) for this study. Their findings support the use of their AI method for SAT and muscle volume estimation from CT images. Ackermans et al. also recently published a study testing the accuracy of a deep learning neural network trained on 3413 CT images for VAT, SAT, and muscle segmentation at the L3 vertebra compared against manual segmentation by a trained investigator [15]. The deep learning neural network was tested on 233 patients. Their results showed a median DSC of 0.93 (range 0.86–0.96) for muscle CSA, 0.95 (range 0.89–0.97) for VAT CSA, and 0.95 (range 0.92–0.96) for SAT CSA between the manual segmentation and newly developed deep learning neural network. These DSC scores reported by Ackermans et al. also aligned with the DSC scores obtained in this study for SAT and muscle CSA as stated above and for VAT CSA (DSC = 0.92). These data also support the neural network developed by Ackermans et al. for CSA body composition analysis of muscle, VAT, and SAT at the L3 landmark. Similar to the AutoMATiCA neural network tested in the present study, the AI-based neural networks by Borrelli et al. and Ackermans et al. provide reliable and quick analysis of body composition parameters at the L3 landmark [14,15] as well as volume estimates [15]. However, an additional benefit of using the test method (AutoMATiCA) developed by Paris et al. [9] that was validated in this study, aside from it being open-source, is that it also provides estimates for IMAT and VAT as well as estimates of tissue density unlike the neural networks developed by Borrelli et al. and Ackermans et al.
5. Conclusions
The test method shows good performance and almost perfect concordance for body composition analysis at L3 per DSC estimates and Bland–Altman plots for muscle, VAT, SAT, and IMAT for most clinical populations evaluated in this study. Care must be taken to assess the density of the CT images from critically ill patients before applying the automated neural network (test method). An important next step would be determining what is it about the CT images from critically ill patients at the L3 region that makes them less compatible with this neural network. Once this is determined, future studies using larger, more diverse clinical populations, particularly for critically ill patients, are needed to confirm the utility, accuracy, and generalizability of the automated neural network (AutoMATiCA) tested herein.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/s22093357/s1, Table S1: DSC by Sex; Table S2: DSC by Race/Ethnic categories; Table S3: DSC by BMI; Table S4: Bland–Altman and proportional bias statistics for females; Table S5: Bland–Altman and proportional bias statistics for males; Table S6: Bland–Altman and proportional bias statistics for Black race/ethnic category; Table S7: Bland–Altman and proportional bias statistics for White race/ethnic category; Table S8: Bland–Altman and proportional bias statistics for Others race/ethnic category.
Author Contributions
Conceptualization, S.L.G.-P.; methodology, S.L.G.-P., Y.Z. and S.P.; validation, S.L.G.-P., Y.Z. and S.P.; formal analysis, Y.Z. and S.L.G.-P.; investigation, S.L.G.-P. and S.P.; resources, S.L.G.-P., J.S., S.P., C.W. and T.G.; data curation, S.L.G.-P. and Y.Z.; writing—original draft preparation, S.L.G.-P.; writing—review and editing, S.L.G.-P., Y.Z., S.P., C.B., C.W., T.G. and J.S.; visualization, S.L.G.-P., Y.Z. and S.P.; supervision, S.L.G.-P., J.S. and S.P.; project administration, S.L.G.-P.; funding acquisition, S.L.G.-P. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the National Cancer Institute 5R01CA204808-03 (S.G.P.).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Rush University (ORA#21042301, 21 April 2021). The original research studies providing anonymized computed tomography data were approved by the research ethics institutional review board (IRB) at Rush University Medical Center.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
We would like to thank Patricia Sheean for providing anonymized CT images for this project.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Martin, L.; Birdsell, L.; Macdonald, N.; Reiman, T.; Clandinin, M.T.; McCargar, L.J.; Murphy, R.; Ghosh, S.; Sawyer, M.B.; Baracos, V.E. Cancer cachexia in the age of obesity: Skeletal muscle depletion is a powerful prognostic factor, independent of body mass index. J. Clin. Oncol. 2013, 31, 1539–1547. [Google Scholar] [CrossRef] [PubMed]
- Prado, C.M.; Lieffers, J.R.; McCargar, L.J.; Reiman, T.; Sawyer, M.B.; Martin, L.; Baracos, V.E. Prevalence and clinical implications of sarcopenic obesity in patients with solid tumours of the respiratory and gastrointestinal tracts: A population-based study. Lancet Oncol. 2008, 9, 629–635. [Google Scholar] [CrossRef]
- Gomez-Perez, S.L.; Chaudhry, V.; Mar, W.; Patel, B.; Fantuzzi, G.; Freels, S.; Braunschweig, C.A. Impact of abdominal adipose depots and race on risk for colorectal cancer: A case-control study. Nutr. Cancer 2017, 69, 573–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braunschweig, C.A.; Sheean, P.M.; Peterson, S.J.; Gomez Perez, S.; Freels, S.; Lateef, O.; Gurka, D.; Fantuzzi, G. Intensive Nutrition in Acute Lung Injury: A Clinical Trial (INTACT). JPEN J. Parenter. Enter. Nutr. 2014, 39, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Peterson, S.J.; Braunschweig, C.A. Prevalence of Sarcopenia and Associated Outcomes in the Clinical Setting. Nutr. Clin. Pract. 2016, 31, 40–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, M.; Caruso, D.; Tuccinardi, D.; Risi, R.; Zerunian, M.; Polici, M.; Pucciarelli, F.; Tarallo, M.; Strigari, L.; Manfrini, S.; et al. Visceral fat shows the strongest association with the need of intensive care in patients with COVID-19. Metabolism 2020, 111, 154319. [Google Scholar] [CrossRef] [PubMed]
- Gualtieri, P.; Falcone, C.; Romano, L.; Macheda, S.; Correale, P.; Arciello, P.; Polimeni, N.; Lorenzo, A. Body Composition Findings by Computed Tomography in SARS-CoV-2 Patients: Increased Risk of Muscle Wasting in Obesity. Int. J. Mol. Sci. 2020, 21, 4670. [Google Scholar] [CrossRef] [PubMed]
- Earthman, C.P. Body Composition Tools for Assessment of Adult Malnutrition at the Bedside: A Tutorial on Research Considerations and Clinical Applications. JPEN J. Parenter. Enter. Nutr. 2015, 39, 787–822. [Google Scholar] [CrossRef] [PubMed]
- Paris, M.T.; Tandon, P.; Heyland, D.K.; Furberg, H.; Premji, T.; Low, G.; Mourtzakis, M. Automated body composition analysis of clinically acquired computed tomography scans using neural networks. Clin. Nutr. 2020, 39, 3049–3055. [Google Scholar] [CrossRef] [PubMed]
- Zou, K.H.; Warfield, S.K.; Bharatha, A.; Tempany, C.M.C.; Kaus, M.R.; Haker, S.J.; Wells, W.M., 3rd; Jolesz, F.A.; Kikinis, R. Statistical validation of image segmentation quality based on a spatial overlap index. Acad. Radiol. 2004, 11, 178–189. [Google Scholar] [CrossRef] [Green Version]
- Mourtzakis, M.; Prado, C.M.; Lieffers, J.R.; Reiman, T.; McCargar, L.J.; Baracos, V.E. A practical and precise approach to quantification of body composition in cancer patients using computed tomography images acquired during routine care. Appl. Physiol. Nutr. Metab. 2008, 33, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Hedayat, A.S.; Sinha, B.; Yang, M. Statistical Methods in Assessing Agreement. J. Am. Stat. Assoc. 2002, 97, 257–270. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986, 1, 307–310. [Google Scholar] [CrossRef]
- Borrelli, P.; Kaboteh, R.; Enqvist, O.; Ulén, J.; Trägårdh, E.; Kjölhede, H.; Edenbrandt, L. Artificial intelligence-aided CT segmentation for body composition analysis: A validation study. Eur. Radiol. Exp. 2021, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Ackermans, L.; Volmer, L.; Wee, L.; Brecheisen, R.; Sánchez-González, P.; Seiffert, A.P.; Gómez, E.J.; Dekker, A.; Ten Bosch, J.A.; Damink, S.M.W.O.; et al. Deep Learning Automated Segmentation for Muscle and Adipose Tissue from Abdominal Computed Tomography in Polytrauma Patients. Sensors 2021, 21, 2083. [Google Scholar] [CrossRef]
- Chandarana, H.; Pisuchpen, N.; Krieger, R.; Dane, B.; Mikheev, A.; Feng, Y.; Kambadakone, A.; Rusinek, H. Association of body composition parameters measured on CT with risk of hospitalization in patients with COVID-19. Eur. J. Radiol. 2021, 145, 110031. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).