Magnetic Nanoparticles Enhanced Surface Plasmon Resonance Biosensor for Rapid Detection of Salmonella Typhimurium in Romaine Lettuce
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Instrument
2.2. Preparation of Antibody-Coupled Magnetic Nanoparticles
2.3. Preparation of Flagellin Extraction
2.4. Preparation of SPR Sensor Surface
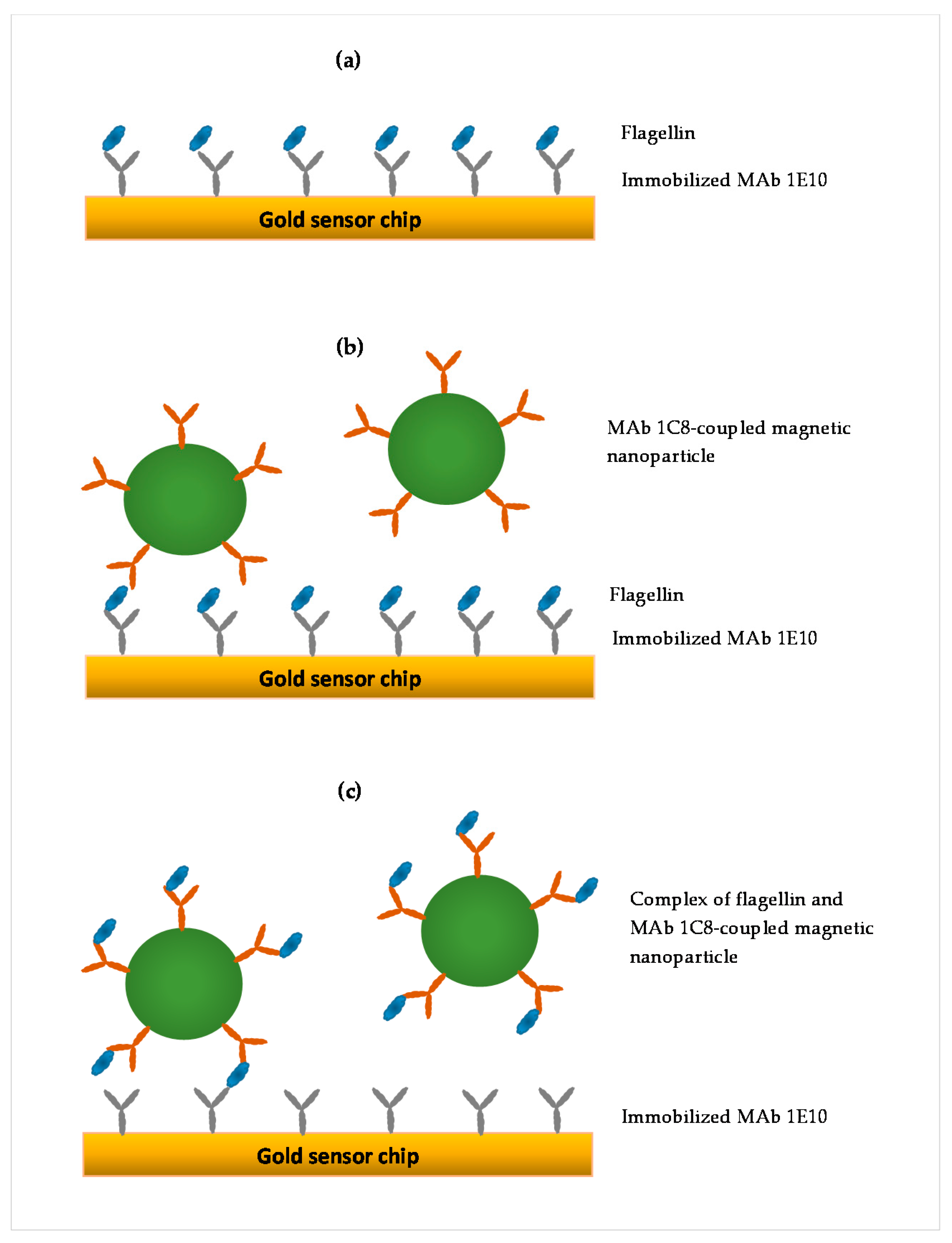
2.5. SPR Assay Formats
2.6. Preparation of Flagellin Sample from Romaine Lettuce
3. Results
3.1. Optimization of Antibody-Coupled Magnetic Nanoparticles
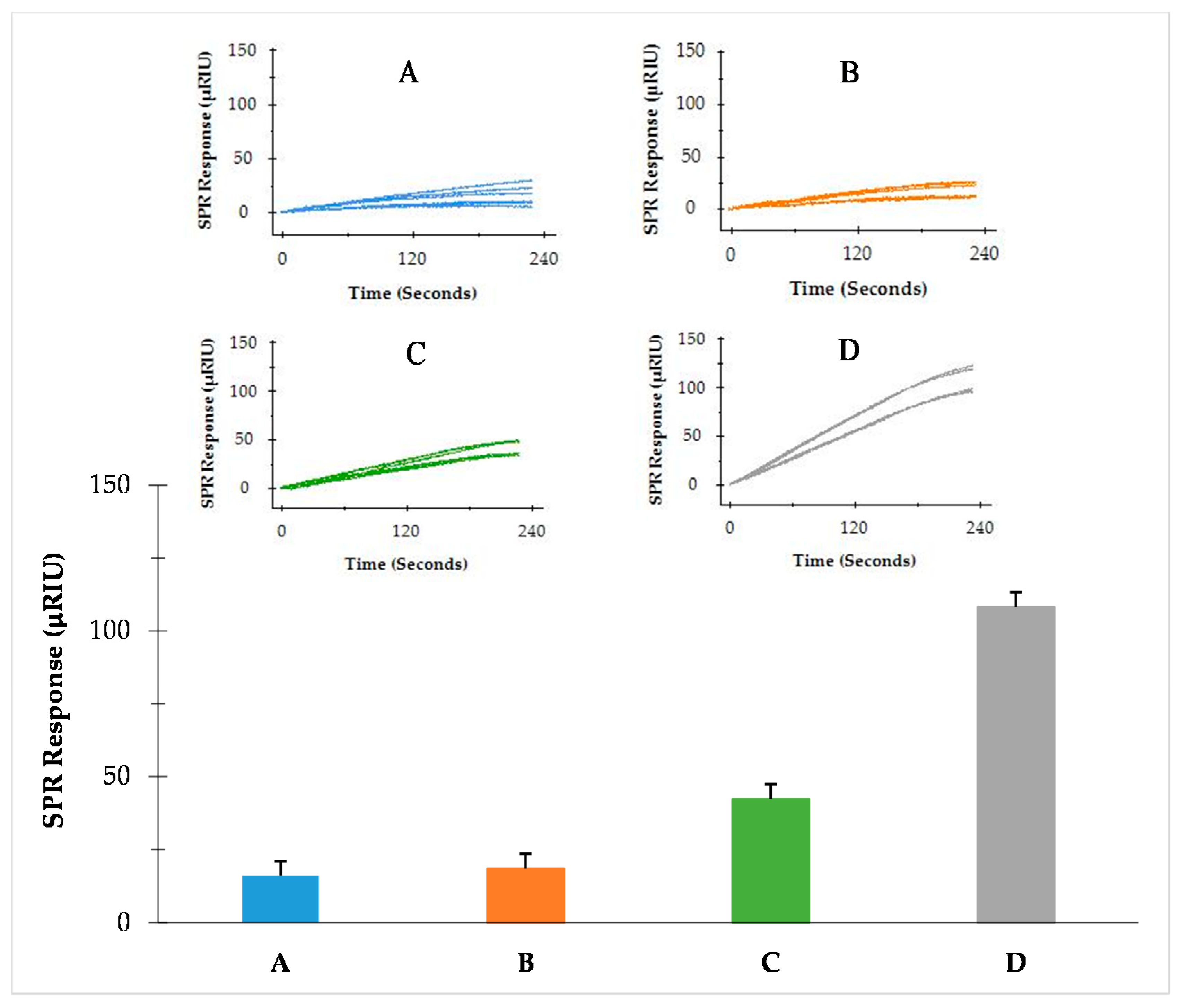
3.2. Comparison of Three SPR Assay Formats
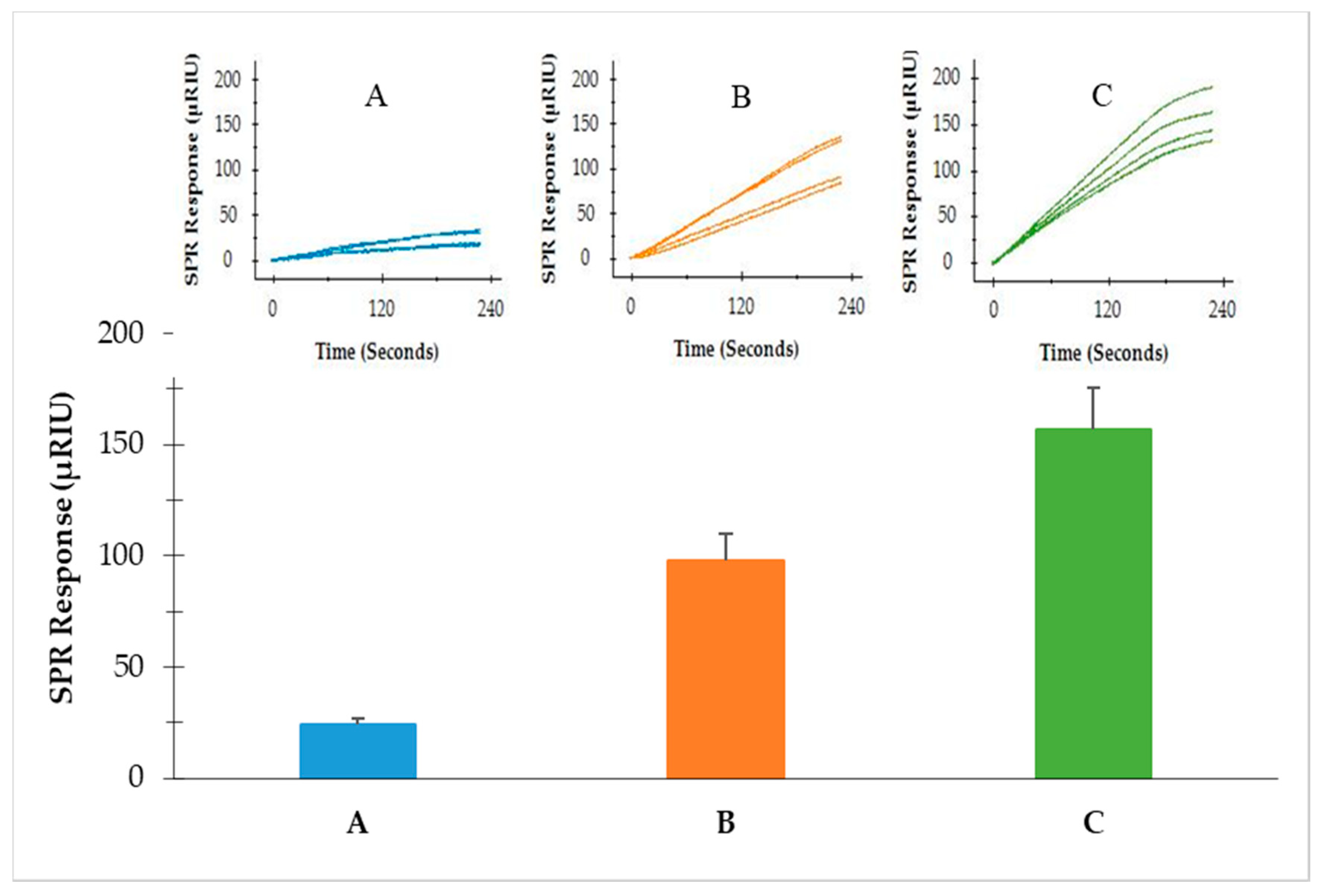
3.3. Detection of S. Typhimurium from Enriched BPW
3.4. Detection of S. Typhimurium from Romaine Lettuce
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States-Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Hoffmann, S.; Batz, M.B.; Morris, J.G. Annual cost of illness and quality-adjusted life year losses in the United States due to 14 foodborne pathogens. J. Food Prot. 2012, 75, 1292–1302. [Google Scholar] [CrossRef]
- Galanis, E.; Wong, D.M.A.L.F.; Patrick, M.E.; Binsztein, N.; Cieslik, A.; Chalermchaikit, T.; Aidara-Kane, A.; Ellis, A.; Angulo, F.J.; Wegener, H.C. Web-based surveillance and global Salmonella distribution, 2000–2002. Emerg. Infect. Dis. 2006, 12, 381–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrew, W.H.; Wang, H.; Jacobson, A.; Hammack, T. Bacteriological Analytical Manual (BAM): Salmonella. Available online: https://www.fda.gov/food/laboratory-methods-food/bacteriological-analytical-manual-bam-chapter-5-salmonella (accessed on 19 August 2019).
- Swaminathan, B.; Feng, P. Rapid detection of food-Borne pathogenic bacteria. Annu. Rev. Microbiol. 1994, 48, 401–426. [Google Scholar] [CrossRef] [PubMed]
- Beumer, R.R.; Brinkman, E.; Rombouts, F.M. Enzyme-linked immunoassays for the detection of Salmonella spp.: A comparison with other methods. Int. J. Food Microbiol. 1991, 12, 363–374. [Google Scholar] [CrossRef]
- Lee, H.A.; Wyatt, G.M.; Bramham, S.; Morgan, M.R. Enzyme-linked immunosorbent assay for Salmonella Typhimurium in food: Feasibility of 1-day Salmonella detection. Appl. Environ. Microbiol. 1990, 56, 1541–1546. [Google Scholar] [CrossRef] [Green Version]
- Valdivieso-Garcia, A.; Riche, E.; Abubakar, O.; Waddell, T.E.; Brooks, B.W. A Double antibody sandwich enzyme-linked immunosorbent assay for the detection of Salmonella using biotinylated monoclonal antibodies. J. Food Prot. 2001, 64, 1166–1171. [Google Scholar] [CrossRef] [PubMed]
- Minnich, S.A.; Hartman, P.A.; Heimsch, R.C. Enzyme immunoassay for detection of Salmonellae in foods. Appl. Environ. Microbiol. 1982, 43, 877–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fluit, A.C.; Widjojoatmodjo, M.N.; Box, A.T.A.; Torensma, R.; Verhoef, J. Rapid detection of Salmonellae in poultry with the magnetic immuno-polymerase chain reaction assay. Appl. Environ. Microbiol. 1993, 59, 1342–1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.-S.; Tsen, H.-Y. Development and use of polymerase chain reaction for the specific detection of Salmonella Typhimurium in stool and food samples. J. Food Prot. 1999, 62, 1103–1110. [Google Scholar] [CrossRef] [PubMed]
- Riyaz-Ul-Hassan, S.; Verma, V.; Qazi, G.N. Rapid detection of Salmonella by polymerase chain reaction. Mol. Cell. Probes 2004, 18, 333–339. [Google Scholar] [CrossRef]
- Cocolin, L.; Manzano, M.; Cantoni, C.; Comi, G. Use of polymerase chain reaction and restriction enzyme analysis to directly detect and identify Salmonella typhimurium in food. J. Appl. Microbiol. 1998, 85, 673–677. [Google Scholar] [CrossRef] [Green Version]
- Kurowski, P.B.; Traub-Dargatz, J.L.; Morley, P.S.; Gentry-Weeks, C.R. Detection of Salmonella spp in fecal specimens by use of real-time polymerase chain reaction assay. Am. J. Vet. Res. 2002, 63, 1265–1268. [Google Scholar] [CrossRef]
- Chen, S.; Yee, A.; Griffiths, M.; Larkin, C.; Yamashiro, C.T.; Behari, R.; Paszko-Kolva, C.; Rahn, K.; De Grandis, S.A. The evaluation of a fluorogenic polymerase chain reaction assay for the detection of Salmonella species in food commodities. Int. J. Food Microbiol. 1997, 35, 239–250. [Google Scholar] [CrossRef]
- Bailey, J.S. Detection of Salmonella cells within 24 to 26 hours in poultry samples with the polymerase chain reaction BAX system. J. Food Prot. 2016, 61, 792–795. [Google Scholar] [CrossRef]
- Abu Al-Soud, W.; Rådström, P. Effects of amplification facilitators on diagnostic PCR in the presence of blood, feces, and meat. J. Clin. Microbiol. 2000, 38, 4463–4470. [Google Scholar] [CrossRef] [Green Version]
- Lantz, P.G.; Al-Soud, W.A.; Knutsson, R.; Hahn-Hägerdal, B.; Rådström, P. Biotechnical use of polymerase chain reaction for microbiological analysis of biological samples. Biotechnol. Annu. Rev. 2000, 5, 87–130. [Google Scholar]
- Rossen, L.; Nørskov, P.; Holmstrøm, K.; Rasmussen, O.F. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int. J. Food Microbiol. 1992, 17, 37–45. [Google Scholar] [CrossRef]
- Homola, J. Present and future of surface plasmon resonance biosensors. Anal. Bioanal. Chem. 2003, 377, 528–539. [Google Scholar] [CrossRef]
- Nguyen, H.; Park, J.; Kang, S.; Kim, M.; Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface plasmon resonance: A versatile technique for biosensor applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef] [Green Version]
- Oh, B.-K.; Kim, Y.-K.; Park, K.W.; Lee, W.H.; Choi, J.-W. Surface plasmon resonance immunosensor for the detection of Salmonella typhimurium. Biosens. Bioelectron. 2004, 19, 1497–1504. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.D.; Ladd, J.; Yu, Q.; Chen, S.; Homola, J.; Jiang, S. Quantitative and simultaneous detection of four foodborne bacterial pathogens with a multi-channel SPR sensor. Biosens. Bioelectron. 2006, 22, 752–758. [Google Scholar] [CrossRef]
- Mazumdar, S.D.; Hartmann, M.; Kämpfer, P.; Keusgen, M. Rapid method for detection of Salmonella in milk by surface plasmon resonance (SPR). Biosens. Bioelectron. 2007, 22, 2040–2046. [Google Scholar] [CrossRef]
- Bokken, G.C.A.M.; Corbee, R.J.; Knapen, F.; Bergwerff, A.A. Immunochemical detection of Salmonella group B, D and E using an optical surface plasmon resonance biosensor. FEMS Microbiol. Lett. 2003, 222, 75–82. [Google Scholar] [CrossRef] [Green Version]
- Lan, Y.B.; Wang, S.Z.; Yin, Y.G.; Hoffmann, W.C.; Zheng, X.Z. Using a surface plasmon resonance biosensor for rapid detection of Salmonella Typhimurium in chicken carcass. J. Bionic Eng. 2008, 5, 239–246. [Google Scholar] [CrossRef]
- Bhandari, D.; Chen, F.-C.; Bridgman, R.C. Detection of Salmonella Typhimurium in romaine lettuce using a surface plasmon resonance biosensor. Biosensors 2019, 9, 94. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhu, Z.; Munir, A.; Zhou, H.S. Fe3O4 nanoparticles-enhanced SPR sensing for ultrasensitive sandwich bio-assay. Talanta 2011, 84, 783–788. [Google Scholar] [CrossRef]
- Pollet, J.; Delport, F.; Janssen, K.P.F.; Tran, D.T.; Wouters, J.; Verbiest, T.; Lammertyn, J. Fast and accurate peanut allergen detection with nanobead enhanced optical fiber SPR biosensor. Talanta 2011, 83, 1436–1441. [Google Scholar] [CrossRef]
- Liang, R.P.; Yao, G.H.; Fan, L.X.; Qiu, J.D. Magnetic Fe3O4 @Au composite-enhanced surface plasmon resonance for ultrasensitive detection of magnetic nanoparticle-enriched α-fetoprotein. Anal. Chim. Acta 2012, 737, 22–28. [Google Scholar] [CrossRef]
- Wang, J.; Munir, A.; Zhu, Z.; Zhou, H.S. Magnetic nanoparticle enhanced surface plasmon resonance sensing and its application for the ultrasensitive detection of magnetic nanoparticle-enriched small molecules. Anal. Chem. 2010, 82, 6782–6789. [Google Scholar] [CrossRef]
- Liu, X.; Hu, Y.; Zheng, S.; Liu, Y.; He, Z.; Luo, F. Surface plasmon resonance immunosensor for fast, highly sensitive, and in situ detection of the magnetic nanoparticles-enriched Salmonella enteritidis. Sens. Actuators B Chem. 2016, 230, 191–198. [Google Scholar] [CrossRef]
- Bhandari, D.; Chen, F.-C.; Hamal, S.; Bridgman, R.C. Kinetic analysis and epitope mapping of monoclonal antibodies to Salmonella Typhimurium flagellin using a surface plasmon resonance biosensor. Antibodies 2019, 8, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soelberg, S.D.; Stevens, R.C.; Limaye, A.P.; Furlong, C.E. Surface plasmon resonance detection using antibody-linked magnetic nanoparticles for analyte capture, purification, concentration, and signal amplification. Anal. Chem. 2009, 81, 2357–2363. [Google Scholar] [CrossRef] [Green Version]
- Torun, Ö.; Boyacı, İ.H.; Temür, E.; Tamer, U. Comparison of sensing strategies in SPR biosensor for rapid and sensitive enumeration of bacteria. Biosens. Bioelectron. 2012, 37, 53–60. [Google Scholar] [CrossRef] [PubMed]


| Steps | Procedures | Minutes |
|---|---|---|
| 1 | Romaine lettuce is washed with deionized water, and the washing liquid is removed and filtered under vacuum to collect solid matters. | 20 |
| 2 | Filter membrane is submerged in glycine-HCl (250 mM, pH 2.0) solution and held on a shaker. | 30 |
| 3 | Solution of pH is adjusted to 7 and supernatant is collected by centrifugation. | 20 |
| 4 | Supernatant is concentrated using an Amicon centrifugal filter. | 20 |
| 5 | Supernatant is exchanged into PBST buffer using an Amicon centrifugal filter. | 40 |
| 6 | Supernatant is collected, and the final volume is adjusted with PBST. | 10 |
| 7 | Supernatant is incubated with antibody-coupled MNPs. | 30 |
| 8 | Complex of flagellin-antibody-coupled MNPs is separated and eluted in PBST. | 20 |
| 9 | SPR assay | 10 |
| Total Time | 200 |
| Direct Assay | Sequential Two-Step Sandwich Assay | Preincubation One-Step Sandwich Assay |
|---|---|---|
| Less sample preparation (total time 150 min) | Less sample preparation (total time 150 min) | Additional incubation (total time 200 min) |
| One-step injection | Two-step injection | One-step injection |
| Signal from flagellin only (no amplification) | Separated signals from flagellin and MNPs (amplification) | Signal from the complex of flagellin and MNPs (amplification) |
| Ratio of amplification to flagellin signal = 1 | Ratio of amplification to flagellin signal = 7.5 | Ratio of amplification to flagellin signal = 14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhandari, D.; Chen, F.-C.; Bridgman, R.C. Magnetic Nanoparticles Enhanced Surface Plasmon Resonance Biosensor for Rapid Detection of Salmonella Typhimurium in Romaine Lettuce. Sensors 2022, 22, 475. https://doi.org/10.3390/s22020475
Bhandari D, Chen F-C, Bridgman RC. Magnetic Nanoparticles Enhanced Surface Plasmon Resonance Biosensor for Rapid Detection of Salmonella Typhimurium in Romaine Lettuce. Sensors. 2022; 22(2):475. https://doi.org/10.3390/s22020475
Chicago/Turabian StyleBhandari, Devendra, Fur-Chi Chen, and Roger C. Bridgman. 2022. "Magnetic Nanoparticles Enhanced Surface Plasmon Resonance Biosensor for Rapid Detection of Salmonella Typhimurium in Romaine Lettuce" Sensors 22, no. 2: 475. https://doi.org/10.3390/s22020475
APA StyleBhandari, D., Chen, F.-C., & Bridgman, R. C. (2022). Magnetic Nanoparticles Enhanced Surface Plasmon Resonance Biosensor for Rapid Detection of Salmonella Typhimurium in Romaine Lettuce. Sensors, 22(2), 475. https://doi.org/10.3390/s22020475






