Lightweight Compound Scaling Network for Nasopharyngeal Carcinoma Segmentation from MR Images
Abstract
:1. Introduction
2. Method
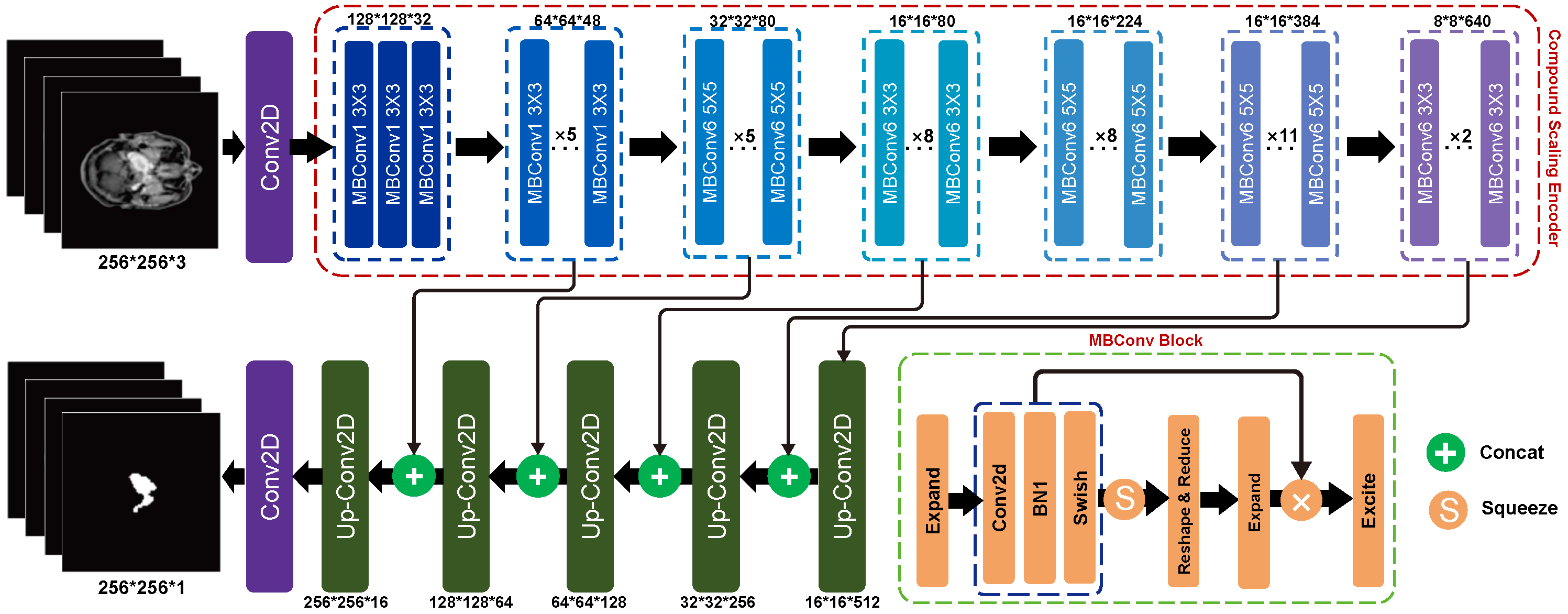
2.1. Compound Scaling Encoder
2.2. UNet-like Decoder
2.3. Evaluation Method
2.3.1. Accuracy Evaluation
2.3.2. Parameters Evaluation
3. Experiments
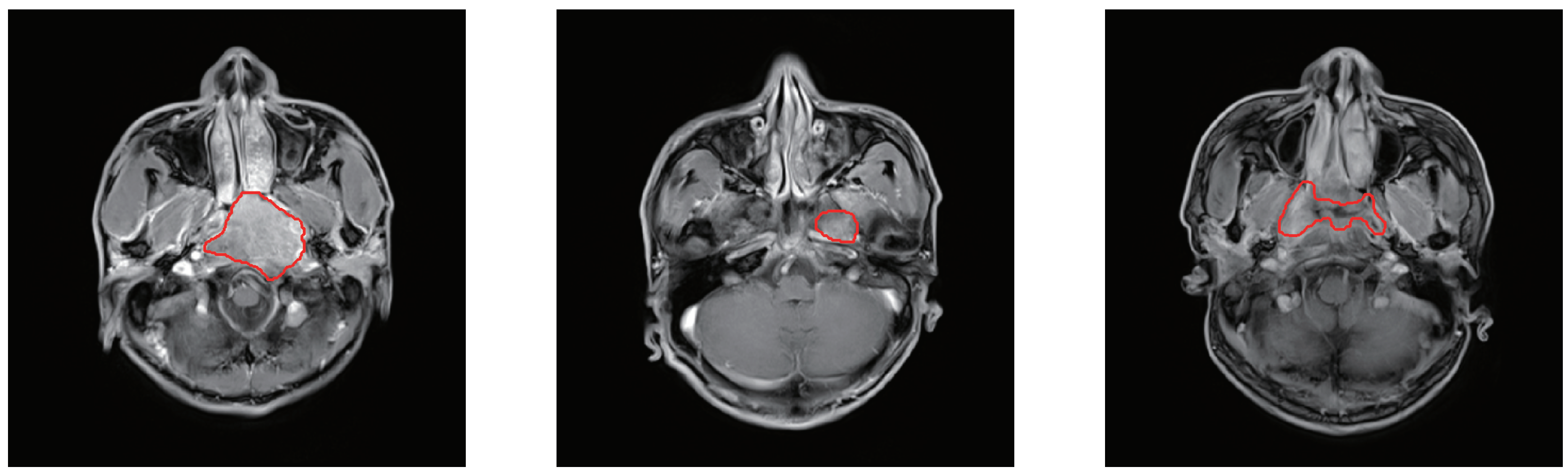
3.1. Dataset Description
3.2. Data Preprocessing
3.3. Implementation Details
4. Result
4.1. Ablation Study
4.2. Comparison with State-of-the-Art Models
4.2.1. Comparison of Accuracy
4.2.2. Comparison of Parameters and FLOPs
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, A.W.; Ng, W.; Chan, Y.; Sze, H.; Chan, C.; Lam, T. The battle against nasopharyngeal cancer. Radiother. Oncol. 2012, 104, 272–278. [Google Scholar] [CrossRef] [PubMed]
- Tsao, S.W.; Lo, K.W.; Huang, D.P. Nasopharyngeal carcinoma. In Epstein-Barr Virus; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Mimi, C.Y.; Yuan, J.M. Epidemiology of nasopharyngeal carcinoma. Semin. Cancer Biol. 2002, 12, 421–429. [Google Scholar]
- Hamid, G.A. Epidemiology and Outcomes of Nasopharyngeal Carcinoma. In Pharynx-Diagnosis and Treatment; IntechOpen: London, UK, 2021. [Google Scholar]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, I.J.; Moskvin, V.; Johnstone, P.A. Analysis of treatment planning time among systems and planners for intensity-modulated radiation therapy. J. Am. Coll. Radiol. 2009, 6, 514–517. [Google Scholar] [CrossRef] [PubMed]
- Tatanun, C.; Ritthipravat, P.; Bhongmakapat, T.; Tuntiyatorn, L. Automatic segmentation of nasopharyngeal carcinoma from CT images: Region growing based technique. In Proceedings of the 2010 2nd International Conference on Signal Processing Systems, Dalian, China, 5–7 July 2010; p. V2-537. [Google Scholar]
- Huang, K.W.; Zhao, Z.Y.; Gong, Q.; Zha, J.; Chen, L.; Yang, R. Nasopharyngeal carcinoma segmentation via HMRF-EM with maximum entropy. In Proceedings of the 2015 37th annual international conference of the IEEE engineering in medicine and biology society (EMBC), Milan, Italy, 25–29 August 2015; pp. 2968–2972. [Google Scholar]
- Wu, B.; Khong, P.L.; Chan, T. Automatic detection and classification of nasopharyngeal carcinoma on PET/CT with support vector machine. Int. J. Comput. Assist. Radiol. Surg. 2012, 7, 635–646. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Chan, K.L.; Xu, P.; Chong, V.F. Nasopharyngeal carcinoma lesion segmentation from MR images by support vector machine. In Proceedings of the 3rd IEEE International Symposium on Biomedical Imaging: Nano to Macro, Arlington, VA, USA, 6–9 April 2006; pp. 1364–1367. [Google Scholar]
- Mohammed, M.A.; Abd Ghani, M.K.; Hamed, R.I.; Ibrahim, D.A.; Abdullah, M.K. Artificial neural networks for automatic segmentation and identification of nasopharyngeal carcinoma. J. Comput. Sci. 2017, 21, 263–274. [Google Scholar] [CrossRef]
- Chanapai, W.; Ritthipravat, P. Adaptive thresholding based on SOM technique for semi-automatic NPC image segmentation. In Proceedings of the 2009 International Conference on Machine Learning and Applications, Miami, FL, USA, 13–15 December 2009; pp. 504–508. [Google Scholar]
- Gao, Z.; Liu, X.; Qi, S.; Wu, W.; Hau, W.K.; Zhang, H. Automatic segmentation of coronary tree in CT angiography images. Int. J. Adapt. Control. Signal Process. 2019, 33, 1239–1247. [Google Scholar] [CrossRef]
- Boudiaf, M.; Kervadec, H.; Masud, Z.I.; Piantanida, P.; Ben Ayed, I.; Dolz, J. Few-shot segmentation without meta-learning: A good transductive inference is all you need? In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 13979–13988. [Google Scholar]
- Cho, J.H.; Mall, U.; Bala, K.; Hariharan, B. Picie: Unsupervised semantic segmentation using invariance and equivariance in clustering. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 16794–16804. [Google Scholar]
- Xu, C.; Xu, L.; Gao, Z.; Zhao, S.; Zhang, H.; Zhang, Y.; Du, X.; Zhao, S.; Ghista, D.; Liu, H.; et al. Direct delineation of myocardial infarction without contrast agents using a joint motion feature learning architecture. Med. Image Anal. 2018, 50, 82–94. [Google Scholar] [CrossRef]
- Douillard, A.; Chen, Y.; Dapogny, A.; Cord, M. Plop: Learning without forgetting for continual semantic segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 4040–4050. [Google Scholar]
- Guzov, V.; Mir, A.; Sattler, T.; Pons-Moll, G. Human poseitioning system (hps): 3d human pose estimation and self-localization in large scenes from body-mounted sensors. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 4318–4329. [Google Scholar]
- Xu, C.; Xu, L.; Ohorodnyk, P.; Roth, M.; Chen, B.; Li, S. Contrast agent-free synthesis and segmentation of ischemic heart disease images using progressive sequential causal GANs. Med. Image Anal. 2020, 62, 101668. [Google Scholar] [CrossRef]
- Mok, T.C.; Chung, A. Fast symmetric diffeomorphic image registration with convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 4644–4653. [Google Scholar]
- Sriram, A.; Zbontar, J.; Murrell, T.; Zitnick, C.L.; Defazio, A.; Sodickson, D.K. GrappaNet: Combining parallel imaging with deep learning for multi-coil MRI reconstruction. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 14315–14322. [Google Scholar]
- Luo, X.; Liao, W.; Chen, J.; Song, T.; Chen, Y.; Zhang, S.; Chen, N.; Wang, G.; Zhang, S. Efficient semi-supervised gross target volume of nasopharyngeal carcinoma segmentation via uncertainty rectified pyramid consistency. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Virtual, 27–1 September–October 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 318–329. [Google Scholar]
- Tao, G.; Li, H.; Liu, L.; Cai, H. Detection-and-Excitation Neural Network Achieves Accurate Nasopharyngeal Carcinoma Segmentation in Multi-modality MR Images. In Proceedings of the 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Houston, TX, USA, 9–12 December 2021; pp. 1063–1068. [Google Scholar]
- Tang, P.; Zu, C.; Hong, M.; Yan, R.; Peng, X.; Xiao, J.; Wu, X.; Zhou, J.; Zhou, L.; Wang, Y. DSU-net: Dense SegU-net for automatic head-and-neck tumour segmentation in MR images. arXiv 2020, arXiv:2006.06278. [Google Scholar]
- Jin, Z.; Li, X.; Shen, L.; Lang, J.; Li, J.; Wu, J.; Xu, P.; Duan, J. Automatic Primary Gross Tumor Volume Segmentation for Nasopharyngeal carcinoma using ResSE-UNet. In Proceedings of the 2020 IEEE 33rd International Symposium on Computer-Based Medical Systems (CBMS), Rochester, MN, USA, 28–30 July 2020; pp. 585–590. [Google Scholar]
- Gao, Z.; Chung, J.; Abdelrazek, M.; Leung, S.; Hau, W.K.; Xian, Z.; Zhang, H.; Li, S. Privileged modality distillation for vessel border detection in intracoronary imaging. IEEE Trans. Med. Imaging 2019, 39, 1524–1534. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Zhang, H.; Zhang, Z.; Liu, M. CNN-based encoder-decoder networks for salient object detection: A comprehensive review and recent advances. Inf. Sci. 2021, 546, 835–857. [Google Scholar] [CrossRef]
- Xu, J.; Zhou, W.; Chen, Z.; Ling, S.; Le Callet, P. Binocular Rivalry Oriented Predictive Autoencoding Network for Blind Stereoscopic Image Quality Measurement. IEEE Trans. Instrum. Meas. 2021, 70, 1–13. [Google Scholar]
- Bang, S.; Park, S.; Kim, H.; Kim, H. Encoder–decoder network for pixel-level road crack detection in black-box images. Comput.-Aided Civ. Infrastruct. Eng. 2019, 34, 713–727. [Google Scholar] [CrossRef]
- Mandal, M.; Vipparthi, S.K. Scene independency matters: An empirical study of scene dependent and scene independent evaluation for CNN-based change detection. IEEE Trans. Intell. Transp. Syst. 2020, 23, 2031–2044. [Google Scholar] [CrossRef]
- Deschaintre, V.; Aittala, M.; Durand, F.; Drettakis, G.; Bousseau, A. Single-image svbrdf capture with a rendering-aware deep network. ACM Trans. Graph. (ToG) 2018, 37, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Sueiras, J.; Ruiz, V.; Sanchez, A.; Velez, J.F. Offline continuous handwriting recognition using sequence to sequence neural networks. Neurocomputing 2018, 289, 119–128. [Google Scholar] [CrossRef]
- Chong, V.; Fan, Y.F. Detection of recurrent nasopharyngeal carcinoma: MR imaging versus CT. Radiology 1997, 202, 463–470. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Cconference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.J.; Heinrich, M.P.; Misawa, K.; Mori, K.; McDonagh, S.G.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Zhang, J.; Gu, L.; Han, G.; Liu, X. AttR2U-Net: A Fully Automated Model for MRI Nasopharyngeal Carcinoma Segmentation Based on Spatial Attention and Residual Recurrent Convolution. Front. Oncol. 2021, 11, 816672. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.; Zu, C.; Hong, M.; Yan, R.; Peng, X.; Xiao, J.; Wu, X.; Zhou, J.; Zhou, L.; Wang, Y. DA-DSUnet: Dual attention-based dense SU-net for automatic head-and-neck tumour segmentation in MRI images. Neurocomputing 2021, 435, 103–113. [Google Scholar] [CrossRef]
- Li, Q.; Xu, Y.; Chen, Z.; Liu, D.; Feng, S.T.; Law, M.; Ye, Y.; Huang, B. Tumor segmentation in contrast-enhanced magnetic resonance imaging for nasopharyngeal carcinoma: Deep learning with convolutional neural network. BioMed Res. Int. 2018, 2018, 9128527. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Wu, X.; Song, Q.; Luo, Y.; Wang, Y.; Zhou, J. Automated nasopharyngeal carcinoma segmentation in magnetic resonance images by combination of convolutional neural networks and graph cut. Exp. Ther. Med. 2018, 16, 2511–2521. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–5 June 2019; pp. 6105–6114. [Google Scholar]
- Huang, Y.; Cheng, Y.; Bapna, A.; Firat, O.; Chen, D.; Chen, M.; Lee, H.; Ngiam, J.; Le, Q.V.; Wu, Y.; et al. Gpipe: Efficient training of giant neural networks using pipeline parallelism. Adv. Neural Inf. Process. Syst. 2019, 32, 103–112. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Bertels, J.; Eelbode, T.; Berman, M.; Vandermeulen, D.; Maes, F.; Bisschops, R.; Blaschko, M.B. Optimizing the dice score and jaccard index for medical image segmentation: Theory and practice. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 92–100. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, Lille, France, 7–9 July 2015; pp. 448–456. [Google Scholar]
- Wu, Y.; He, K. Group normalization. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2015, arXiv:1412.6980. [Google Scholar]
- Smith, L.N. Cyclical learning rates for training neural networks. In Proceedings of the 2017 IEEE Winter Conference on Applications of Computer Vision (WACV), Santa Rosa, CA, USA, 24–31 March 2017; pp. 464–472. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Xu, G.; Wu, X.; Zhang, X.; He, X. Levit-unet: Make faster encoders with transformer for medical image segmentation. arXiv 2021, arXiv:2107.08623. [Google Scholar] [CrossRef]
- Li, Y.; Peng, H.; Dan, T.; Hu, Y.; Tao, G.; Cai, H. Coarse-to-fine Nasopharyngeal carcinoma Segmentation in MRI via Multi-stage Rendering. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, Korea, 16–19 December 2020; pp. 623–628. [Google Scholar]
- Wu, C.; Zhang, H.; Chen, J.; Gao, Z.; Zhang, P.; Muhammad, K.; Del Ser, J. Vessel-GAN: Angiographic reconstructions from myocardial CT perfusion with explainable generative adversarial networks. Future Gener. Comput. Syst. 2022, 130, 128–139. [Google Scholar] [CrossRef]
- Graham, B.; El-Nouby, A.; Touvron, H.; Stock, P.; Joulin, A.; Jegou, H.; Douze, M. LeViT: A Vision Transformer in ConvNet’s Clothing for Faster Inference. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021; pp. 12259–12269. [Google Scholar]
- Islam, M.A.; Kalash, M.; Bruce, N.D. Revisiting salient object detection: Simultaneous detection, ranking, and subitizing of multiple salient objects. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7142–7150. [Google Scholar]
- Guo, S.; Xu, L.; Feng, C.; Xiong, H.; Gao, Z.; Zhang, H. Multi-level semantic adaptation for few-shot segmentation on cardiac image sequences. Med. Image Anal. 2021, 73, 102170. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.W.; Sultani, W.; Shah, M. Dogfight: Detecting drones from drones videos. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 7067–7076. [Google Scholar]
- Salehi, M.; Sadjadi, N.; Baselizadeh, S.; Rohban, M.H.; Rabiee, H.R. Multiresolution knowledge distillation for anomaly detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 14902–14912. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. Shufflenet: An extremely efficient convolutional neural network for mobile devices. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 6848–6856. [Google Scholar]
- Gao, Z.; Wu, S.; Liu, Z.; Luo, J.; Zhang, H.; Gong, M.; Li, S. Learning the implicit strain reconstruction in ultrasound elastography using privileged information. Med. Image Anal. 2019, 58, 101534. [Google Scholar] [CrossRef]
- Kim, E.; Kim, S.; Seo, M.; Yoon, S. XProtoNet: Diagnosis in chest radiography with global and local explanations. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 15719–15728. [Google Scholar]
- Gamper, J.; Rajpoot, N. Multiple instance captioning: Learning representations from histopathology textbooks and articles. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 16549–16559. [Google Scholar]
- Gao, Z.; Xiong, H.; Liu, X.; Zhang, H.; Ghista, D.; Wu, W.; Li, S. Robust estimation of carotid artery wall motion using the elasticity-based state-space approach. Med. Image Anal. 2017, 37, 1–21. [Google Scholar] [CrossRef]
- Lee, H.J.; Kim, J.U.; Lee, S.; Kim, H.G.; Ro, Y.M. Structure boundary preserving segmentation for medical image with ambiguous boundary. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 4817–4826. [Google Scholar]
- Mahapatra, D.; Bozorgtabar, B.; Shao, L. Pathological retinal region segmentation from oct images using geometric relation based augmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 9611–9620. [Google Scholar]
- Roth, H.R.; Lu, L.; Farag, A.; Shin, H.C.; Liu, J.; Turkbey, E.B.; Summers, R.M. Deeporgan: Multi-level deep convolutional networks for automated pancreas segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 556–564. [Google Scholar]
- Gotra, A.; Sivakumaran, L.; Chartrand, G.; Vu, K.N.; Vandenbroucke-Menu, F.; Kauffmann, C.; Kadoury, S.; Gallix, B.; de Guise, J.A.; Tang, A. Liver segmentation: Indications, techniques and future directions. Insights Imaging 2017, 8, 377–392. [Google Scholar] [CrossRef]
- Skourt, B.A.; El Hassani, A.; Majda, A. Lung CT image segmentation using deep neural networks. Procedia Comput. Sci. 2018, 127, 109–113. [Google Scholar] [CrossRef]



| Models | Depth Scaling Coefficient | Width Scaling Coefficient | Resolution Scaling Coefficient |
|---|---|---|---|
| LW-UNet 0 | 1.0 | 1.0 | 1.0 |
| LW-UNet 1 | 1.4 | 1.2 | 1.3 |
| LW-UNet 2 | 2.2 | 1.6 | 2.0 |
| LW-UNet 3 | 3.1 | 2.0 | 2.7 |
| LW-UNet 4 | 3.6 | 2.2 | 3.0 |
| LW-UNet 5 | 4.1 | 2.4 | 3.4 |
| Models | DSC | IoU | JC | SE | PC | SP |
|---|---|---|---|---|---|---|
| UNet | 0.769 ± 0.063 | 0.618 ±0.058 | 0.632 ±0.076 | 0.858 ± 0.076 | 0.713 ± 0.089 | 0.996 ± 0.002 |
| LW-UNet-0 | 0.696 ± 0.035 | 0.516 ± 0.043 | 0.542 ± 0.044 | 0.674 ± 0.035 | 0.623 ± 0.067 | 0.986 ± 0.001 |
| LW-UNet-1 | 0.771 ± 0.041 | 0.621 ± 0.058 | 0.634 ± 0.058 | 0.801 ± 0.058 | 0.765 ± 0.059 | 0.997 ± 0.001 |
| LW-UNet-2 | 0.796 ± 0.060 | 0.685 ±0.035 | 0.685 ±0.052 | 0.815 ± 0.048 | 0.767 ±0.053 | 0.998 ± 0.001 |
| LW-UNet-3 (Our) | 0.813 ± 0.039 | 0.696± 0.055 | 0.695 ± 0.055 | 0.824 ± 0.044 | 0.787± 0.043 | 0.998 ± 0.001 |
| LW-UNet-4 | 0.815 ± 0.075 | 0.698 ± 0.043 | 0.699 ± 0.043 | 0.814 ± 0.011 | 0.787± 0.056 | 0.998 ± 0.001 |
| LW-UNet-5 | 0.806 ± 0.054 | 0.688 ± 0.027 | 0.689 ± 0.076 | 0.820 ± 0.058 | 0.774± 0.084 | 0.998 ± 0.001 |
| Models | DSC | IoU | JC | SE | PC | SP |
|---|---|---|---|---|---|---|
| LW-UNet-0 | 0.632 ± 0.081 | 0.443 ± 0.028 | 0.482 ± 0.056 | 0.561 ± 0.048 | 0.545 ± 0.059 | 0.976 ± 0.001 |
| LW-UNet-0 (with dataset augmentation) | 0.696 ± 0.035 | 0.516 ± 0.043 | 0.542 ± 0.044 | 0.674 ± 0.035 | 0.623 ± 0.067 | 0.986 ± 0.001 |
| LW-UNet-1 | 0.698 ± 0.071 | 0.513 ± 0.098 | 0.553 ± 0.078 | 0.668 ± 0.038 | 0.626 ± 0.079 | 0.984 ± 0.002 |
| LW-UNet-1 (with dataset augmentation) | 0.771 ± 0.041 | 0.621 ± 0.058 | 0.634 ± 0.058 | 0.801 ± 0.058 | 0.765 ± 0.059 | 0.997 ± 0.001 |
| LW-UNet-2 | 0.718 ± 0.053 | 0.561 ± 0.076 | 0.561 ±0.055 | 0.736 ± 0.088 | 0.647 ±0.043 | 0.988 ± 0.001 |
| LW-UNet-2 (with dataset augmentation) | 0.796 ± 0.060 | 0.685 ±0.035 | 0.685 ±0.052 | 0.815 ± 0.048 | 0.767 ±0.053 | 0.998 ± 0.001 |
| LW-UNet-3 | 0.735 ± 0.053 | 0.574 ± 0.068 | 0.583 ±0.096 | 0.775 ± 0.064 | 0.686 ± 0.085 | 0.995 ± 0.003 |
| LW-UNet-3 (with dataset augmentation) | 0.813 ± 0.039 | 0.696± 0.055 | 0.695 ± 0.055 | 0.824 ± 0.044 | 0.787± 0.043 | 0.998 ± 0.001 |
| Category | Models | Parameters (M) | FLOPs (G) |
|---|---|---|---|
| Ablation Study Models | LW-UNet 5 | 7.36 | 9.01 |
| LW-UNet 4 | 5.32 | 8.38 | |
| LW-UNet 3 (Our) | 3.55 | 7.51 | |
| LW-UNet 2 | 2.22 | 4.75 | |
| LW-UNet 1 | 1.27 | 2.99 | |
| LW-UNet 0 | 0.85 | 1.85 | |
| UNet | 34.53 | 65.47 | |
| State-of-the-art Models | DeepLabV3 | 15.31 | 16.14 |
| Att-UNet | 34.88 | 66.57 | |
| FCN32 | 14.72 | 20.07 | |
| RendUNet | 45.80 | 47.58 | |
| FastTransNet | 29.85 | 20.50 | |
| TransNet | 105.28 | 24.64 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Han, G.; Liu, X. Lightweight Compound Scaling Network for Nasopharyngeal Carcinoma Segmentation from MR Images. Sensors 2022, 22, 5875. https://doi.org/10.3390/s22155875
Liu Y, Han G, Liu X. Lightweight Compound Scaling Network for Nasopharyngeal Carcinoma Segmentation from MR Images. Sensors. 2022; 22(15):5875. https://doi.org/10.3390/s22155875
Chicago/Turabian StyleLiu, Yi, Guanghui Han, and Xiujian Liu. 2022. "Lightweight Compound Scaling Network for Nasopharyngeal Carcinoma Segmentation from MR Images" Sensors 22, no. 15: 5875. https://doi.org/10.3390/s22155875
APA StyleLiu, Y., Han, G., & Liu, X. (2022). Lightweight Compound Scaling Network for Nasopharyngeal Carcinoma Segmentation from MR Images. Sensors, 22(15), 5875. https://doi.org/10.3390/s22155875






