Photoacoustic-MR Image Registration Based on a Co-Sparse Analysis Model to Compensate for Brain Shift
Abstract
:1. Introduction
2. Materials and Methods
2.1. Brain-Mimicking Phantom Data
2.2. Murine Brain Data
2.3. Inducing Brain Deformation
2.4. PA-MR Image Registration Framework
2.5. Co-Sparse Analysis Model
- The rows of have the unit Euclidean norm;
- The operator has full rank, i.e., it has the maximal number of linear independent rows.
- The rows of the operator are not trivially linearly dependent.
2.6. Multi-Modal Image Registration Algorithm
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Orringer, D.A.; Golby, A.; Jolesz, F. Neuronavigation in the surgical management of brain tumors: Current and future trends. Expert Rev. Med. Devices 2012, 9, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Gerard, I.J.; Kersten-Oertel, M.; Petrecca, K.; Sirhan, D.; Hall, J.A.; Collins, D.L. Brain shift in neuronavigation of brain tumors: A review. Med. Image Anal. 2017, 35, 403–420. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Rivaz, H.; Chabanas, M.; Fortin, M.; Machado, I.; Ou, Y.; Heinrich, M.P.; Schnabel, J.A.; Zhong, X.; Maier, A. Evaluation of MRI to ultrasound registration methods for brain shift correction: The CuRIOUS2018 Challenge. IEEE Trans. Med. Imaging 2019, 39, 777–786. [Google Scholar] [CrossRef] [Green Version]
- Gerard, I.J.; Kersten-Oertel, M.; Hall, J.A.; Sirhan, D.; Collins, D.L. Brain Shift in Neuronavigation of Brain Tumors: An Updated Review of Intra-Operative Ultrasound Applications. Front. Oncol. 2021, 10, 3390. [Google Scholar] [CrossRef]
- Mitsui, T.; Fujii, M.; Tsuzaka, M.; Hayashi, Y.; Asahina, Y.; Wakabayashi, T. Skin shift and its effect on navigation accuracy in image-guided neurosurgery. Radiol. Phys. Technol. 2011, 4, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.L.; Maurer, C.R.; Maciunas, R.J.; Maciunas, R.J.; Barwise, J.A.; Fitzpatrick, J.M.; Wang, M.Y. Measurement of intraoperative brain surface deformation under a craniotomy. Neurosurgery 1998, 43, 514–526. [Google Scholar] [CrossRef]
- Hammoud, M.A.; Ligon, B.L.; Elsouki, R.; Shi, W.M.; Schomer, D.F.; Sawaya, R. Use of intraoperative ultrasound for localizing tumors and determining the extent of resection: A comparative study with magnetic resonance imaging. J. Neurosurg. 1996, 84, 737–741. [Google Scholar] [CrossRef]
- Škrinjar, O.; Nabavi, A.; Duncan, J. Model-driven brain shift compensation. Med. Image Anal. 2002, 6, 361–373. [Google Scholar] [CrossRef]
- Wittek, A.; Kikinis, R.; Warfield, S.K.; Miller, K. Brain shift computation using a fully nonlinear biomechanical model. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Palm Springs, CA, USA, 26–29 October 2005; Springer: Berlin/Heidelberg, Germany, 2005; pp. 583–590. [Google Scholar]
- Miga, M.I.; Sun, K.; Chen, I.; Clements, L.W.; Pheiffer, T.S.; Simpson, A.L.; Thompson, R.C.J.I. Clinical evaluation of a model-updated image-guidance approach to brain shift compensation: Experience in 16 cases. Int. J. Comput. Assist. Radiol. Surg. 2016, 11, 1467–1474. [Google Scholar] [CrossRef] [Green Version]
- Grunert, P.; Müller-Forell, W.; Darabi, K.; Reisch, R.; Busert, C.; Hopf, N.; Perneczky, A.J.C.A.S. Basic principles and clinical applications of neuronavigation and intraoperative computed tomography. Comput. Aided Surg. 1998, 3, 166–173. [Google Scholar] [CrossRef]
- Nimsky, C.; Ganslandt, O.; Cerny, S.; Hastreiter, P.; Greiner, G.; Fahlbusch, R.J.N. Quantification of, visualization of, and compensation for brain shift using intraoperative magnetic resonance imaging. Neurosurgery 2000, 47, 1070–1080. [Google Scholar] [CrossRef]
- Kuhnt, D.; Bauer, M.H.; Nimsky, C.J.C.R.i.B.E. Brain shift compensation and neurosurgical image fusion using intraoperative MRI: Current status and future challenges. Crit. Rev. Biomed. Eng. 2012, 40, 175–185. [Google Scholar] [CrossRef]
- Clatz, O.; Delingette, H.; Talos, I.-F.; Golby, A.J.; Kikinis, R.; Jolesz, F.A.; Ayache, N.; Warfield, S.K. Robust nonrigid registration to capture brain shift from intraoperative MRI. IEEE Trans. Med. Imaging 2005, 24, 1417–1427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valdés, P.A.; Fan, X.; Ji, S.; Harris, B.T.; Paulsen, K.D.; Roberts, D.W. Estimation of brain deformation for volumetric image updating in protoporphyrin IX fluorescence-guided resection. Stereotact. Funct. Neurosurg. 2010, 88, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Trobaugh, J.W.; Richard, W.D.; Smith, K.R.; Bucholz, R.D. Frameless stereotactic ultrasonography: Method and applications. Comput. Med. Imaging Graph. 1994, 18, 235–246. [Google Scholar] [CrossRef]
- Roche, A.; Pennec, X.; Rudolph, M.; Auer, D.; Malandain, G.; Ourselin, S.; Auer, L.M.; Ayache, N. Generalized correlation ratio for rigid registration of 3D ultrasound with MR images. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; pp. 567–577. [Google Scholar]
- Koivukangas, J.; Ylitalo, J.; Alasaarela, E.; Tauriainen, A. Three-dimensional ultrasound imaging of brain for neurosurgery. Ann. Clin. Res. 1986, 18, 65–72. [Google Scholar] [PubMed]
- Farnia, P.; Ahmadian, A.; Shabanian, T.; Serej, N.D.; Alirezaie, J. Brain-shift compensation by non-rigid registration of intra-operative ultrasound images with preoperative MR images based on residual complexity. Int. J. Comput. Assist. Radiol. Surg. 2015, 10, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Bayer, S.; Maier, A.; Ostermeier, M.; Fahrig, R. Intraoperative imaging modalities and compensation for brain shift in tumor resection surgery. Int. J. Biomed. Imaging 2017, 2017, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Farnia, P.; Mohammadi, M.; Najafzadeh, E.; Alimohamadi, M.; Makkiabadi, B.; Ahmadian, A. High-quality photoacoustic image reconstruction based on deep convolutional neural network: Towards intra-operative photoacoustic imaging. Biomed. Phys. Eng. Express 2020, 6, 045019. [Google Scholar] [CrossRef]
- Pramanik, M.; Ku, G.; Li, C.; Wang, L.V. Design and evaluation of a novel breast cancer detection system combining both thermoacoustic (TA) and photoacoustic (PA) tomography. Med. Phys. 2008, 35, 2218–2223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehrmohammadi, M.; Joon Yoon, S.; Yeager, D.; Emelianov, S.Y. Photoacoustic imaging for cancer detection and staging. Curr. Mol. Imaging 2013, 2, 89–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Najafzadeh, E.; Ghadiri, H.; Alimohamadi, M.; Farnia, P.; Mehrmohammadi, M.; Ahmadian, A. Application of multi-wavelength technique for photoacoustic imaging to delineate tumor margins during maximum-safe resection of glioma: A preliminary simulation study. J. Clin. Neurosci. 2019, 70, 242–246. [Google Scholar] [CrossRef] [PubMed]
- Arabpou, S.; Najafzadeh, E.; Farnia, P.; Ahmadian, A.; Ghadiri, H.; Akhoundi, M.S.A. Detection of Early Stages Dental Caries Using Photoacoustic Signals: The Simulation Study. Front. Biomed. Technol. 2019, 6, 35–40. [Google Scholar] [CrossRef]
- Moore, C.; Bai, Y.; Hariri, A.; Sanchez, J.B.; Lin, C.-Y.; Koka, S.; Sedghizadeh, P.; Chen, C.; Jokerst, J.V. Photoacoustic imaging for monitoring periodontal health: A first human study. Photoacoustics 2018, 12, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; John, S.; Ghalehnovi, M.; Kabbani, L.; Kennedy, N.A.; Mehrmohammadi, M. Photoacoustic Imaging for Image-guided endovenous Laser Ablation procedures. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Petrova, E.; Brecht, H.; Motamedi, M.; Oraevsky, A.; Ermilov, S.A. In Vivo optoacoustic temperature imaging for image-guided cryotherapy of prostate cancer. Phys. Med. Biol. 2018, 63, 064002. [Google Scholar] [CrossRef]
- Eddins, B.; Bell, M.A.L. Design of a multifiber light delivery system for photoacoustic-guided surgery. J. Biomed. Opt. 2017, 22, 041011. [Google Scholar] [CrossRef]
- Wang, L.V.; Hu, S. Photoacoustic tomography: In Vivo imaging from organelles to organs. Science 2012, 335, 1458–1462. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.V.; Yao, J. A practical guide to photoacoustic tomography in the life sciences. Nat. Methods 2016, 13, 627. [Google Scholar] [CrossRef]
- Attia, A.B.E.; Balasundaram, G.; Moothanchery, M.; Dinish, U.; Bi, R.; Ntziachristos, V.; Olivo, M. A review of clinical photoacoustic imaging: Current and future trends. Photoacoustics 2019, 16, 100144. [Google Scholar] [CrossRef]
- Beard, P. Biomedical photoacoustic imaging. Interface Focus 2011, 1, 602–631. [Google Scholar] [CrossRef]
- Rosencwaig, A.; Gersho, A. Theory of the photoacoustic effect with solids. J. Appl. Phys. 1976, 47, 64–69. [Google Scholar] [CrossRef]
- Zackrisson, S.; Van De Ven, S.; Gambhir, S. Light in and sound out: Emerging translational strategies for photoacoustic imaging. Cancer Res. 2014, 74, 979–1004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, M.; Wang, L.V. Photoacoustic imaging in biomedicine. Rev. Sci. Instrum. 2006, 77, 041101. [Google Scholar] [CrossRef] [Green Version]
- Farnia, P.; Najafzadeh, E.; Hariri, A.; Lavasani, S.N.; Makkiabadi, B.; Ahmadian, A.; Jokerst, J.V. Dictionary learning technique enhances signal in LED-based photoacoustic imaging. Biomed. Opt. Express 2020, 11, 2533–2547. [Google Scholar] [CrossRef] [PubMed]
- Hoelen, C.; De Mul, F.; Pongers, R.; Dekker, A. Three-dimensional photoacoustic imaging of blood vessels in tissue. Opt. Lett. 1998, 23, 648–650. [Google Scholar] [CrossRef]
- Raumonen, P.; Tarvainen, T. Segmentation of vessel structures from photoacoustic images with reliability assessment. Biomed. Opt. Express 2018, 9, 2887–2904. [Google Scholar] [CrossRef] [Green Version]
- Najafzadeh, E.; Ghadiri, H.; Alimohamadi, M.; Farnia, P.; Mehrmohammadi, M.; Ahmadian, A. Evaluation of multi-wavelengths LED-based photoacoustic imaging for maximum safe resection of glioma: A proof of concept study. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 1053–1062. [Google Scholar] [CrossRef]
- Karthikesh, M.S.; Yang, X.J.E.B. Photoacoustic image-guided interventions. Exp. Biol. Med. 2019, 245, 330–341. [Google Scholar] [CrossRef] [Green Version]
- Han, S.H.J.N. Review of photoacoustic imaging for imaging-guided spinal surgery. Neurospine 2018, 15, 306–322. [Google Scholar] [CrossRef]
- Kubelick, K.P.; Emelianov, S.Y. A Trimodal Ultrasound, Photoacoustic and Magnetic Resonance Imaging Approach for Longitudinal Post-operative Monitoring of Stem Cells in the Spinal Cord. Ultrasound Med. Biol. 2020, 46, 3468–3474. [Google Scholar] [CrossRef]
- Iversen, D.H.; Wein, W.; Lindseth, F.; Unsgård, G.; Reinertsen, I. Automatic intraoperative correction of brain shift for accurate neuronavigation. World Neurosurg. 2018, 120, e1071–e1078. [Google Scholar] [CrossRef]
- Reinertsen, I.; Descoteaux, M.; Siddiqi, K.; Collins, D.L. Validation of vessel-based registration for correction of brain shift. Med. Image Anal. 2007, 11, 374–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.J.-S.; Reinertsen, I.; Coupé, P.; Yan, C.X.; Mercier, L.; Del Maestro, D.R.; Collins, D.L. Validation of a hybrid Doppler ultrasound vessel-based registration algorithm for neurosurgery. Int. J. Comput. Assist. Radiol. Surg. 2012, 7, 667–685. [Google Scholar] [CrossRef] [Green Version]
- Farnia, P.; Ahmadian, A.; Khoshnevisan, A.; Jaberzadeh, A.; Serej, N.D.; Kazerooni, A.F. An efficient point based registration of intra-operative ultrasound images with MR images for computation of brain shift; A phantom study. In Proceedings of the Engineering in Medicine and Biology Society, EMBC, 33rd Annual International Conference of the IEEE, EMBC, Boston, MA, USA, 30 August–3 Septmber 2011; pp. 8074–8077. [Google Scholar]
- Arbel, T.; Morandi, X.; Comeau, R.M.; Collins, D.L. Automatic non-linear MRI-ultrasound registration for the correction of intra-operative brain deformations. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; pp. 913–922. [Google Scholar]
- Ji, S.; Hartov, A.; Roberts, D.; Paulsen, K. Mutual-information-corrected tumor displacement using intraoperative ultrasound for brain shift compensation in image-guided neurosurgery. In Proceedings of the SPIE 6918, Medical Imaging 2008: Visualization, Image-Guided Procedures, and Modeling, San Diego, CA, USA, 17 March 2008; p. 69182H. [Google Scholar]
- Wein, W.; Ladikos, A.; Fuerst, B.; Shah, A.; Sharma, K.; Navab, N. Global registration of ultrasound to MRI using the LC 2 metric for enabling neurosurgical guidance. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Nagoya, Japan, 22–26 September 2013; pp. 34–41. [Google Scholar]
- Coupé, P.; Hellier, P.; Morandi, X.; Barillot, C. 3D rigid registration of intraoperative ultrasound and preoperative MR brain images based on hyperechogenic structures. J. Biomed. Imaging 2012, 2012, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivaz, H.; Karimaghaloo, Z.; Collins, D.L. Self-similarity weighted mutual information: A new nonrigid image registration metric. Med. Image Anal. 2014, 18, 343–358. [Google Scholar] [CrossRef] [Green Version]
- Rivaz, H.; Chen, S.J.-S.; Collins, D.L. Automatic deformable MR-ultrasound registration for image-guided neurosurgery. IEEE Trans. Med. Imaging 2015, 34, 366–380. [Google Scholar] [CrossRef] [PubMed]
- Machado, I.; Toews, M.; George, E.; Unadkat, P.; Essayed, W.; Luo, J.; Teodoro, P.; Carvalho, H.; Martins, J.; Golland, P. Deformable MRI-ultrasound registration using correlation-based attribute matching for brain shift correction: Accuracy and generality in multi-site data. NeuroImage 2019, 202, 116094. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, Y.; Blum, R.S.; Han, J.; Tao, D. Sparse representation based multi-sensor image fusion for multi-focus and multi-modality images: A review. Inf. Fusion 2018, 40, 57–75. [Google Scholar] [CrossRef]
- Farnia, P.; Ahmadian, A.; Shabanian, T.; Serej, N.D.; Alirezaie, J. A hybrid method for non-rigid registration of intra-operative ultrasound images with pre-operative MR images. In Proceedings of the Engineering in Medicine and Biology Society (EMBC), 2014 36th Annual International Conference of the IEEE, Chicago, IL, USA, 26–30 August 2014; pp. 5562–5565. [Google Scholar]
- Farnia, P.; Makkiabadi, B.; Ahmadian, A.; Alirezaie, J. Curvelet based residual complexity objective function for non-rigid registration of pre-operative MRI with intra-operative ultrasound images. In Proceedings of the Engineering in Medicine and Biology Society (EMBC), 2016 IEEE 38th Annual International Conference, Orlando, FL, USA, 16–20 August 2016; pp. 1167–1170. [Google Scholar]
- Huang, K.; Aviyente, S. Sparse representation for signal classification. In Proceedings of the Advances in Neural Information Processing Systems, San Francisco, CA, USA, 30 November–3 December 1992; pp. 609–616. [Google Scholar]
- Roozgard, A.; Barzigar, N.; Verma, P.; Cheng, S. 3D-SCoBeP: 3D medical image registration using sparse coding and belief propagation. Int. J. Diagn. Imaging 2014, 2, 54. [Google Scholar] [CrossRef]
- Nam, S.; Davies, M.E.; Elad, M.; Gribonval, R. The cosparse analysis model and algorithms. Appl. Comput. Harmon. Anal. 2013, 34, 30–56. [Google Scholar] [CrossRef]
- Zhou, N.; Jiang, H.; Gong, L.; Xie, X. Double-image compression and encryption algorithm based on co-sparse representation and random pixel exchanging. Opt. Lasers Eng. 2018, 110, 72–79. [Google Scholar] [CrossRef]
- Kiechle, M.; Hawe, S.; Kleinsteuber, M. A joint intensity and depth co-sparse analysis model for depth map super-resolution. In Proceedings of the IEEE international conference on computer vision, Sydney, NSW, Australia, 1–8 December 2013; pp. 1545–1552. [Google Scholar]
- Kiechle, M.; Habigt, T.; Hawe, S.; Kleinsteuber, M. A bimodal co-sparse analysis model for image processing. Int. J. Comput. Vis. 2015, 114, 233–247. [Google Scholar] [CrossRef] [Green Version]
- Han, C.; Zhang, H.; Gao, C.; Jiang, C.; Sang, N.; Zhang, L. A Remote Sensing Image Fusion Method Based on the Analysis Sparse Model. IEEE J. Sel. Top. Appl. Earth Obs. Remote. Sens. 2016, 9, 439–453. [Google Scholar] [CrossRef]
- Gao, R.; Vorobyov, S.A.; Zhao, H. Image fusion with cosparse analysis operator. IEEE Signal Process. Lett. 2017, 24, 943–947. [Google Scholar] [CrossRef] [Green Version]
- Farnia, P.; Najafzadeh, E.; Ahmadian, A.; Makkiabadi, B.; Alimohamadi, M.; Alirezaie, J. Co-sparse analysis model based image registration to compensate brain shift by using intra-operative ultrasound imaging. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 17–21 July 2018; pp. 1–4. [Google Scholar]
- Ren, W.; Skulason, H.; Schlegel, F.; Rudin, M.; Klohs, J.; Ni, R. Automated registration of magnetic resonance imaging and optoacoustic tomography data for experimental studies. Neurophotonics 2019, 6, 025001. [Google Scholar] [CrossRef] [Green Version]
- Gehrung, M.; Tomaszewski, M.; McIntyre, D.; Disselhorst, J.; Bohndiek, S. Co-Registration of Optoacoustic Tomography and Magnetic Resonance Imaging Data from Murine Tumour Models. Photoacoustics 2020, 18, 100147. [Google Scholar] [CrossRef] [PubMed]
- Surry, K.; Austin, H.; Fenster, A.; Peters, T. Poly (vinyl alcohol) cryogel phantoms for use in ultrasound and MR imaging. Phys. Med. Biol. 2004, 49, 5529. [Google Scholar] [CrossRef]
- Myronenko, A.; Song, X. Intensity-based image registration by minimizing residual complexity. IEEE Trans. Med. Imaging 2010, 29, 1882–1891. [Google Scholar] [CrossRef]
- Ou, Y.; Akbari, H.; Bilello, M.; Da, X.; Davatzikos, C. Comparative evaluation of registration algorithms in different brain databases with varying difficulty: Results and insights. IEEE Trans. Med. Imaging 2014, 33, 2039–2065. [Google Scholar] [CrossRef] [Green Version]
- Hawe, S.; Kleinsteuber, M.; Diepold, K. Analysis operator learning and its application to image reconstruction. IEEE Trans. Image Process. 2013, 22, 2138–2150. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Kang, Z.; Yang, M.; Xiong, X.; Peng, C.; Xiao, M.J.S. Image denoising via improved dictionary learning with global structure and local similarity preservations. Symmetry 2018, 10, 167. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Fu, Y.; Li, H.; Zou, J. Dictionary learning method for joint sparse representation-based image fusion. Opt. Eng. 2013, 52, 057006. [Google Scholar] [CrossRef]
- Najafzadeh, E.; Farnia, P.; Lavasani, S.N.; Basij, M.; Yan, Y.; Ghadiri, H.; Ahmadian, A.; Mehrmohammadi, M. Photoacoustic image improvement based on a combination of sparse coding and filtering. J. Biomed. Opt. 2020, 25, 106001. [Google Scholar] [CrossRef] [PubMed]
- Manwar, R.; Hosseinzadeh, M.; Hariri, A.; Kratkiewicz, K.; Noei, S.; Avanaki, M.R.N. Photoacoustic signal enhancement: Towards utilization of low energy laser diodes in real-time photoacoustic imaging. Sensors 2018, 18, 3498. [Google Scholar] [CrossRef] [Green Version]
- Singh, M.K.A. LED-Based Photoacoustic Imaging: From Bench to Bedside; Springer Nature: Singapore, 2020. [Google Scholar]
- Agrawal, S.; Kuniyil Ajith Singh, M.; Johnstonbaugh, K.; Han, D.C.; Pameijer, C.R.; Kothapalli, S.-R. Photoacoustic imaging of human vasculature using LED versus laser illumination: A comparison study on tissue phantoms and In Vivo humans. Sensors 2021, 21, 424. [Google Scholar] [CrossRef]
- Hariri, A.; Alipour, K.; Mantri, Y.; Schulze, J.P.; Jokerst, J.V. Deep learning improves contrast in low-fluence photoacoustic imaging. Biomed. Opt. Express 2020, 11, 3360–3373. [Google Scholar] [CrossRef] [PubMed]


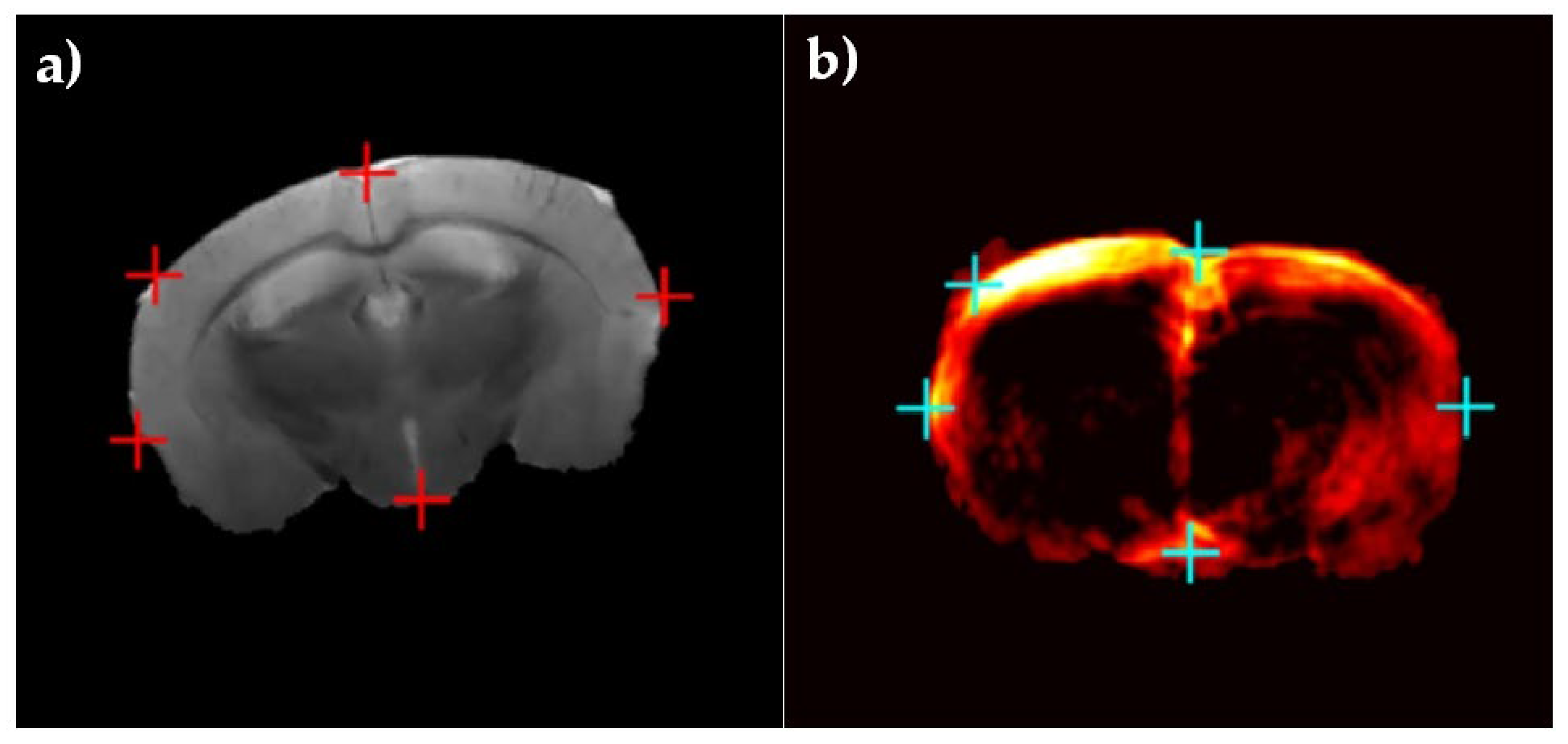


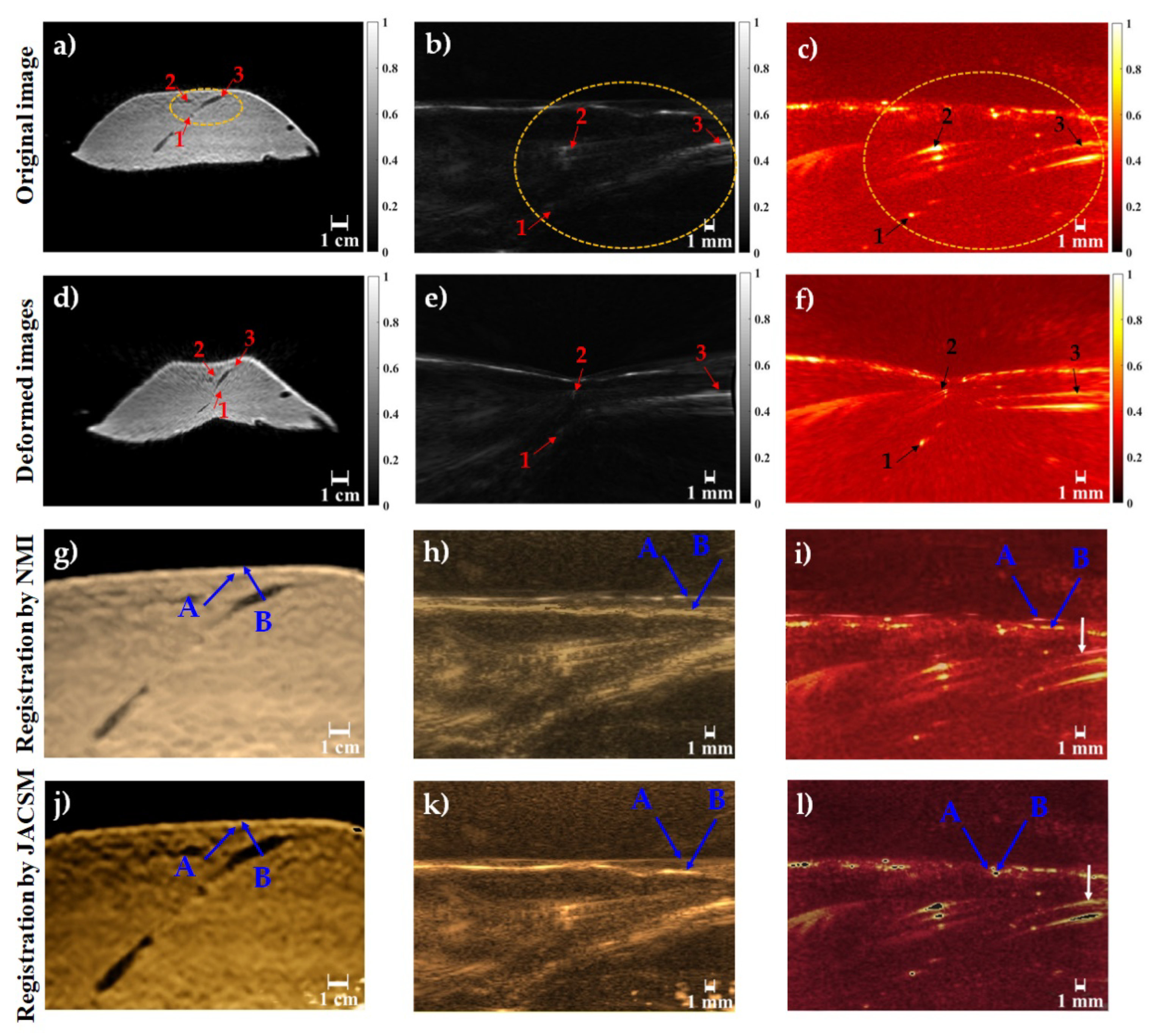
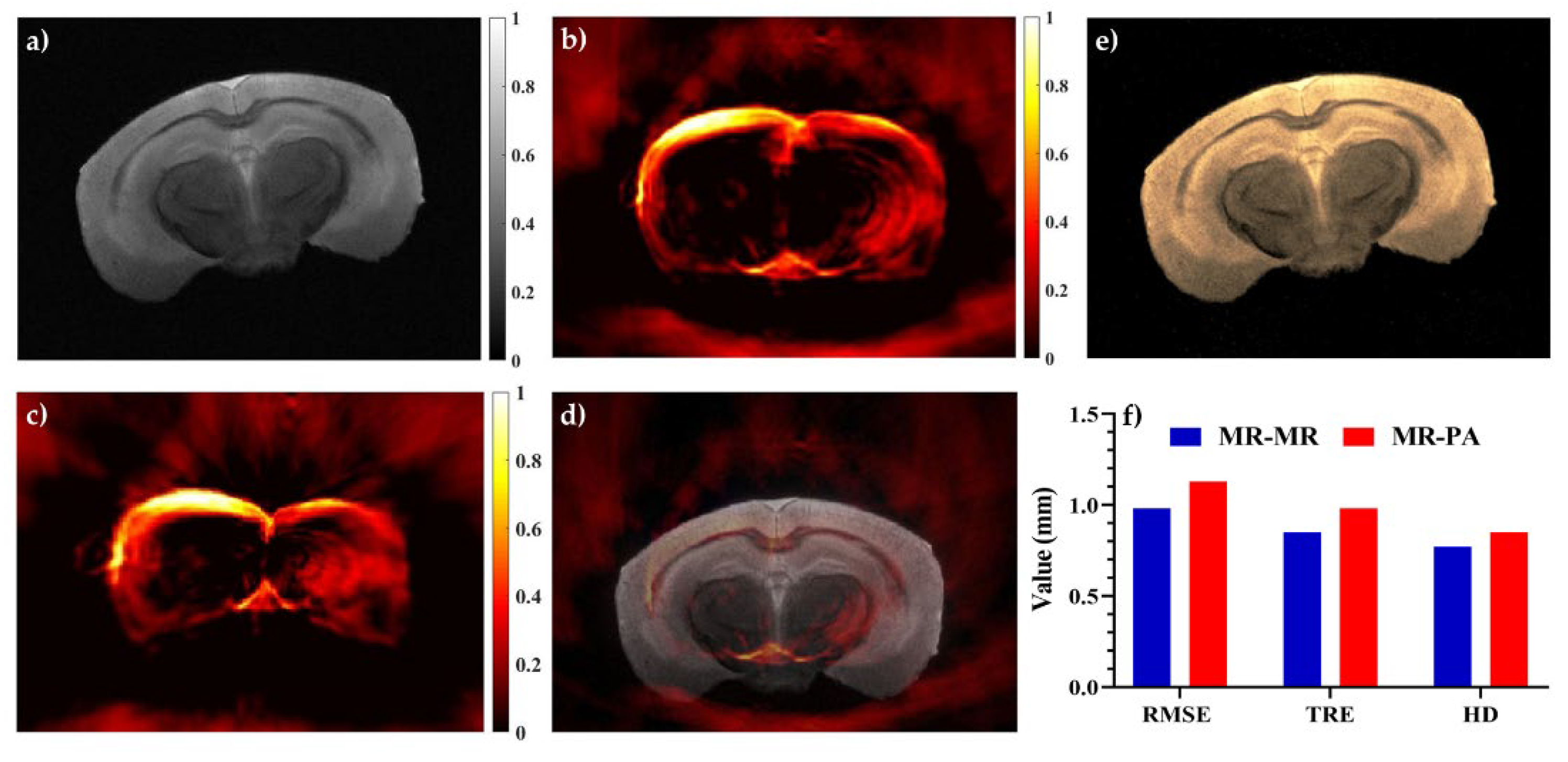
| Multimodal Registration | RMSE (Mean ± Std) | TRE (Mean ± Std) Number of Targets: 3 | HD (Mean ± Std) | |
|---|---|---|---|---|
| MR-MR | JACSM | 0.62 ± 0.04 | 0.32 ± 0.03 0.51 ± 0.04 | 0.21 ± 0.03 0.46 ± 0.07 |
| NMI | 0.98 ± 0.09 | |||
| US-MR | JACSM | 1.17 ± 0.13 1.87 ± 0.15 | 0.96 ± 0.08 1.58 ± 0.11 | 0.51 ± 0.03 1.23 ± 0.13 |
| NMI | ||||
| PA-MR | JACSM | 0.73 ± 0.05 | 0.58 ± 0.04 | 0.32 ± 0.04 |
| NMI | 1.18 ± 0.09 | 0.96 ± 0.08 | 0.68 ± 0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farnia, P.; Makkiabadi, B.; Alimohamadi, M.; Najafzadeh, E.; Basij, M.; Yan, Y.; Mehrmohammadi, M.; Ahmadian, A. Photoacoustic-MR Image Registration Based on a Co-Sparse Analysis Model to Compensate for Brain Shift. Sensors 2022, 22, 2399. https://doi.org/10.3390/s22062399
Farnia P, Makkiabadi B, Alimohamadi M, Najafzadeh E, Basij M, Yan Y, Mehrmohammadi M, Ahmadian A. Photoacoustic-MR Image Registration Based on a Co-Sparse Analysis Model to Compensate for Brain Shift. Sensors. 2022; 22(6):2399. https://doi.org/10.3390/s22062399
Chicago/Turabian StyleFarnia, Parastoo, Bahador Makkiabadi, Maysam Alimohamadi, Ebrahim Najafzadeh, Maryam Basij, Yan Yan, Mohammad Mehrmohammadi, and Alireza Ahmadian. 2022. "Photoacoustic-MR Image Registration Based on a Co-Sparse Analysis Model to Compensate for Brain Shift" Sensors 22, no. 6: 2399. https://doi.org/10.3390/s22062399
APA StyleFarnia, P., Makkiabadi, B., Alimohamadi, M., Najafzadeh, E., Basij, M., Yan, Y., Mehrmohammadi, M., & Ahmadian, A. (2022). Photoacoustic-MR Image Registration Based on a Co-Sparse Analysis Model to Compensate for Brain Shift. Sensors, 22(6), 2399. https://doi.org/10.3390/s22062399






