Early Detection of Fusarium oxysporum Infection in Processing Tomatoes (Solanum lycopersicum) and Pathogen–Soil Interactions Using a Low-Cost Portable Electronic Nose and Machine Learning Modeling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant and Pathogen Preparation
2.2. Physiological Measurements
2.3. Low-Cost Electronic Nose Measurements
2.4. Parallel Soil Experiment
2.5. Statistical Analysis and Machine Learning Models
3. Results
4. Discussion
4.1. Physiological Response of Tomato Plants to F. oxysporum Infection
4.2. Production of Plant Volatile Compounds in Response to F. oxysporum Infection
4.3. Response of Soil Samples to F. oxysporum Inoculation
4.4. Development of Machine Learning Models
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Callaghan, S.E.; Burgess, L.W.; Ades, P.; Mann, E.; Morrison, A.; Tesoriero, L.A.; Taylor, P.W.J. Progress in understanding soil-borne pathogens associated with poor growth of tomato plants in the Australian Processing Tomato Industry. Annu. Process. Tomato Grow. Mag. 2018, 39, 10–13. [Google Scholar]
- Callaghan, S.E.; Burgess, L.W.; Ades, P.; Mann, E.; Morrison, A.; Tesoriero, L.A.; Taylor, P.W.J. Association of soil-borne pathogens with poor root growth of processing tomato plants. Annu. Process. Tomato Grow. Mag. 2019, 40, 11–15. [Google Scholar]
- Callaghan, S.E.; Burgess, L.W.; Ades, P.; Mann, E.; Morrison, A.; Tesoriero, L.A.; Taylor, P.W.J. Identification of soilborne fungal pathogens associated with poor growth of tomato plants. Annu. Process. Tomato Grow. Mag. 2017, 38, 11–13. [Google Scholar]
- Testen, A.L.; Martínez, M.B.; Madrid, A.J.; Deblais, L.; Taylor, C.G.; Paul, P.A.; Miller, S.A. On-Farm Evaluations of Anaerobic Soil Disinfestation and Grafting for Management of a Widespread Soilborne Disease Complex in Protected Culture Tomato Production. Phytopathology 2021, 111, 954–965. [Google Scholar] [CrossRef]
- Wu, P.H.; Tsay, T.T.; Chen, P. Evaluation of Streptomyces saraciticas as Soil Amendments for Controlling Soil-Borne Plant Pathogens. Plant Pathol. J. 2021, 37, 596–606. [Google Scholar] [CrossRef]
- Bakker, P.; Berendsen, R.L.; Van Pelt, J.A.; Vismans, G.; Yu, K.; Li, E.; Van Bentum, S.; Poppeliers, S.W.M.; Sanchez Gil, J.J.; Zhang, H.; et al. The Soil-Borne Identity and Microbiome-Assisted Agriculture: Looking Back to the Future. Mol. Plant 2020, 13, 1394–1401. [Google Scholar] [CrossRef]
- Cui, S.; Wang, J.; Yang, L.; Wu, J.; Wang, X. Qualitative and quantitative analysis on aroma characteristics of ginseng at different ages using E-nose and GC-MS combined with chemometrics. J. Pharm. Biomed. Anal. 2015, 102, 64–77. [Google Scholar] [CrossRef]
- Genzardi, D.; Greco, G.; Núñez-Carmona, E.; Sberveglieri, V. Real Time Monitoring of Wine Vinegar Supply Chain through MOX Sensors. Sensors 2022, 22, 6247. [Google Scholar] [CrossRef]
- Magro, C.; Gonçalves, O.C.; Morais, M.; Ribeiro, P.A.; Sério, S.; Vieira, P.; Raposo, M. Volatile Organic Compound Monitoring during Extreme Wildfires: Assessing the Potential of Sensors Based on LbL and Sputtering Films. Sensors 2022, 22, 6677. [Google Scholar] [CrossRef]
- Tothill, I.E. Biosensors developments and potential applications in the agricultural diagnosis sector. Comput. Electron. Agric. 2001, 30, 205–218. [Google Scholar] [CrossRef]
- Borowik, P.; Adamowicz, L.; Tarakowski, R.; Wacławik, P.; Oszako, T.; Ślusarski, S.; Tkaczyk, M. Development of a Low-Cost Electronic Nose for Detection of Pathogenic Fungi and Applying It to Fusarium oxysporum and Rhizoctonia solani. Sensors 2021, 21, 5868. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, S.; Tongson, E.; Unnithan, R.R.; Gonzalez Viejo, C. Early Detection of Aphid Infestation and Insect-Plant Interaction Assessment in Wheat Using a Low-Cost Electronic Nose (E-Nose), Near-Infrared Spectroscopy and Machine Learning Modeling. Sensors 2021, 21, 5948. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, S.; Gonzalez Viejo, C. Novel use of e-noses for digital agriculture, food and beverage applications. In Nanotechnology-Based E-Noses: Fundamentals and Emerging Applications; Elsevier Science: Amsterdam, The Netherlands, 2023. [Google Scholar]
- Wilson, A.D. Diverse applications of electronic-nose technologies in agriculture and forestry. Sensors 2013, 13, 2295–2348. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Liu, Y.; Li, Q. Recent Progress in Smart Electronic Nose Technologies Enabled with Machine Learning Methods. Sensors 2021, 21, 7620. [Google Scholar] [CrossRef]
- Cozzolino, R.; Cefola, M.; Laurino, C.; Pellicano, M.P.; Palumbo, M.; Stocchero, M.; Pace, B. Electronic-Nose as Non-destructive Tool to Discriminate “Ferrovia” Sweet Cherries Cold Stored in Air or Packed in High CO(2) Modified Atmospheres. Front. Nutr. 2021, 8, 720092. [Google Scholar] [CrossRef]
- Bonah, E.; Huang, X.; Aheto, J.H.; Osae, R. Application of electronic nose as a non-invasive technique for odor fingerprinting and detection of bacterial foodborne pathogens: A review. J. Food Sci. Technol. 2020, 57, 1977–1990. [Google Scholar] [CrossRef]
- Pallottino, F.; Costa, C.; Antonucci, F.; Strano, M.C.; Calandra, M.; Solaini, S.; Menesatti, P. Electronic nose application for determination of Penicillium digitatum in Valencia oranges. J. Sci. Food Agric. 2012, 92, 2008–2012. [Google Scholar] [CrossRef]
- Gonzalez Viejo, C.; Tongson, E.; Fuentes, S. Integrating a Low-Cost Electronic Nose and Machine Learning Modelling to Assess Coffee Aroma Profile and Intensity. Sensors 2021, 21, 2016. [Google Scholar] [CrossRef]
- Domènech-Gil, G.; Puglisi, D. A Virtual Electronic Nose for the Efficient Classification and Quantification of Volatile Organic Compounds. Sensors 2022, 22, 7340. [Google Scholar] [CrossRef]
- Labanska, M.; van Amsterdam, S.; Jenkins, S.; Clarkson, J.P.; Covington, J.A. Preliminary Studies on Detection of Fusarium Basal Rot Infection in Onions and Shallots Using Electronic Nose. Sensors 2022, 22, 5453. [Google Scholar] [CrossRef]
- Palacín, J.; Rubies, E.; Clotet, E. Classification of Three Volatiles Using a Single-Type eNose with Detailed Class-Map Visualization. Sensors 2022, 22, 5262. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.P.; Sayago, I.; Sanjurjo, J.L.; Perez-Coello, M.S.; Díaz-Maroto, M.C. Rapid and Non-Destructive Analysis of Corky Off-Flavors in Natural Cork Stoppers by a Wireless and Portable Electronic Nose. Sensors 2022, 22, 4687. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez Viejo, C.; Fuentes, S. Digital Assessment and Classification of Wine Faults Using a Low-Cost Electronic Nose, Near-Infrared Spectroscopy and Machine Learning Modelling. Sensors 2022, 22, 2303. [Google Scholar] [CrossRef]
- Vijayarani, S.; Dhayanand, S.; Phil, M. Kidney disease prediction using SVM and ANN algorithms. Int. J. Comput. Bus. Res. 2015, 6, 1–12. [Google Scholar]
- De Luca, G. Advantages and Disadvantages of Neural Networks Against SVMs. Available online: https://www.baeldung.com/cs/ml-ann-vs-svm (accessed on 4 November 2022).
- Prem. Which Is Better—Random Forest vs Support Vector Machine vs Neural Network. Available online: https://www.iunera.com/kraken/fabric/random-forest-vs-support-vector-machine-vs-neural-network/ (accessed on 4 November 2022).
- Gonzalez Viejo, C.; Torrico, D.; Dunshea, F.; Fuentes, S. Development of Artificial Neural Network Models to Assess Beer Acceptability Based on Sensory Properties Using a Robotic Pourer: A Comparative Model Approach to Achieve an Artificial Intelligence System. Beverages 2019, 5, 33. [Google Scholar] [CrossRef] [Green Version]
- Ramsey, M.D.; O’Brien, R.G.; Pegg, K.G. Fusarium oxysporum associated with wilt and root rot of tomato in Queensland; races and vegetative compatibility groups. Anim. Prod. Sci. 1992, 32, 651–655. [Google Scholar] [CrossRef]
- Ma, L.J.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; O’Donnell, K.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef] [Green Version]
- Abeysekara, N.S.; Swaminathan, S.; Desai, N.; Guo, L.; Bhattacharyya, M.K. The plant immunity inducer pipecolic acid accumulates in the xylem sap and leaves of soybean seedlings following Fusarium virguliforme infection. Plant Sci. 2016, 243, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Roberts, P.D.; Meru, G.; McGovern, R.J.; Datnoff, L.E. Fusarium Crown and Root Rot of Tomato in Florida: PP52/PG082, rev. 12/2021. EDIS 2021, 2021, 54. [Google Scholar] [CrossRef]
- Ozbay, N.; Newman, S.E. Fusarium Crown and Root Rot of Tomato and Control Methods. Plant Pathol. J. 2004, 3, 9–18. [Google Scholar] [CrossRef] [Green Version]
- Vitale, A.; Rocco, M.; Arena, S.; Giuffrida, F.; Cassaniti, C.; Scaloni, A.; Lomaglio, T.; Guarnaccia, V.; Polizzi, G.; Marra, M.; et al. Tomato susceptibility to Fusarium crown and root rot: Effect of grafting combination and proteomic analysis of tolerance expression in the rootstock. Plant Physiol. Biochem. 2014, 83, 207–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szczechura, W.; Staniaszek, M.; Habdas, H. Fusarium Oxysporum F. Sp. Radicis-Lycopersici—The Cause of Fusarium Crown and Root Rot in Tomato Cultivation. J. Plant Prot. Res. 2013, 53, 172–176. [Google Scholar] [CrossRef]
- Abdalla, M.; Ahmed, M.A.; Cai, G.; Wankmüller, F.; Schwartz, N.; Litig, O.; Javaux, M.; Carminati, A. Stomatal closure during water deficit is controlled by below-ground hydraulics. Ann. Bot. 2022, 129, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Tombesi, S.; Nardini, A.; Frioni, T.; Soccolini, M.; Zadra, C.; Farinelli, D.; Poni, S.; Palliotti, A. Stomatal closure is induced by hydraulic signals and maintained by ABA in drought-stressed grapevine. Sci. Rep. 2015, 5, 12449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osakabe, Y.; Osakabe, K.; Shinozaki, K.; Tran, L.-S. Response of plants to water stress. Front. Plant Sci. 2014, 5, 86. [Google Scholar] [CrossRef] [Green Version]
- Muir, C.D. A Stomatal Model of Anatomical Tradeoffs Between Gas Exchange and Pathogen Colonization. Front. Plant Sci. 2020, 11, 518991. [Google Scholar] [CrossRef]
- Gudesblat, G.E.; Torres, P.S.; Vojnov, A.A. Stomata and pathogens: Warfare at the gates. Plant Signal. Behav. 2009, 4, 1114–1116. [Google Scholar] [CrossRef]
- Ye, W.; Munemasa, S.; Shinya, T.; Wu, W.; Ma, T.; Lu, J.; Kinoshita, T.; Kaku, H.; Shibuya, N.; Murata, Y. Stomatal immunity against fungal invasion comprises not only chitin-induced stomatal closure but also chitosan-induced guard cell death. Proc. Natl. Acad. Sci. USA 2020, 117, 20932–20942. [Google Scholar] [CrossRef]
- Ali, S.S.; Nugent, B.; Mullins, E.; Doohan, F.M. Insights from the fungus Fusarium oxysporum point to high affinity glucose transporters as targets for enhancing ethanol production from lignocellulose. PLoS ONE 2013, 8, e54701. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.S.; Nugent, B.; Mullins, E.; Doohan, F.M. Fungal-mediated consolidated bioprocessing: The potential of Fusarium oxysporum for the lignocellulosic ethanol industry. AMB Express 2016, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Anasontzis, G.E.; Christakopoulos, P. Challenges in ethanol production with Fusarium oxysporum through consolidated bioprocessing. Bioengineered 2014, 5, 393–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Y.; Wu, P.; Xue, J.; Zhang, M.; Wei, X. Cosmosporasides F-H, three new sugar alcohol conjugated acyclic sesquiterpenes from a Fusarium oxysporum fungus. Nat. Prod. Res. 2022, 36, 3420–3428. [Google Scholar] [CrossRef] [PubMed]
- Nugent, B.; Ali, S.S.; Mullins, E.; Doohan, F.M. A Major Facilitator Superfamily Peptide Transporter From Fusarium oxysporum Influences Bioethanol Production From Lignocellulosic Material. Front. Microbiol. 2019, 10, 295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deans, R.M.; Brodribb, T.J.; Busch, F.A.; Farquhar, G.D. Optimization can provide the fundamental link between leaf photosynthesis, gas exchange and water relations. Nat. Plants 2020, 6, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- Ďurkovič, J.; Bubeníková, T.; Gužmerová, A.; Fleischer, P.; Kurjak, D.; Čaňová, I.; Lukáčik, I.; Dvořák, M.; Milenković, I. Effects of Phytophthora Inoculations on Photosynthetic Behaviour and Induced Defence Responses of Plant Volatiles in Field-Grown Hybrid Poplar Tolerant to Bark Canker Disease. J. Fungi 2021, 7, 969. [Google Scholar] [CrossRef] [PubMed]
- Sage, R.F. Acclimation of photosynthesis to increasing atmospheric CO2: The gas exchange perspective. Photosynth. Res. 1994, 39, 351–368. [Google Scholar] [CrossRef]
- Kuzyakov, Y.; Cheng, W. Photosynthesis controls of CO2 efflux from maize rhizosphere. Plant Soil 2004, 263, 85–99. [Google Scholar] [CrossRef]
- Stover, R.H.; Freiberg, S.R. Effect of Carbon Dioxide on Multiplication of Fusarium in Soil. Nature 1958, 181, 788–789. [Google Scholar] [CrossRef]
- Gougoulias, C.; Clark, J.M.; Shaw, L.J. The role of soil microbes in the global carbon cycle: Tracking the below-ground microbial processing of plant-derived carbon for manipulating carbon dynamics in agricultural systems. J. Sci. Food Agric. 2014, 94, 2362–2371. [Google Scholar] [CrossRef] [Green Version]
- Bardgett, R.D.; Freeman, C.; Ostle, N.J. Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2008, 2, 805–814. [Google Scholar] [CrossRef] [Green Version]
- Carney, K.M.; Hungate, B.A.; Drake, B.G.; Megonigal, J.P. Altered soil microbial community at elevated CO(2) leads to loss of soil carbon. Proc. Natl. Acad. Sci. USA 2007, 104, 4990–4995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welke, J.E.; Hernandes, K.C.; Nicolli, K.P.; Barbará, J.A.; Biasoto, A.C.T.; Zini, C.A. Role of gas chromatography and olfactometry to understand the wine aroma: Achievements denoted by multidimensional analysis. J. Sep. Sci. 2021, 44, 135–168. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.H.; Kuo, C.H. Gas chromatography-mass spectrometry-based analytical strategies for fatty acid analysis in biological samples. J. Food Drug Anal. 2020, 28, 60–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, C.; Gonçalves, R.; Tiritan, M.E. Separation of Enantiomers Using Gas Chromatography: Application in Forensic Toxicology, Food and Environmental Analysis. Crit. Rev. Anal. Chem. 2021, 51, 787–811. [Google Scholar] [CrossRef]
- Beć, K.B.; Grabska, J.; Huck, C.W. Near-Infrared Spectroscopy in Bio-Applications. Molecules 2020, 25, 2948. [Google Scholar] [CrossRef]


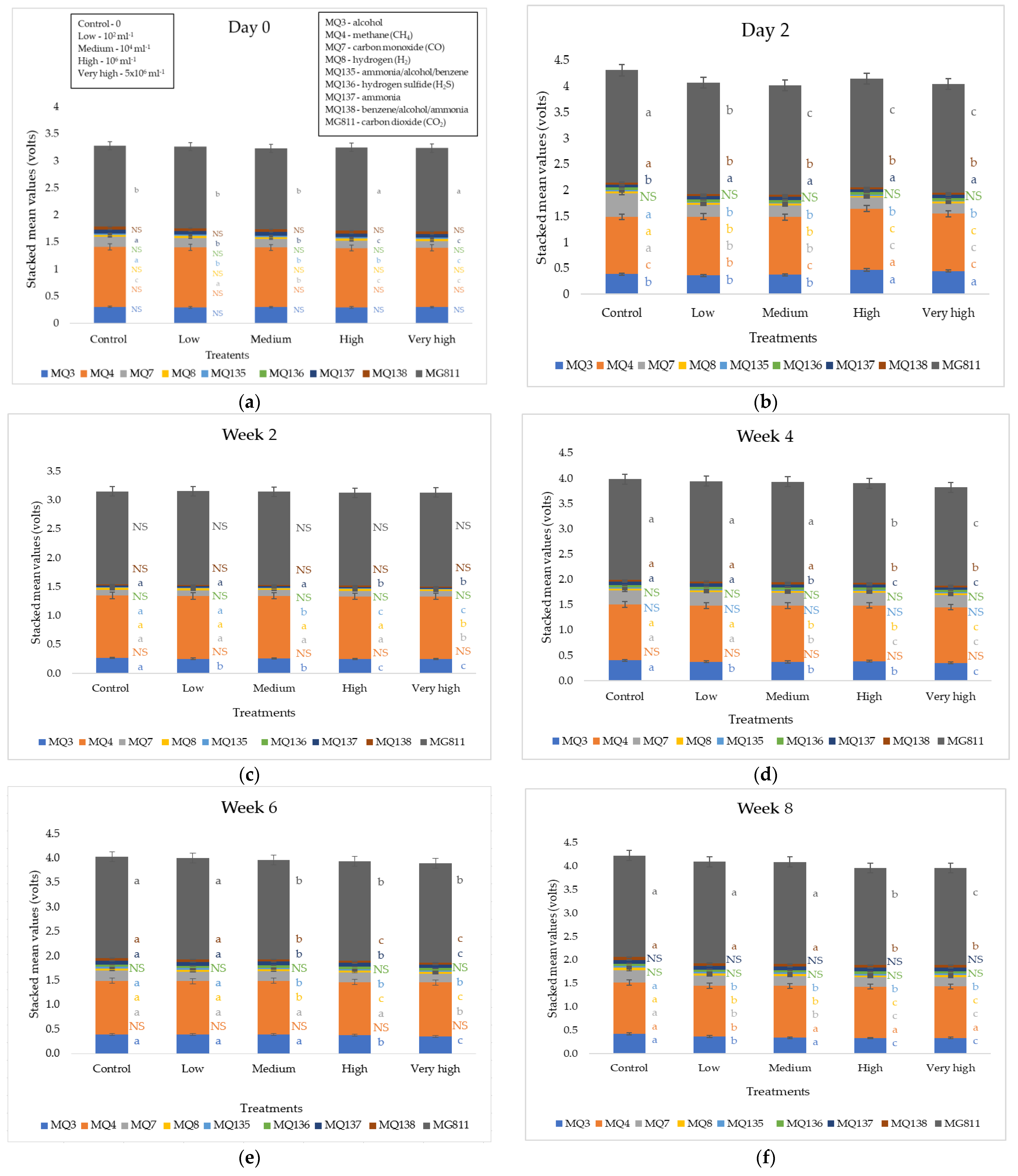



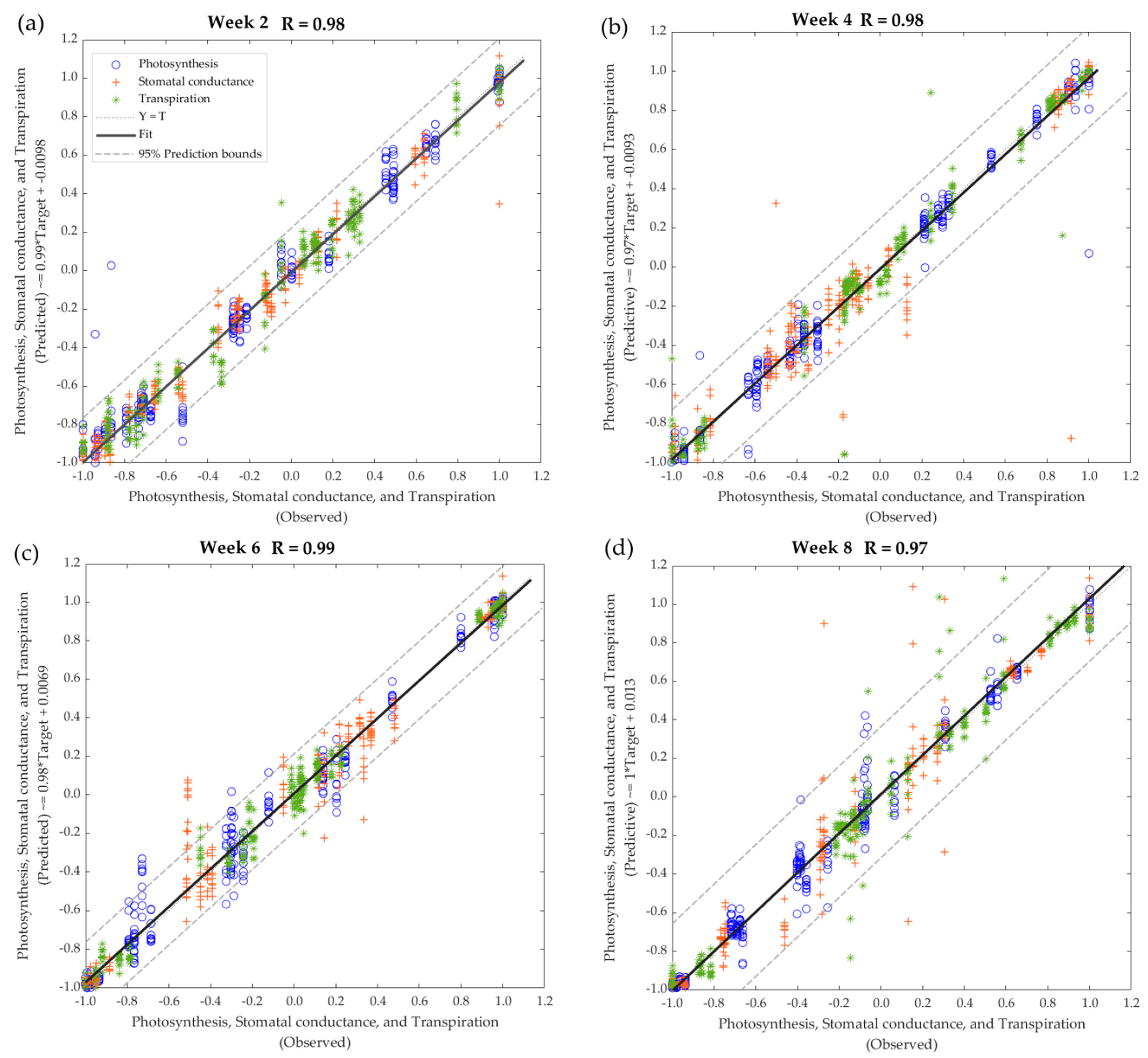
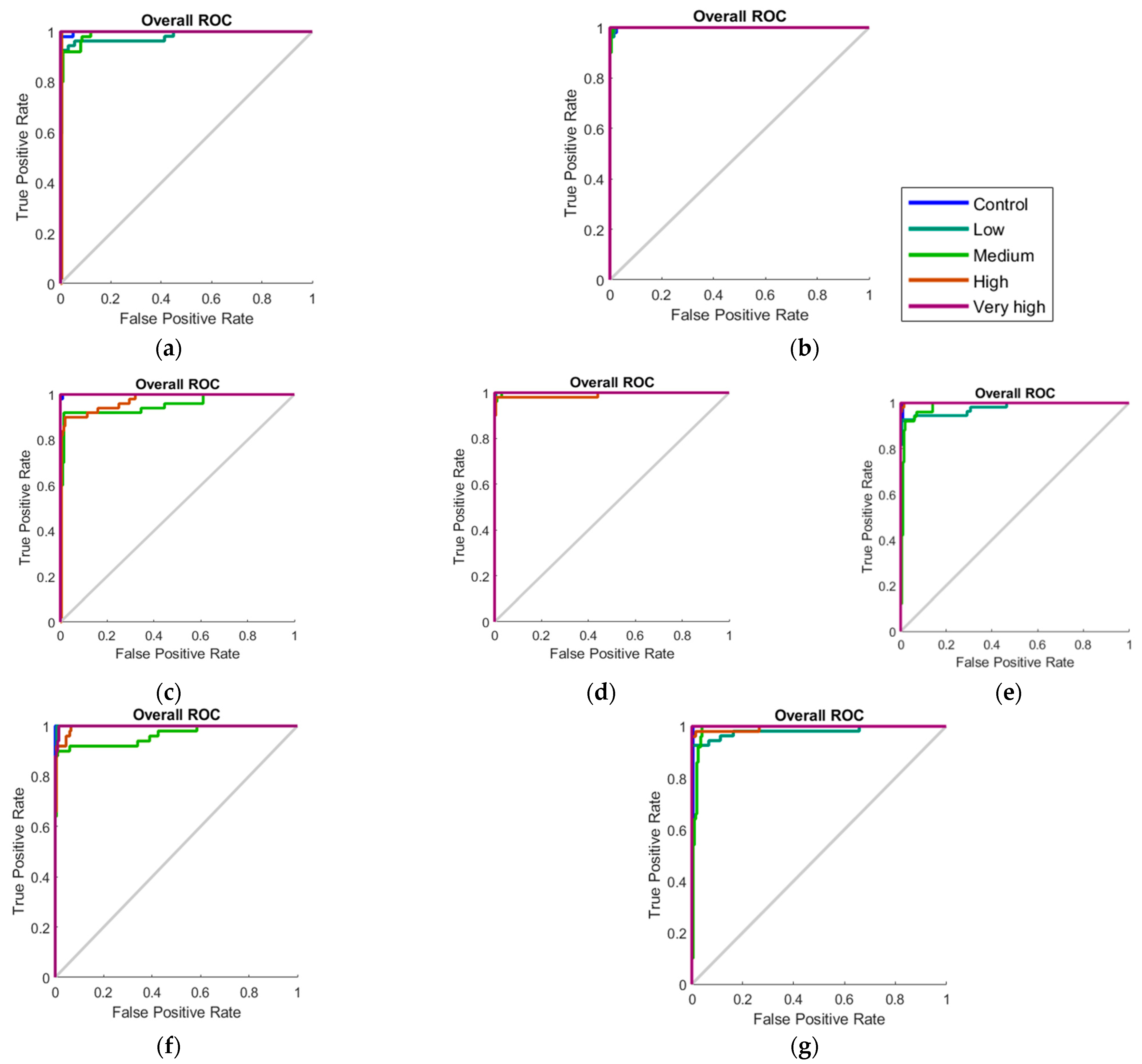
| Treatment | Photosynthesis (μmol CO2 m−2 s−1) | Stomatal Conductance (mol H2O m−2 s−1) | Transpiration (mmol H2O m−2 s−1) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Measurements | W2 | W4 | W6 | W8 | W2 | W4 | W6 | W8 | W2 | W4 | W6 | W8 |
| Control | 14.86 a | 14.17 a | 17.84 a | 15.42 a | 1.03 a | 0.94 a | 0.93 a | 0.98 a | 5.19 ab | 6.60 a | 6.46 a | 5.61 a |
| ±0.57 | ±0.11 | ±0.43 | ±0.07 | ±0.12 | ±0.01 | ±0.01 | ±0.04 | ±0.42 | ±0.13 | ±0.05 | ±0.08 | |
| 102 (low) | 10.98 b | 10.42 b | 11.77 b | 9.41 b | 0.77 ab | 0.44 bc | 0.68 b | 0.57 b | 4.68 b | 4.55 b | 4.20 b | 4.47 b |
| ±0.84 | ±0.16 | ±0.54 | ±0.14 | ±0.05 | ±0.23 | ±0.02 | ±0.27 | ±0.15 | ±0.36 | ±0.09 | ±0.14 | |
| 104 (medium) | 6.32 c | 9.75 b | 8.28 bc | 6.60 c | 0.37 bc | 0.38 bc | 0.59 b | 0.47 bc | 3.31 c | 4.11 b | 3.89 bc | 3.41 c |
| ±0.48 | ±0.13 | ±0.11 | ±0.08 | ±0.05 | ±0.23 | ±0.02 | ±0.04 | ±0.20 | ±0.12 | ±0.03 | ±0.11 | |
| 106 (high) | 4.89 cd | 6.03 d | 4.43 cd | 3.55 dc | 0.36 bc | 0.31 bc | 0.34 bc | 0.23 c | 2.92 cd | 3.45 c | 3.07 c | 3.05 c |
| ±0.42 | ±0.23 | ±0.16 | ±0.14 | ±0.03 | ±0.10 | ±0.01 | ±0.03 | ±0.18 | ±0.15 | ±0.13 | ±0.04 | |
| 5 × 106 (very high) | 2.29 d | 2.26 d | 2.49 d | 1.07 d | 0.20 c | 0.16 c | 0.13 c | 0.11 c | 2.56 d | 2.40 d | 1.39 d | 1.27 d |
| ±0.41 | ±0.16 | ±0.10 | ±0.07 | ±0.02 | ±0.01 | ±0.02 | ±0.01 | ±0.20 | ±0.09 | ±0.11 | ±0.09 | |
| Stage | Samples | Observations | R | b | Performance (MSE) |
|---|---|---|---|---|---|
| Model 1—All treatments, Week 2 | |||||
| Training | 175 | 525 | 0.99 | 0.98 | <0.01 |
| Testing | 75 | 225 | 0.95 | 0.99 | 0.02 |
| Overall | 250 | 750 | 0.97 | 0.99 | - |
| Model 2—All treatments, Week 4 | |||||
| Training | 175 | 525 | 0.99 | 0.99 | <0.01 |
| Testing | 75 | 225 | 0.93 | 0.93 | 0.02 |
| Overall | 250 | 750 | 0.98 | 0.97 | - |
| Model 3—All treatments, Week 6 | |||||
| Training | 175 | 525 | 0.99 | 0.98 | <0.01 |
| Testing | 75 | 225 | 0.98 | 0.97 | 0.01 |
| Overall | 250 | 750 | 0.99 | 0.98 | - |
| Model 4—All treatments, Week 8 | |||||
| Training | 175 | 525 | 0.99 | 0.99 | <0.01 |
| Testing | 75 | 225 | 0.91 | 1.00 | 0.02 |
| Overall | 250 | 750 | 0.97 | 1.00 | - |
| Stage | Samples | Accuracy | Error | Performance (MSE) |
|---|---|---|---|---|
| Model 5—Day 0 + Day 3 | ||||
| Training | 350 | 99.40% | 0.60% | <0.01 |
| Testing | 150 | 85.30% | 14.70% | 0.02 |
| Overall | 500 | 95.20% | 4.80% | - |
| Model 6—Week 2 | ||||
| Training | 175 | 98.30% | 1.70% | <0.01 |
| Testing | 75 | 93.30% | 6.70% | 0.02 |
| Overall | 250 | 96.80% | 3.20% | - |
| Model 7—Week 4 | ||||
| Training | 175 | 98.30% | 1.70% | <0.01 |
| Testing | 75 | 85.30% | 14.70% | 0.01 |
| Overall | 250 | 94.40% | 5.60% | - |
| Model 8—Week 6 | ||||
| Training | 175 | 97.70% | 2.30% | <0.01 |
| Testing | 75 | 94.70% | 5.30% | 0.01 |
| Overall | 250 | 96.80% | 3.20% | - |
| Model 9—Week 8 | ||||
| Training | 175 | 99.40% | 0.60% | 0.01 |
| Testing | 75 | 84.00% | 16.00% | 0.02 |
| Overall | 250 | 94.80% | 5.20% | - |
| Stage | Samples | Accuracy | Error | Performance (MSE) |
|---|---|---|---|---|
| Model 10—Baseline + Day 2 | ||||
| Training | 350 | 98.30% | 1.70% | <0.01 |
| Testing | 150 | 91.35% | 8.65% | 0.02 |
| Overall | 500 | 96.22% | 3.78% | - |
| Model 11—Weeks 1–4 | ||||
| Training | 700 | 97.13% | 2.87% | <0.01 |
| Testing | 300 | 89.40% | 10.60% | 0.02 |
| Overall | 1000 | 94.81% | 5.19% | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, H.; Gonzalez Viejo, C.; Vaghefi, N.; Taylor, P.W.J.; Tongson, E.; Fuentes, S. Early Detection of Fusarium oxysporum Infection in Processing Tomatoes (Solanum lycopersicum) and Pathogen–Soil Interactions Using a Low-Cost Portable Electronic Nose and Machine Learning Modeling. Sensors 2022, 22, 8645. https://doi.org/10.3390/s22228645
Feng H, Gonzalez Viejo C, Vaghefi N, Taylor PWJ, Tongson E, Fuentes S. Early Detection of Fusarium oxysporum Infection in Processing Tomatoes (Solanum lycopersicum) and Pathogen–Soil Interactions Using a Low-Cost Portable Electronic Nose and Machine Learning Modeling. Sensors. 2022; 22(22):8645. https://doi.org/10.3390/s22228645
Chicago/Turabian StyleFeng, Hanyue, Claudia Gonzalez Viejo, Niloofar Vaghefi, Paul W. J. Taylor, Eden Tongson, and Sigfredo Fuentes. 2022. "Early Detection of Fusarium oxysporum Infection in Processing Tomatoes (Solanum lycopersicum) and Pathogen–Soil Interactions Using a Low-Cost Portable Electronic Nose and Machine Learning Modeling" Sensors 22, no. 22: 8645. https://doi.org/10.3390/s22228645
APA StyleFeng, H., Gonzalez Viejo, C., Vaghefi, N., Taylor, P. W. J., Tongson, E., & Fuentes, S. (2022). Early Detection of Fusarium oxysporum Infection in Processing Tomatoes (Solanum lycopersicum) and Pathogen–Soil Interactions Using a Low-Cost Portable Electronic Nose and Machine Learning Modeling. Sensors, 22(22), 8645. https://doi.org/10.3390/s22228645









