Estimation of Soil Organic Carbon Content in the Ebinur Lake Wetland, Xinjiang, China, Based on Multisource Remote Sensing Data and Ensemble Learning Algorithms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Soil Data Source
2.3. Environmental Variables
2.3.1. Topographic Variables
2.3.2. Remote Sensing Variables and Processing
2.3.3. Index Variables for Remote Sensing Data
2.3.4. Climate Variables
2.4. Modeling Techniques
2.4.1. Random Forest
2.4.2. Gradient Boosted Decision Tree
2.4.3. Extreme Gradient Boosting
2.5. Modeling Strategy and Validation Metrics
3. Results
3.1. Descriptive Statistics
3.2. Analysis of the Importance of Variables
3.3. Comparison of the Performances of Different Models
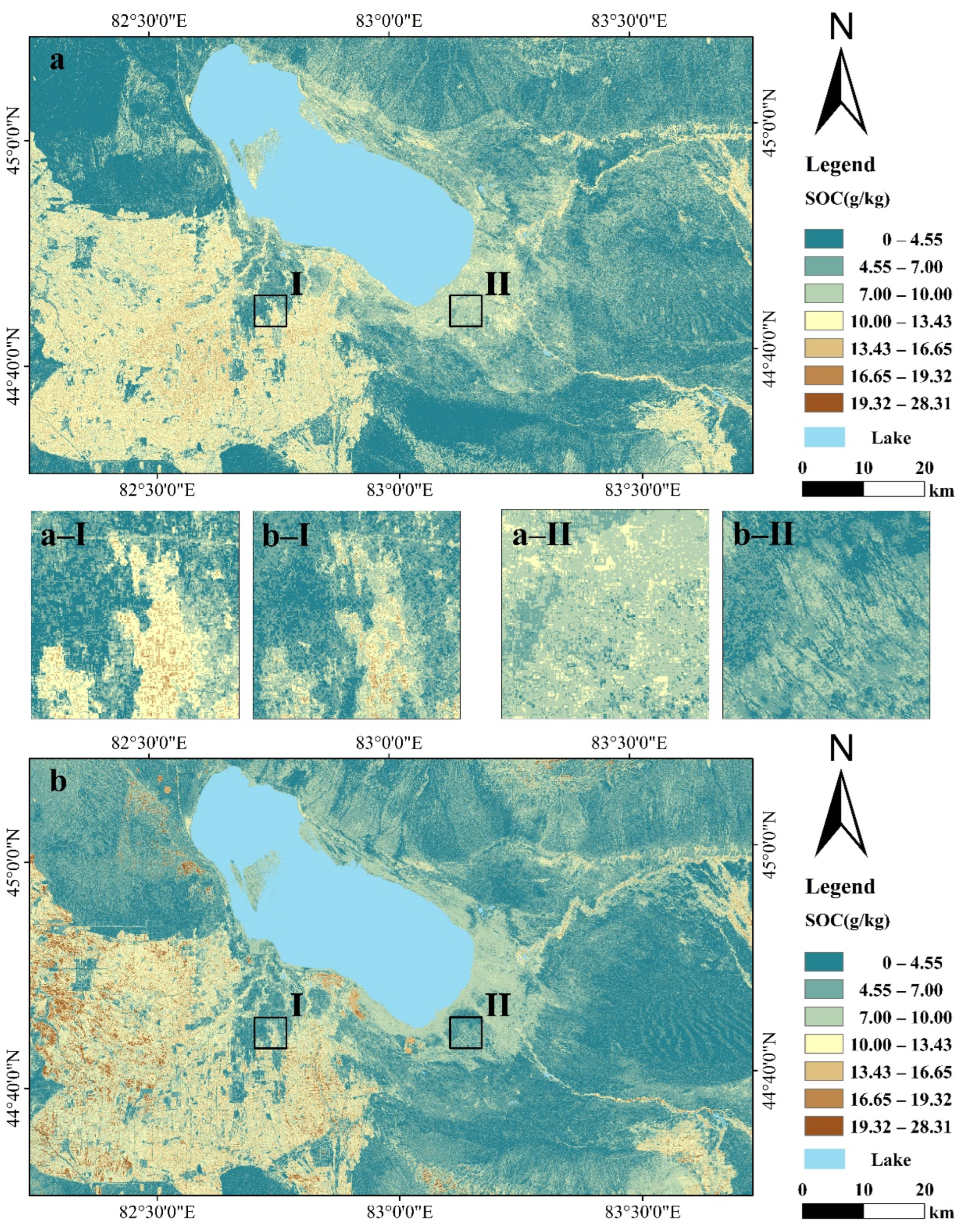
3.4. SOC Spatial Predictions from Landsat 8 and Sentinel-2A
4. Discussion
4.1. Comparison of Models Based on Landsat 8 and Sentinel-2A Data
4.2. Analysis of Environmental Variables
4.3. Uncertainty Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chabbi, A.; Lehmann, J.; Ciais, P.; Loescher, H.W.; Cotrufo, M.F.; Don, A.; SanClements, M.; Schipper, L.; Six, J.; Smith, P.; et al. Aligning agriculture and climate policy. Nat. Clim. Change 2017, 7, 307–309. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Pham, T.D.; Nguyen, C.T.; Delfos, J.; Archibald, R.; Dang, K.B.; Hoang, N.B.; Guo, W.; Ngo, H.H. A novel intelligence approach based active and ensemble learning for agricultural soil organic carbon prediction using multispectral and SAR data fusion. Sci. Total Environ. 2021, 804, 150187. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Geng, Y.; Ji, C.; Xu, X.; Wang, H.; Pan, J.; Bumberger, J.; Haase, D.; Lausch, A. Prediction of soil organic carbon and the C:N ratio on a national scale using machine learning and satellite data: A comparison between Sentinel-2, Sentinel-3 and Landsat-8 images. Sci. Total Environ. 2021, 755, 142661. [Google Scholar] [CrossRef]
- Zhao, D.; Zhu, Y.; Wu, S.; Lu, Q. Simulated response of soil organic carbon density to climate change in the Northern Tibet permafrost region. Geoderma 2022, 405, 115455. [Google Scholar] [CrossRef]
- Stockmann, U.; Adams, M.A.; Crawford, J.W.; Field, D.J.; Henakaarchchi, N.; Jenkins, M.; Minasny, B.; McBratney, A.B.; Courcelles, V.d.R.d.; Singh, K.; et al. The knowns, known unknowns and unknowns of sequestration of soil organic carbon. Agric. Ecosyst. Environ. 2013, 164, 80–99. [Google Scholar] [CrossRef]
- Smith, D.M.; Scaife, A.A.; Hawkins, E.; Bilbao, R.; Boer, G.J.; Caian, M.; Caron, L.P.; Danabasoglu, G.; Delworth, T.; Doblas-Reyes, F.J.; et al. Predicted Chance That Global Warming Will Temporarily Exceed 1.5 °C. Geophys. Res. Lett. 2018, 45, 11895–11903. [Google Scholar] [CrossRef] [Green Version]
- Amelung, W.; Bossio, D.; de Vries, W.; Kogel-Knabner, I.; Lehmann, J.; Amundson, R.; Bol, R.; Collins, C.; Lal, R.; Leifeld, J.; et al. Towards a global-scale soil climate mitigation strategy. Nat. Commun. 2020, 11, 5427. [Google Scholar] [CrossRef]
- Bossio, D.A.; Cook-Patton, S.C.; Ellis, P.W.; Fargione, J.; Sanderman, J.; Smith, P.; Wood, S.; Zomer, R.J.; von Unger, M.; Emmer, I.M.; et al. The role of soil carbon in natural climate solutions. Nat. Sustain. 2020, 3, 391–398. [Google Scholar] [CrossRef]
- Hui, D.; Forkuor, G.; Hounkpatin, O.K.L.; Welp, G.; Thiel, M. High Resolution Mapping of Soil Properties Using Remote Sensing Variables in South-Western Burkina Faso: A Comparison of Machine Learning and Multiple Linear Regression Models. PLoS ONE 2017, 12, e0170478. [Google Scholar] [CrossRef]
- Loiseau, T.; Chen, S.; Mulder, V.L.; Román Dobarco, M.; Richer-de-Forges, A.C.; Lehmann, S.; Bourennane, H.; Saby, N.P.A.; Martin, M.P.; Vaudour, E.; et al. Satellite data integration for soil clay content modelling at a national scale. Int. J. Appl. Earth Obs. Geoinf. 2019, 82, 101905. [Google Scholar] [CrossRef]
- McBratney, A.B.; Mendonça Santos, M.L.; Minasny, B. On digital soil mapping. Geoderma 2003, 117, 3–52. [Google Scholar] [CrossRef]
- Lamichhane, S.; Kumar, L.; Wilson, B. Digital soil mapping algorithms and covariates for soil organic carbon mapping and their implications: A review. Geoderma 2019, 352, 395–413. [Google Scholar] [CrossRef]
- Kang, Y.; Li, X.; Mao, D.; Wang, Z.; Liang, M. Combining Artificial Neural Network and Ordinary Kriging to Predict Wetland Soil Organic Carbon Concentration in China’s Liao River Basin. Sensors 2020, 20, 7005. [Google Scholar] [CrossRef] [PubMed]
- Jeong, G.; Oeverdieck, H.; Park, S.J.; Huwe, B.; Ließ, M. Spatial soil nutrients prediction using three supervised learning methods for assessment of land potentials in complex terrain. Catena 2017, 154, 73–84. [Google Scholar] [CrossRef]
- Camera, C.; Zomeni, Z.; Noller, J.S.; Zissimos, A.M.; Christoforou, I.C.; Bruggeman, A. A high resolution map of soil types and physical properties for Cyprus: A digital soil mapping optimization. Geoderma 2017, 285, 35–49. [Google Scholar] [CrossRef]
- Doetterl, S.; Stevens, A.; Six, J.; Merckx, R.; Van Oost, K.; Casanova Pinto, M.; Casanova-Katny, A.; Muñoz, C.; Boudin, M.; Zagal Venegas, E.; et al. Soil carbon storage controlled by interactions between geochemistry and climate. Nat. Geosci. 2015, 8, 780–783. [Google Scholar] [CrossRef]
- Viscarra Rossel, R.A.; Walvoort, D.J.J.; McBratney, A.B.; Janik, L.J.; Skjemstad, J.O. Visible, near infrared, mid infrared or combined diffuse reflectance spectroscopy for simultaneous assessment of various soil properties. Geoderma 2006, 131, 59–75. [Google Scholar] [CrossRef]
- Angelopoulou, T.; Tziolas, N.; Balafoutis, A.; Zalidis, G.; Bochtis, D. Remote Sensing Techniques for Soil Organic Carbon Estimation: A Review. Remote Sens. 2019, 11, 676. [Google Scholar] [CrossRef] [Green Version]
- Castaldi, F. Sentinel-2 and Landsat-8 Multi-Temporal Series to Estimate Topsoil Properties on Croplands. Remote Sens. 2021, 13, 3345. [Google Scholar] [CrossRef]
- Gholizadeh, A.; Žižala, D.; Saberioon, M.; Borůvka, L. Soil organic carbon and texture retrieving and mapping using proximal, airborne and Sentinel-2 spectral imaging. Remote Sens. Environ. 2018, 218, 89–103. [Google Scholar] [CrossRef]
- Bartsch, A.; Widhalm, B.; Kuhry, P.; Hugelius, G.; Palmtag, J.; Siewert, M.B. Can C-band synthetic aperture radar be used to estimate soil organic carbon storage in tundra? Biogeosciences 2016, 13, 5453–5470. [Google Scholar] [CrossRef] [Green Version]
- Doetterl, S.; Stevens, A.; van Oost, K.; Quine, T.A.; van Wesemael, B. Spatially-explicit regional-scale prediction of soil organic carbon stocks in cropland using environmental variables and mixed model approaches. Geoderma 2013, 204–205, 31–42. [Google Scholar] [CrossRef]
- Meersmans, J.; De Ridder, F.; Canters, F.; De Baets, S.; Van Molle, M. A multiple regression approach to assess the spatial distribution of Soil Organic Carbon (SOC) at the regional scale (Flanders, Belgium). Geoderma 2008, 143, 1–13. [Google Scholar] [CrossRef]
- Kumar, S.; Lal, R.; Liu, D.; Rafiq, R. Estimating the spatial distribution of organic carbon density for the soils of Ohio, USA. J. Geogr. Sci. 2013, 23, 280–296. [Google Scholar] [CrossRef]
- Lark, R.M. Soil–landform relationships at within-field scales: An investigation using continuous classification. Geoderma 1999, 92, 141–165. [Google Scholar] [CrossRef]
- Li, X.; Ding, J.; Liu, J.; Ge, X.; Zhang, J. Digital Mapping of Soil Organic Carbon Using Sentinel Series Data: A Case Study of the Ebinur Lake Watershed in Xinjiang. Remote Sens. 2021, 13, 769. [Google Scholar] [CrossRef]
- Pouladi, N.; Møller, A.B.; Tabatabai, S.; Greve, M.H. Mapping soil organic matter contents at field level with Cubist, Random Forest and kriging. Geoderma 2019, 342, 85–92. [Google Scholar] [CrossRef]
- Li, S.; Shi, Z.; Chen, S.; Ji, W.; Zhou, L.; Yu, W.; Webster, R. In Situ Measurements of Organic Carbon in Soil Profiles Using vis-NIR Spectroscopy on the Qinghai–Tibet Plateau. Environ. Sci. Technol. 2015, 49, 4980–4987. [Google Scholar] [CrossRef]
- Dotto, A.C.; Dalmolin, R.S.D.; ten Caten, A.; Grunwald, S. A systematic study on the application of scatter-corrective and spectral-derivative preprocessing for multivariate prediction of soil organic carbon by Vis-NIR spectra. Geoderma 2018, 314, 262–274. [Google Scholar] [CrossRef]
- Hobley, E.; Wilson, B.; Wilkie, A.; Gray, J.; Koen, T. Drivers of soil organic carbon storage and vertical distribution in Eastern Australia. Plant Soil 2015, 390, 111–127. [Google Scholar] [CrossRef]
- Ru, F.; Yin, A.; Jin, J.; Zhang, X.; Yang, X.; Zhang, M.; Gao, C. Prediction of cadmium enrichment in reclaimed coastal soils by classification and regression tree. Estuar. Coast. Shelf Sci. 2016, 177, 1–7. [Google Scholar] [CrossRef]
- Wang, B.; Waters, C.; Orgill, S.; Gray, J.; Cowie, A.; Clark, A.; Liu, D.L. High resolution mapping of soil organic carbon stocks using remote sensing variables in the semi-arid rangelands of eastern Australia. Sci. Total Environ. 2018, 630, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Grimm, R.; Behrens, T.; Märker, M.; Elsenbeer, H. Soil organic carbon concentrations and stocks on Barro Colorado Island—Digital soil mapping using Random Forests analysis. Geoderma 2008, 146, 102–113. [Google Scholar] [CrossRef]
- Pham, T.D.; Yokoya, N.; Nguyen, T.T.T.; Le, N.N.; Ha, N.T.; Xia, J.; Takeuchi, W.; Pham, T.D. Improvement of Mangrove Soil Carbon Stocks Estimation in North Vietnam Using Sentinel-2 Data and Machine Learning Approach. GIScience Remote Sens. 2020, 58, 68–87. [Google Scholar] [CrossRef]
- Bond-Lamberty, B.; Hengl, T.; Mendes de Jesus, J.; Heuvelink, G.B.M.; Ruiperez Gonzalez, M.; Kilibarda, M.; Blagotić, A.; Shangguan, W.; Wright, M.N.; Geng, X.; et al. SoilGrids250m: Global gridded soil information based on machine learning. PLoS ONE 2017, 12, e0169748. [Google Scholar] [CrossRef] [Green Version]
- Zhou, T.; Geng, Y.; Chen, J.; Liu, M.; Haase, D.; Lausch, A. Mapping soil organic carbon content using multi-source remote sensing variables in the Heihe River Basin in China. Ecol. Indic. 2020, 114, 106288. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Liang, Z.; Chen, S.; Yang, Y.; Zhou, Y.; Shi, Z. High-resolution three-dimensional mapping of soil organic carbon in China: Effects of SoilGrids products on national modeling. Sci. Total Environ. 2019, 685, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Abuduwaili, J.; Lei, J.; Wu, G.; Gui, D. Wind erosion of saline playa sediments and its ecological effects in Ebinur Lake, Xinjiang, China. Environ. Earth Sci. 2010, 63, 241–250. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, M.; Li, F. Soil respiration in typical plant communities in the wetland surrounding the high-salinity Ebinur Lake. Front. Earth Sci. 2018, 12, 611–624. [Google Scholar] [CrossRef]
- Wang, J.; Ding, J.; Abulimiti, A.; Cai, L. Quantitative estimation of soil salinity by means of different modeling methods and visible-near infrared (VIS-NIR) spectroscopy, Ebinur Lake Wetland, Northwest China. PeerJ 2018, 6, e4703. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Lv, G.; Qin, L.; Chang, S.; Yang, M.; Yang, J.; Yang, X. Effects of Simulated Nitrogen Deposition on Soil Respiration in a Populus euphratica Community in the Ebinur Lake Area, a Desert Ecosystem of Northwestern China. PLoS ONE 2015, 10, e0137827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Zhang, F.; Kung, H.-t.; Johnson, V.C. New methods for improving the remote sensing estimation of soil organic matter content (SOMC) in the Ebinur Lake Wetland National Nature Reserve (ELWNNR) in northwest China. Remote Sens. Environ. 2018, 218, 104–118. [Google Scholar] [CrossRef]
- Zhou, T.; Zhao, M.; Sun, C.; Pan, J. Exploring the Impact of Seasonality on Urban Land-Cover Mapping Using Multi-Season Sentinel-1A and GF-1 WFV Images in a Subtropical Monsoon-Climate Region. ISPRS Int. J. Geo-Inf. 2017, 7, 3. [Google Scholar] [CrossRef] [Green Version]
- Castaldi, F.; Hueni, A.; Chabrillat, S.; Ward, K.; Buttafuoco, G.; Bomans, B.; Vreys, K.; Brell, M.; van Wesemael, B. Evaluating the capability of the Sentinel 2 data for soil organic carbon prediction in croplands. ISPRS J. Photogramm. Remote Sens. 2019, 147, 267–282. [Google Scholar] [CrossRef]
- Liang, Z.; Chen, S.; Yang, Y.; Zhao, R.; Shi, Z.; Viscarra Rossel, R.A. National digital soil map of organic matter in topsoil and its associated uncertainty in 1980’s China. Geoderma 2019, 335, 47–56. [Google Scholar] [CrossRef]
- Rouse, J.; Haas, R.; Schell, J.; Deering, D. Monitoring Vegetation Systems in the Great Plains with ERTS. NASA Spec. Publ. 1974, 351, 309. [Google Scholar]
- Huete, A.R.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Chen, D.; Chang, N.; Xiao, J.; Zhou, Q.; Wu, W. Mapping dynamics of soil organic matter in croplands with MODIS data and machine learning algorithms. Sci. Total Environ. 2019, 669, 844–855. [Google Scholar] [CrossRef]
- Ren, H.; Zhou, G. Estimating green biomass ratio with remote sensing in arid grasslands. Ecol. Indic. 2019, 98, 568–574. [Google Scholar] [CrossRef]
- Bagheri, N. Application of aerial remote sensing technology for detection of fire blight infected pear trees. Comput. Electron. Agric. 2020, 168, 105147. [Google Scholar] [CrossRef]
- Venancio, L.P.; Mantovani, E.C.; do Amaral, C.H.; Usher Neale, C.M.; Gonçalves, I.Z.; Filgueiras, R.; Campos, I. Forecasting corn yield at the farm level in Brazil based on the FAO-66 approach and soil-adjusted vegetation index (SAVI). Agric. Water Manag. 2019, 225, 105779. [Google Scholar] [CrossRef]
- Villarreal, M.L.; Norman, L.M.; Buckley, S.; Wallace, C.S.A.; Coe, M.A. Multi-index time series monitoring of drought and fire effects on desert grasslands. Remote Sens. Environ. 2016, 183, 186–197. [Google Scholar] [CrossRef] [Green Version]
- Semeraro, T.; Mastroleo, G.; Pomes, A.; Luvisi, A.; Gissi, E.; Aretano, R. Modelling fuzzy combination of remote sensing vegetation index for durum wheat crop analysis. Comput. Electron. Agric. 2019, 156, 684–692. [Google Scholar] [CrossRef]
- Escadafal, R. Remote sensing of arid soil surface color with Landsat thematic mapper. Adv. Space Res. 1989, 9, 159–163. [Google Scholar] [CrossRef]
- Mathieu, R.; Pouget, M.; Cervelle, B.; Escadafal, R. Relationships between Satellite-Based Radiometric Indices Simulated Using Laboratory Reflectance Data and Typic Soil Color of an Arid Environment. Remote Sens. Environ. 1998, 66, 17–28. [Google Scholar] [CrossRef]
- Yue, T.-X.; Zhao, N.; Ramsey, R.D.; Wang, C.-L.; Fan, Z.-M.; Chen, C.-F.; Lu, Y.-M.; Li, B.-L. Climate change trend in China, with improved accuracy. Clim. Change 2013, 120, 137–151. [Google Scholar] [CrossRef]
- Efron, B.; Tibshirani, R.J. An Introduction to the Bootstrap; CRC Press: Boca Raton, FL, USA, 1994. [Google Scholar]
- Heung, B.; Bulmer, C.E.; Schmidt, M.G. Predictive soil parent material mapping at a regional-scale: A Random Forest approach. Geoderma 2014, 214–215, 141–154. [Google Scholar] [CrossRef]
- Zhang, Z.; Ding, J.; Zhu, C.; Chen, X.; Wang, J.; Han, L.; Ma, X.; Xu, D. Bivariate empirical mode decomposition of the spatial variation in the soil organic matter content: A case study from NW China. CATENA 2021, 206, 105572. [Google Scholar] [CrossRef]
- Friedman, J.H. Stochastic gradient boosting. Comput. Stat. Data Anal. 2002, 38, 367–378. [Google Scholar] [CrossRef]
- Chen, S.; Liang, Z.; Webster, R.; Zhang, G.; Zhou, Y.; Teng, H.; Hu, B.; Arrouays, D.; Shi, Z. A high-resolution map of soil pH in China made by hybrid modelling of sparse soil data and environmental covariates and its implications for pollution. Sci. Total Environ. 2019, 655, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Grunwald, S.; Myers, D.B.; Kim, J.; Harris, W.G.; Comerford, N.B. Holistic environmental soil-landscape modeling of soil organic carbon. Environ. Model. Softw. 2014, 57, 202–215. [Google Scholar] [CrossRef]
- Davis, E.; Wang, C.; Dow, K. Comparing Sentinel-2 MSI and Landsat 8 OLI in soil salinity detection: A case study of agricultural lands in coastal North Carolina. Int. J. Remote Sens. 2019, 40, 6134–6153. [Google Scholar] [CrossRef]
- Guo, L.; Fu, P.; Shi, T.; Chen, Y.; Zeng, C.; Zhang, H.; Wang, S. Exploring influence factors in mapping soil organic carbon on low-relief agricultural lands using time series of remote sensing data. Soil Tillage Res. 2021, 210, 104982. [Google Scholar] [CrossRef]
- Castaldi, F.; Chabrillat, S.; Don, A.; van Wesemael, B. Soil Organic Carbon Mapping Using LUCAS Topsoil Database and Sentinel-2 Data: An Approach to Reduce Soil Moisture and Crop Residue Effects. Remote Sens. 2019, 11, 2121. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Z.; Chen, Z.; Xu, Y.; Ren, C.; Yang, G.; Han, X.; Ren, G.; Feng, Y. Relationship between Soil Organic Carbon Stocks and Clay Content under Different Climatic Conditions in Central China. Forests 2018, 9, 598. [Google Scholar] [CrossRef] [Green Version]
- Zhao, P.; Lu, D.; Wang, G.; Liu, L.; Li, D.; Zhu, J.; Yu, S. Forest aboveground biomass estimation in Zhejiang Province using the integration of Landsat TM and ALOS PALSAR data. Int. J. Appl. Earth Obs. Geoinf. 2016, 53, 1–15. [Google Scholar] [CrossRef]
- Wang, K.; Qi, Y.; Guo, W.; Zhang, J.; Chang, Q. Retrieval and Mapping of Soil Organic Carbon Using Sentinel-2A Spectral Images from Bare Cropland in Autumn. Remote Sens. 2021, 13, 1072. [Google Scholar] [CrossRef]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Yu, H.; Zha, T.; Zhang, X.; Nie, L.; Ma, L.; Pan, Y. Spatial distribution of soil organic carbon may be predominantly regulated by topography in a small revegetated watershed. Catena 2020, 188, 104459. [Google Scholar] [CrossRef]
- Yoo, K.; Amundson, R.; Heimsath, A.M.; Dietrich, W.E. Spatial patterns of soil organic carbon on hillslopes: Integrating geomorphic processes and the biological C cycle. Geoderma 2006, 130, 47–65. [Google Scholar] [CrossRef]
- Mashalaba, L.; Galleguillos, M.; Seguel, O.; Poblete-Olivares, J. Predicting spatial variability of selected soil properties using digital soil mapping in a rainfed vineyard of central Chile. Geoderma Reg. 2020, 22, e00289. [Google Scholar] [CrossRef]
- Bao, Y.; Ustin, S.; Meng, X.; Zhang, X.; Guan, H.; Qi, B.; Liu, H. A regional-scale hyperspectral prediction model of soil organic carbon considering geomorphic features. Geoderma 2021, 403, 115263. [Google Scholar] [CrossRef]
- Schwanghart, W.; Jarmer, T. Linking spatial patterns of soil organic carbon to topography—A case study from south-eastern Spain. Geomorphology 2011, 126, 252–263. [Google Scholar] [CrossRef]
- Dharumarajan, S.; Kalaiselvi, B.; Suputhra, A.; Lalitha, M.; Vasundhara, R.; Kumar, K.S.A.; Nair, K.M.; Hegde, R.; Singh, S.K.; Lagacherie, P. Digital soil mapping of soil organic carbon stocks in Western Ghats, South India. Geoderma Reg. 2021, 25, e00387. [Google Scholar] [CrossRef]
- Adhikari, K.; Hartemink, A.E.; Minasny, B.; Bou Kheir, R.; Greve, M.B.; Greve, M.H. Digital mapping of soil organic carbon contents and stocks in Denmark. PLoS ONE 2014, 9, e105519. [Google Scholar] [CrossRef] [PubMed]
- Jobbágy, E.G.; Jackson, R.B. The vertical distribution of soil organic carbon and its relation to climate and vegetation. Ecol. Appl. 2000, 10, 423–436. [Google Scholar] [CrossRef]
- Yang, R.-M.; Guo, W.-W. Using time-series Sentinel-1 data for soil prediction on invaded coastal wetlands. Environ. Monit. Assess. 2019, 191, 462. [Google Scholar] [CrossRef]
- Yang, R.-M.; Guo, W.-W. Modelling of soil organic carbon and bulk density in invaded coastal wetlands using Sentinel-1 imagery. Int. J. Appl. Earth Obs. Geoinf. 2019, 82, 101906. [Google Scholar] [CrossRef]
- Zhou, T.; Geng, Y.; Chen, J.; Pan, J.; Haase, D.; Lausch, A. High-resolution digital mapping of soil organic carbon and soil total nitrogen using DEM derivatives, Sentinel-1 and Sentinel-2 data based on machine learning algorithms. Sci. Total Environ. 2020, 729, 138244. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, X.; Wu, W.; Liu, H. Prediction of Soil Organic Carbon under Different Land Use Types Using Sentinel-1/-2 Data in a Small Watershed. Remote Sens. 2021, 13, 1229. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Atkinson, P.M.; Yao, H. Predicting soil organic carbon content in Spain by combining Landsat TM and ALOS PALSAR images. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102182. [Google Scholar] [CrossRef]
- Ibrahem Ahmed Osman, A.; Najah Ahmed, A.; Chow, M.F.; Feng Huang, Y.; El-Shafie, A. Extreme gradient boosting (Xgboost) model to predict the groundwater levels in Selangor Malaysia. Ain Shams Eng. J. 2021, 12, 1545–1556. [Google Scholar] [CrossRef]
- Matinfar, H.R.; Maghsodi, Z.; Mousavi, S.R.; Rahmani, A. Evaluation and Prediction of Topsoil organic carbon using Machine learning and hybrid models at a Field-scale. Catena 2021, 202, 105258. [Google Scholar] [CrossRef]
- Ahirwal, J.; Nath, A.; Brahma, B.; Deb, S.; Sahoo, U.K.; Nath, A.J. Patterns and driving factors of biomass carbon and soil organic carbon stock in the Indian Himalayan region. Sci. Total Environ. 2021, 770, 145292. [Google Scholar] [CrossRef]
- Song, X.-D.; Wu, H.-Y.; Ju, B.; Liu, F.; Yang, F.; Li, D.-C.; Zhao, Y.-G.; Yang, J.-L.; Zhang, G.-L. Pedoclimatic zone-based three-dimensional soil organic carbon mapping in China. Geoderma 2020, 363, 114145. [Google Scholar] [CrossRef]


| Satellite Sensor Name | Band Name | Spectral Position (nm) | Central Wavelength (nm) | Original Resolution (m) |
|---|---|---|---|---|
| Sentinel-2A/MSI | B2-Blue | 458–523 | 490 | 10 |
| B3-Green | 543–578 | 560 | 10 | |
| B4-Red | 650–680 | 665 | 10 | |
| B5-Red Edge 1 | 698–713 | 705 | 20 | |
| B6-Red Edge 2 | 733–748 | 740 | 20 | |
| B7-Red Edge 3 | 773–793 | 783 | 20 | |
| B8-NIR | 785–900 | 842 | 10 | |
| B8A-Red Edge 4 | 855–875 | 865 | 20 | |
| B11-SWIR1 | 1565–1655 | 1610 | 20 | |
| B12-SWIR2 | 2100–2280 | 2190 | 20 | |
| Landsat 8/OLI | B2-Blue | 450–515 | 483 | 30 |
| B3-Green | 525–600 | 560 | 30 | |
| B4-Red | 630–680 | 660 | 30 | |
| B5-NIR | 845–885 | 865 | 30 | |
| B6-SWIR1 | 1560–1660 | 1650 | 30 | |
| B7-SWIR2 | 2100–2300 | 2220 | 30 |
| Date | Sensor Mode | Polarization | Direction |
|---|---|---|---|
| 26 July 2017 | IW | VV | Ascending |
| 26 July 2017 | IW | VH | Ascending |
| Index | Formula | Sentinel-2A MSI Equation | Landsat 8 OIL Equation |
|---|---|---|---|
| NDVI | |||
| VI | |||
| VI | |||
| VI | |||
| TVI | |||
| SAVI | |||
| SATVI | |||
| NBR2 | |||
| BI | |||
| BI2 | |||
| RI | |||
| CI | |||
| LSWI | |||
| MSI |
| Index | Formula | Sentinel-2 MSI Equation |
|---|---|---|
| NDVI re1 | ||
| NDVI re2 | ||
| NDVI re3 | ||
| EVI re1 | ||
| EVI re2 | ||
| EVI re3 | ||
| DVI re1 | ||
| DVI re2 | ||
| DVI re3 | ||
| RVI re1 | ||
| RVI re2 | ||
| RVI re3 | ||
| TVI re1 | ||
| TVI re2 | ||
| TVI re3 | ||
| SAVI re1 | ||
| SAVI re2 | ||
| SAVI re3 | ||
| SATVI re1 | ||
| SATVI re2 | ||
| SATVI re3 | ||
| BI re1 | ||
| BI re2 | ||
| BI re3 | ||
| BI2 re1 | ||
| BI2 re2 | ||
| BI2 re3 | ||
| RI re1 | ||
| RI re2 | ||
| RI re3 | ||
| CI re1 | ||
| CI re2 | ||
| CI re3 | ||
| NDWI re1 | ||
| MSI re1 |
| Model Name | Variable Combinations |
|---|---|
| Model A-I | Landsat 8(6band) |
| Model A-II | Landsat 8(6band) + Spectral index |
| Model A-III | Landsat 8(6band) + Spectral index + Climate variables + Topographic variables |
| Model A-IV | Landsat 8(6band) + Spectral index + Climate variables + Topographic variables + Sentinel-1A |
| Model B-I | Sentinel-2A(6band) |
| Model B-II | Sentinel-2A(6band) + Spectral index |
| Model B-III | Sentinel-2A(6band) + Spectral index + Climate variables + Topographic variables |
| Model B-IV | Sentinel-2A(6band) + Spectral index + Climate variables + Topographic variables + Sentinel-1A |
| Model C-I | Sentinel-2A(10band) |
| Model C-II | Sentinel-2A(10band) + Spectral index + Red-edge index |
| Model C-III | Sentinel-2A(10band) + Spectral index + Red-edge index + Climate variables + Topographic variables |
| Model C-IV | Sentinel-2A(10band) + Spectral index + Red-edge index + Climate variables + Topographic variables + Sentinel-1A |
| Dataset | Sample Size | Minimum (g/kg) | Maximum (g/kg) | Median (g/kg) | Mean (g/kg) | Standard Deviation (g/kg) |
|---|---|---|---|---|---|---|
| Whole dataset | 95 | 1.487 | 25.015 | 7.715 | 7.723 | 4.380 |
| Training dataset | 66 | 1.487 | 25.015 | 7.889 | 7.905 | 4.582 |
| Validation dataset | 29 | 1.699 | 17.245 | 7.036 | 7.310 | 3.926 |
| Modeling Technique | Model Name | All Variables | Top 5 Variables | ||||
|---|---|---|---|---|---|---|---|
| RMSE (g/kg) | RPIQ | RMSE (g/kg) | RPIQ | ||||
| RF | Model A-I | 0.583 | 2.781 | 1.711 | 0.606 | 2.474 | 1.924 |
| Model A-II | 0.633 | 2.692 | 1.768 | 0.648 | 2.343 | 2.031 | |
| Model A-III | 0.627 | 2.640 | 1.803 | 0.661 | 2.299 | 2.070 | |
| Model A-IV | 0.681 | 2.447 | 1.945 | 0.709 | 2.141 | 2.223 | |
| Model B-I | 0.615 | 2.660 | 1.789 | 0.624 | 2.617 | 1.818 | |
| Model B-II | 0.632 | 2.502 | 1.902 | 0.655 | 2.426 | 1.962 | |
| Model B-III | 0.569 | 2.537 | 1.876 | 0.685 | 2.179 | 2.184 | |
| Model B-IV | 0.701 | 2.401 | 1.982 | 0.736 | 2.067 | 2.303 | |
| Model C-I | 0.615 | 2.596 | 1.833 | 0.654 | 2.252 | 1.885 | |
| Model C-II | 0.693 | 2.405 | 1.979 | 0.694 | 2.230 | 2.135 | |
| Model C-III | 0.640 | 2.387 | 1.994 | 0.713 | 2.148 | 2.216 | |
| Model C-IV | 0.705 | 2.106 | 2.260 | 0.744 | 2.005 | 2.374 | |
| GBDT | Model A-I | 0.531 | 2.630 | 1.810 | 0.641 | 2.393 | 1.976 |
| Model A-II | 0.689 | 2.463 | 1.933 | 0.695 | 2.237 | 2.128 | |
| Model A-III | 0.670 | 2.369 | 2.009 | 0.704 | 2.236 | 2.129 | |
| Model A-IV | 0.671 | 2.374 | 2.004 | 0.723 | 2.132 | 2.233 | |
| Model B-I | 0.626 | 2.483 | 1.917 | 0.654 | 2.367 | 2.010 | |
| Model B-II | 0.649 | 2.364 | 2.014 | 0.699 | 2.244 | 2.121 | |
| Model B-III | 0.681 | 2.229 | 2.135 | 0.713 | 2.110 | 2.255 | |
| Model B-IV | 0.708 | 2.132 | 2.232 | 0.753 | 2.057 | 2.334 | |
| Model C-I | 0.659 | 2.347 | 2.028 | 0.682 | 2.238 | 2.126 | |
| Model C-II | 0.663 | 2.370 | 2.008 | 0.708 | 2.190 | 2.174 | |
| Model C-III | 0.687 | 2.267 | 2.100 | 0.727 | 2.084 | 2.284 | |
| Model C-IV | 0.751 | 2.104 | 2.262 | 0.772 | 1.965 | 2.423 | |
| XGBoost | Model A-I | 0.600 | 2.483 | 1.917 | 0.637 | 2.327 | 2.045 |
| Model A-II | 0.677 | 2.394 | 1.988 | 0.702 | 2.155 | 2.209 | |
| Model A-III | 0.693 | 2.420 | 1.966 | 0.726 | 2.124 | 2.241 | |
| Model A-IV | 0.701 | 2.291 | 2.077 | 0.759 | 2.003 | 2.376 | |
| Model B-I | 0.685 | 2.236 | 2.129 | 0.701 | 2.175 | 2.188 | |
| Model B-II | 0.693 | 2.342 | 2.033 | 0.722 | 2.242 | 2.223 | |
| Model B-III | 0.712 | 2.111 | 2.254 | 0.754 | 1.987 | 2.395 | |
| Model B-IV | 0.735 | 2.037 | 2.337 | 0.788 | 1.921 | 2.477 | |
| Model C-I | 0.694 | 2.290 | 2.079 | 0.727 | 2.119 | 2.246 | |
| Model C-II | 0.715 | 2.161 | 2.203 | 0.749 | 2.000 | 2.380 | |
| Model C-III | 0.726 | 2.028 | 2.347 | 0.786 | 1.830 | 2.600 | |
| Model C-IV | 0.771 | 1.899 | 2.506 | 0.804 | 1.771 | 2.687 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, B.; Ding, J.; Ge, X.; Li, X.; Han, L.; Wang, Z. Estimation of Soil Organic Carbon Content in the Ebinur Lake Wetland, Xinjiang, China, Based on Multisource Remote Sensing Data and Ensemble Learning Algorithms. Sensors 2022, 22, 2685. https://doi.org/10.3390/s22072685
Xie B, Ding J, Ge X, Li X, Han L, Wang Z. Estimation of Soil Organic Carbon Content in the Ebinur Lake Wetland, Xinjiang, China, Based on Multisource Remote Sensing Data and Ensemble Learning Algorithms. Sensors. 2022; 22(7):2685. https://doi.org/10.3390/s22072685
Chicago/Turabian StyleXie, Boqiang, Jianli Ding, Xiangyu Ge, Xiaohang Li, Lijing Han, and Zheng Wang. 2022. "Estimation of Soil Organic Carbon Content in the Ebinur Lake Wetland, Xinjiang, China, Based on Multisource Remote Sensing Data and Ensemble Learning Algorithms" Sensors 22, no. 7: 2685. https://doi.org/10.3390/s22072685
APA StyleXie, B., Ding, J., Ge, X., Li, X., Han, L., & Wang, Z. (2022). Estimation of Soil Organic Carbon Content in the Ebinur Lake Wetland, Xinjiang, China, Based on Multisource Remote Sensing Data and Ensemble Learning Algorithms. Sensors, 22(7), 2685. https://doi.org/10.3390/s22072685







