Four-Class Classification of Neuropsychiatric Disorders by Use of Functional Near-Infrared Spectroscopy Derived Biomarkers
Abstract
1. Introduction
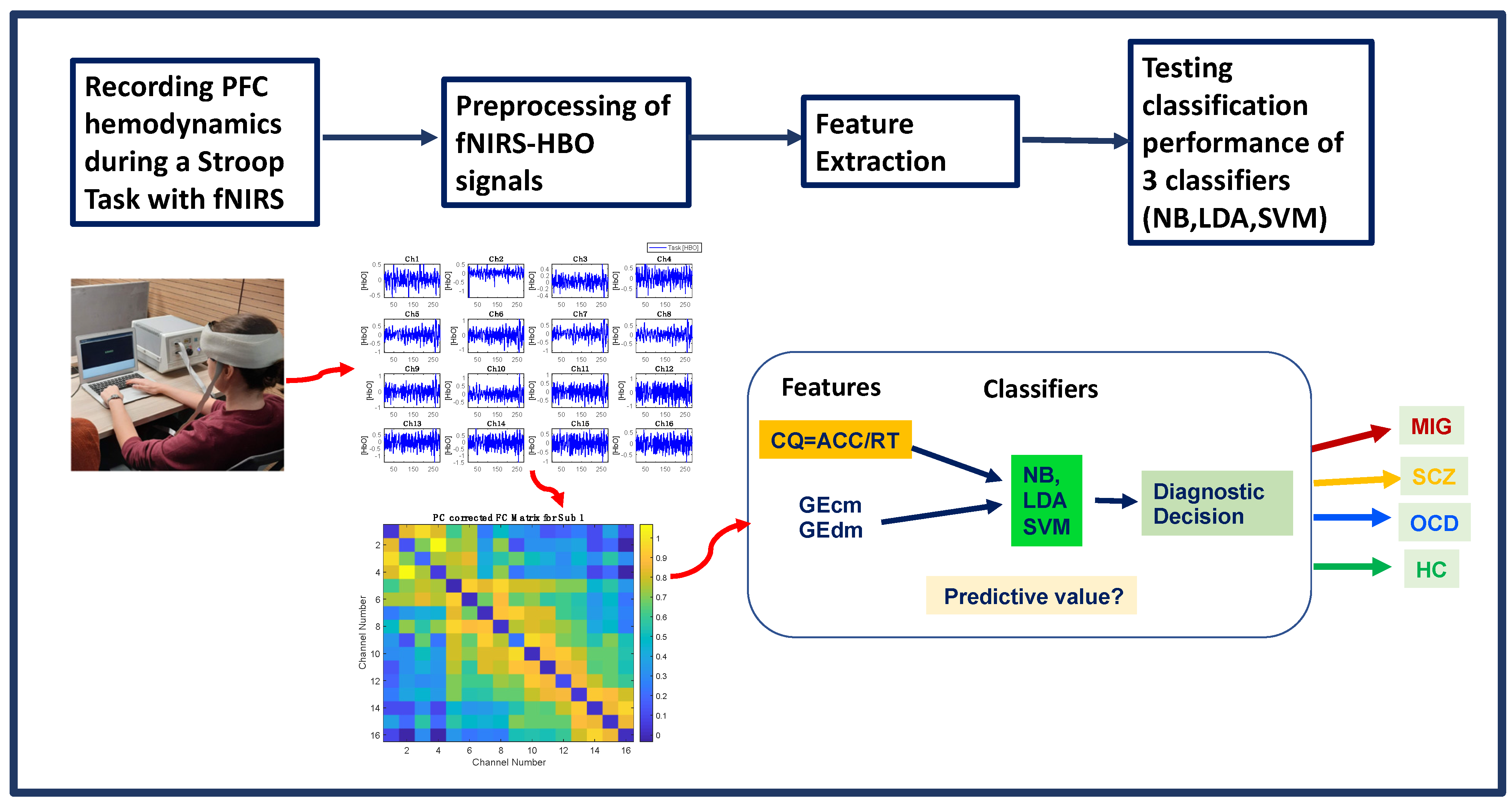
2. Materials and Methods
2.1. Subjects
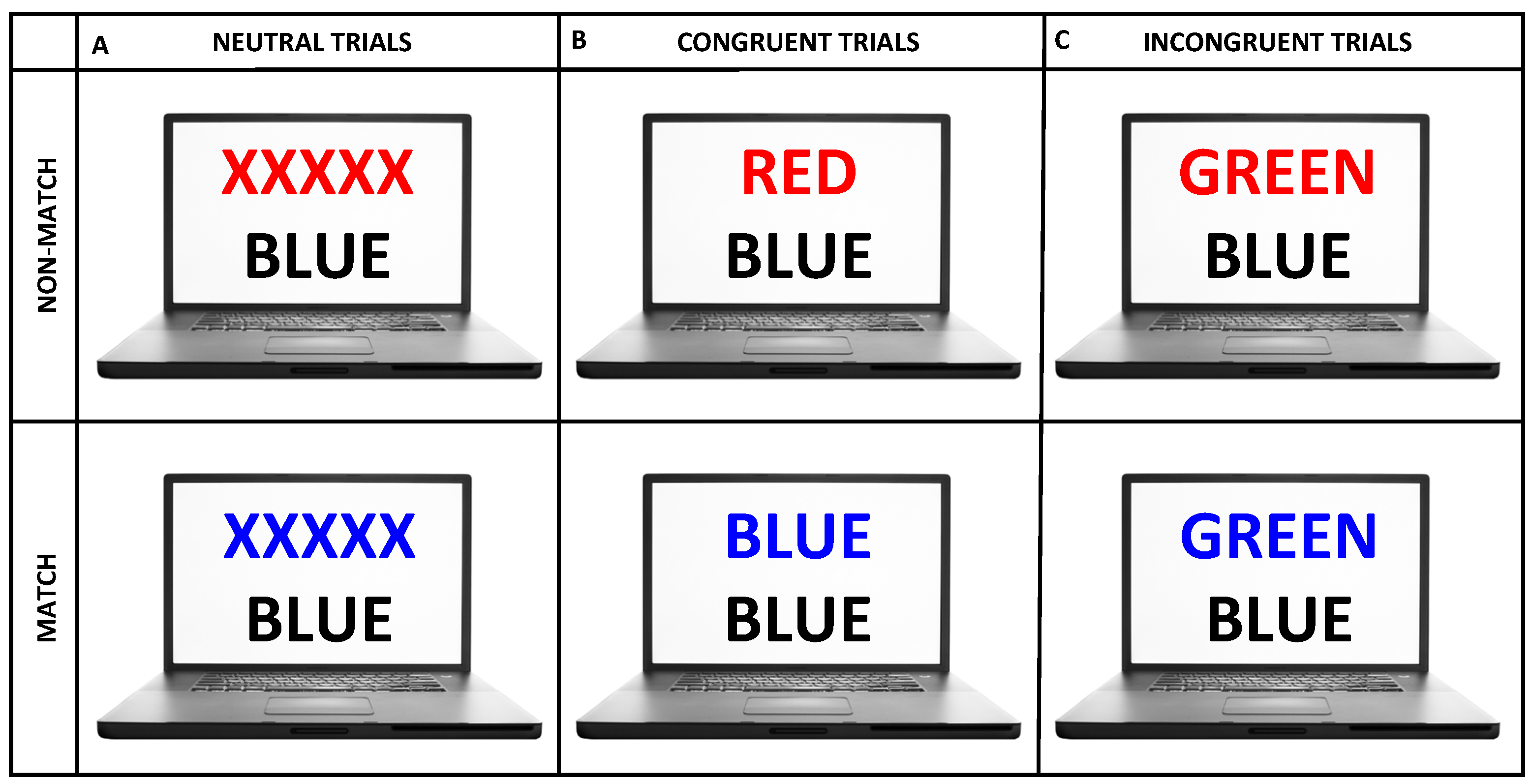
2.2. Experimental Protocol
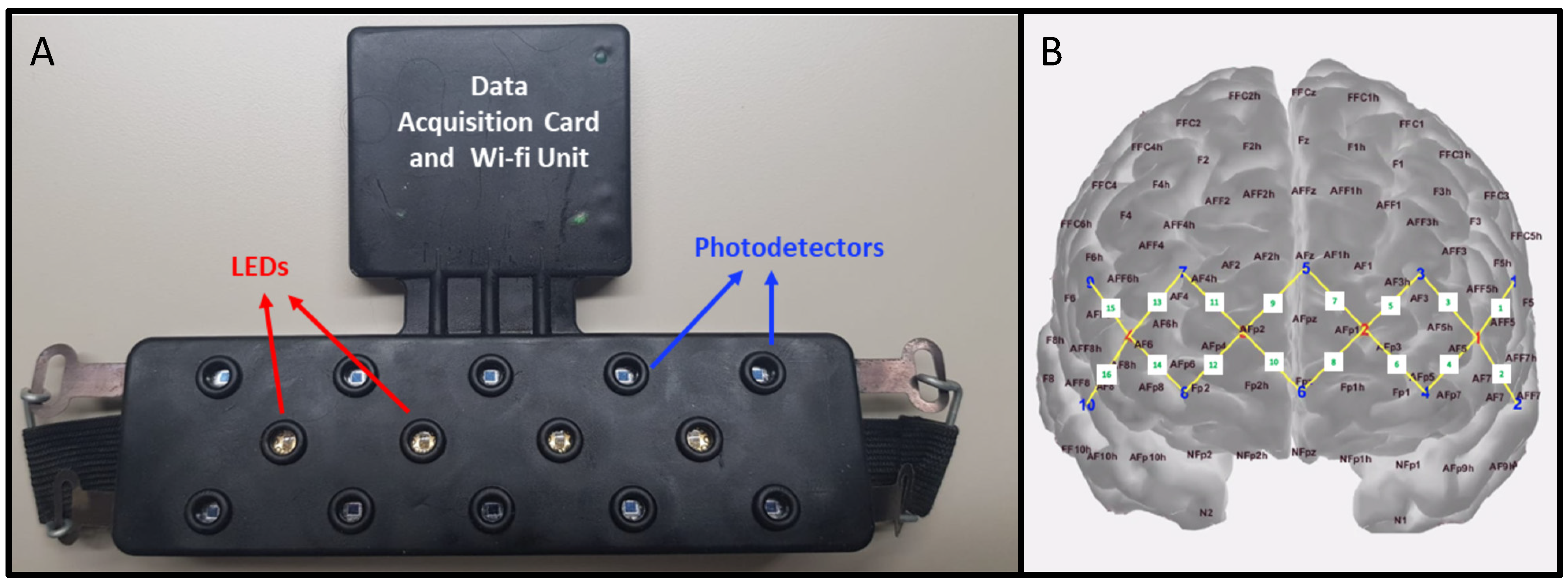
2.3. fNIRS Data Acquisition
2.4. Data Analysis
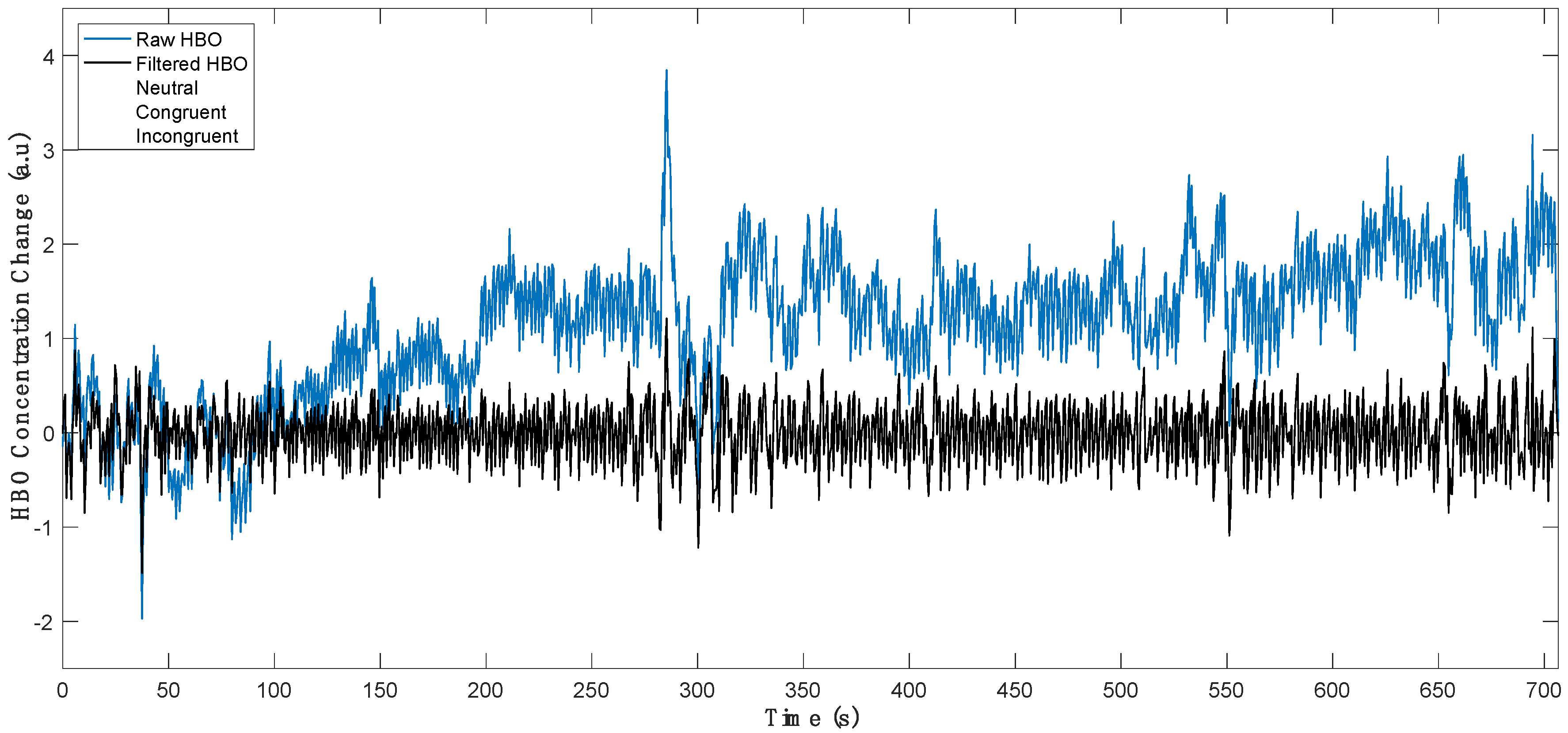
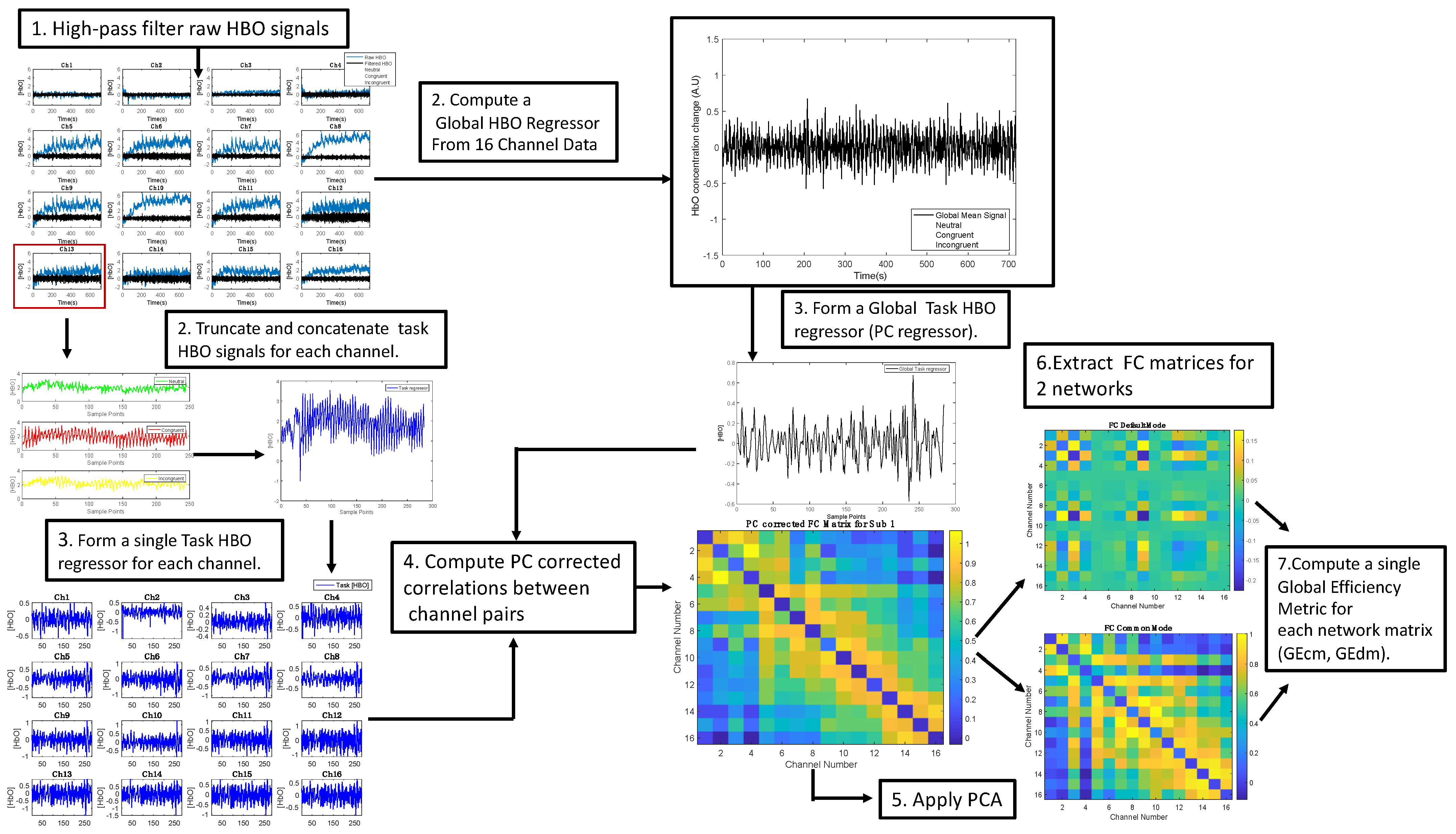
2.4.1. Processing of fNIRS Signals
2.4.2. Computation of Cognitive Quotient and Global Efficiency Features
2.4.3. Classification Methods
2.4.4. Performance Evaluation
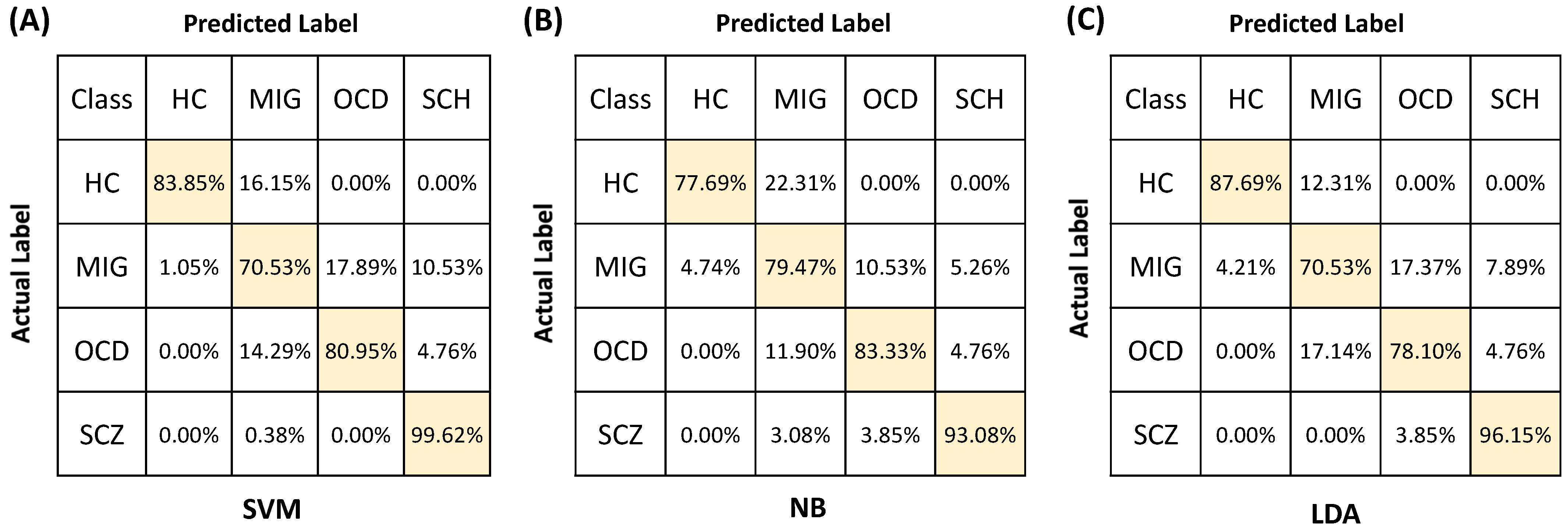
3. Results
4. Discussion
4.1. Comparison of the Classification Performances of LDA, NB, and SVM
4.2. Potential of the Proposed Methodology for Differential Diagnosis
4.3. Limitations of the Study and Recommendations for Future Work
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klöppel, S.; Abdulkadir, A.; Jack, C.R.; Koutsouleris, N.; Mourão-Miranda, J.; Vemuri, P. Diagnostic neuroimaging across diseases. Neuroimage 2012, 61, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Singh, I.; Rose, N. Biomarkers in psychiatry. Nature 2009, 460, 202–207. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. International Statistical Classification of Diseases, 10th ed.; World Health Organization: Geneva, Switzerland, 1992. [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 4th ed.; American Psychiatric Press: Washington, DC, USA, 1994. [Google Scholar]
- Azechi, M.; Iwase, M.; Ikezawa, K.; Takahashi, H.; Canuet, L.; Kurimoto, R.; Nakahachi, T.; Ishii, R.; Fukumoto, M.; Ohi, K.; et al. Discriminant analysis in schizophrenia and healthy subjects using prefrontal activation during frontal lobe tasks: A near-infrared spectroscopy. Schizophr. Res. 2010, 117, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Aboraya, A.; Rankin, E.; France, C.; El-Missiry, A.; John, C. The Reliability of Psychiatric Diagnosis Revisited: The Clinician’s Guide to Improve the Reliability of Psychiatric Diagnosis. Psychiatry 2006, 3, 41–50. [Google Scholar]
- Ward, C.H.; Beck, A.T.; Mendelson, M.; Mock, J.E.; Erbaugh, J.K. The psychiatric nomenclature. Reasons for diagnostic disagreement. Arch. Gen. Psychiatry 1962, 7, 198–205. [Google Scholar] [CrossRef]
- Drevets, W.C. Neuroimaging studies of mood disorders. Biol. Psychiatry 2000, 48, 813–829. [Google Scholar] [CrossRef]
- Okada, G.; Okamoto, Y.; Yamashita, H.; Ueda, K.; Takami, H.; Yamawaki, S. Attenuated prefrontal activation during a verbal fluency task in remitted major depression. Psychiatry Clin. Neurosci. 2009, 63, 423–425. [Google Scholar] [CrossRef]
- Weyandt, L.; Swentosky, A.; Gudmundsdottir, B.G. Neuroimaging and ADHD: fMRI, PET, DTI Findings, and Methodological Limitations. Dev. Neuropsychol. 2013, 38, 211–225. [Google Scholar] [CrossRef]
- Ferrari, M.; Quaresima, V. A brief review on the history of human functional near-infrared spectroscopy (fNIRS) development and fields of application. Neuroimage 2012, 63, 921–935. [Google Scholar] [CrossRef]
- Scholkmann, F.; Kleiser, S.; Metz, A.J.; Zimmermann, R.; Pavia, J.M.; Wolf, U.; Wolf, M. A review on continuous wave functional near-infrared spectroscopy and imaging instrumentation and methodology. Neuroimage 2014, 85, 6–27. [Google Scholar] [CrossRef]
- Ehlis, A.-C.; Schneider, S.; Dresler, T.; Fallgatter, A.J. Application of functional near-infrared spectroscopy in psychiatry. Neuroimage 2014, 85, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Yeung, M.K.; Lin, J. Probing depression, schizophrenia, and other psychiatric disorders using fNIRS and the verbal fluency test: A systematic review and meta-analysis. J. Psychiatr. Res. 2021, 140, 416–435. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Chen, L.; Gao, R.; Bogdan, I.I.M.; Yang, J.; Wang, S.; Dong, W.; Quan, W.; Dang, W.; Yu, X. Automatic schizophrenic discrimination on fNIRS by using complex brain network analysis and SVM. BMC Med. Inform. Decis. Mak. 2017, 17 (Suppl. S3), 166. [Google Scholar] [CrossRef] [PubMed]
- Garrity, A.G.; Pearlson, G.D.; McKiernan, K.; Lloyd, D.; Kiehl, K.A.; Calhoun, V.D. Aberrant “Default Mode” Functional Connectivity in Schizophrenia. Am. J. Psychiatry 2007, 164, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Woodward, T.S.; Leong, K.; Sanford, N.; Tipper, C.M.; Lavigne, K.M. Altered balance of functional brain networks in Schizophrenia. Psychiatry Res. Neuroimaging 2016, 248, 94–104. [Google Scholar] [CrossRef]
- Sharma, A.; Weisbrod, M.; Kaiser, S.; Markela-Lerenc, J.; Bender, S. Deficits in fronto-posterior interactions point to inefficient resource allocation in schizophrenia. Acta Psychiatr. Scand. 2010, 123, 125–135. [Google Scholar] [CrossRef]
- Venkataraman, A.; Whitford, T.J.; Westin, C.-F.; Golland, P.; Kubicki, M. Whole brain resting state functional connectivity abnormalities in schizophrenia. Schizophr. Res. 2012, 139, 7–12. [Google Scholar] [CrossRef]
- Narayanaswamy, J.; Hazari, N.; Venkatasubramanian, G. Neuroimaging findings in obsessive–compulsive disorder: A narrative review to elucidate neurobiological underpinnings. Indian J. Psychiatry 2019, 61 (Suppl. S1), S9–S29. [Google Scholar] [CrossRef]
- Fajnerova, I.; Gregus, D.; Francova, A.; Noskova, E.; Koprivova, J.; Stopkova, P.; Hlinka, J.; Horacek, J. Functional Connectivity Changes in Obsessive–Compulsive Disorder Correspond to Interference Control and Obsessions Severity. Front. Neurol. 2020, 11, 568. [Google Scholar] [CrossRef]
- Erdoğan, S.B.; Yükselen, G.; Yegül, M.M.; Usanmaz, R.; Kıran, E.; Derman, O.; Akın, A. Identification of impulsive adolescents with a functional near infrared spectroscopy (fNIRS) based decision support system. J. Neural Eng. 2021, 18, 056043. [Google Scholar] [CrossRef]
- Çiftçi, K.; Sankur, B.; Kahya, Y.P.; Akin, A. Multilevel Statistical Inference from Functional Near-Infrared Spectroscopy Data during Stroop Interference. IEEE Trans. Biomed. Eng. 2008, 55, 2212–2220. [Google Scholar] [CrossRef][Green Version]
- Aydore, S.; Mihcak, M.K.; Ciftci, K.; Akin, A.; Mihak, M.; Cifti, K. On Temporal Connectivity of PFC via Gauss–Markov Modeling of fNIRS Signals. IEEE Trans. Biomed. Eng. 2009, 57, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Dadgostar, M.; Setarehdan, S.K.; Shahzadi, S.; Akin, A. Functional connectivity of the PFC via partial correlation. Optik 2016, 127, 4748–4754. [Google Scholar] [CrossRef]
- Einalou, Z.; Maghooli, K.; Setarehdan, S.K.; Akin, A. Effective channels in classification and functional connectivity pattern of prefrontal cortex by functional near infrared spectroscopy signals. Optik 2016, 127, 3271–3275. [Google Scholar] [CrossRef]
- Einalou, Z.; Maghooli, K.; Setarehdan, S.K.; Akin, A. Graph theoretical approach to functional connectivity in prefrontal cortex via fNIRS. Neurophotonics 2017, 4, 041407. [Google Scholar] [CrossRef] [PubMed]
- Akin, A. Partial correlation-based functional connectivity analysis for functional near-infrared spectroscopy signals. J. Biomed. Opt. 2017, 22, 126003. [Google Scholar] [CrossRef]
- Akın, A. fNIRS-derived neurocognitive ratio as a biomarker for neuropsychiatric diseases. Neurophotonics 2021, 8, 035008. [Google Scholar] [CrossRef]
- Zysset, S.; Müller, K.; Lohmann, G.; von Cramon, D. Color-Word Matching Stroop Task: Separating Interference and Response Conflict. Neuroimage 2001, 13, 29–36. [Google Scholar] [CrossRef]
- Erdogan, S.B.; Özsarfati, E.; Dilek, B.; Sogukkanlı Kadak, K.; Hanoglu, L.; Akin, A. Classification of motor imagery and execution signals with population-level feature sets: Implications for probe design in fNIRS based BCI. J. Neural Eng. 2019, 16, 026029. [Google Scholar] [CrossRef]
- Akgül, C.B.; Akin, A.; Sankur, B. Extraction of cognitive activity-related waveforms from functional near-infrared spectroscopy signals. Med. Biol. Eng. Comput. 2006, 44, 945–958. [Google Scholar] [CrossRef][Green Version]
- Erdoğan, S.B.; Yucel, M.; Akin, A. Analysis of task-evoked systemic interference in fNIRS measurements: Insights from fMRI. Neuroimage 2014, 87, 490–504. [Google Scholar] [CrossRef] [PubMed]
- Pollonini, L.; Bortfeld, H.; Oghalai, J.S. PHOEBE: A method for real time mapping of optodes-scalp coupling in functional near-infrared spectroscopy. Biomed. Opt. Express 2016, 7, 5104–5119. [Google Scholar] [CrossRef] [PubMed]
- Jobsis, F.F. Noninvasive, infrared monitoring of cerebral and myocardial oxygen sufficiency and circulatory parameters. Science 1977, 198, 1264–1267. [Google Scholar] [CrossRef]
- Firbank, M.; Eiji, O.; Delpy, D.T. A theoretical study of the signal contribution of regions of the adult head to near-infrared spectroscopy studies of visual evoked responses. Neuroimage 1998, 8, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Cope, M.; Delpy, D.T. System for long-term measurement of cerebral blood and tissue oxygenation on newborn infants by near infra-red transillumination. Med. Biol. Eng. Comput. 1988, 26, 289–294. [Google Scholar] [CrossRef]
- Delpy, D.T.; Cope, M.; Zee, P.V.D.; Arridge, S.; Wray Sand Wyatt, J. Estimation of optical pathlength through tissue from direct time of flight measurement. Phys. Med. Biol. 1988, 33, 1433–1442. [Google Scholar] [CrossRef]
- Boas, D.A.; Dale, A.M.; Franceschini, M.A. Diffuse optical imaging of brain activation: Approaches to optimizing image sensitivity, resolution, and accuracy. Neuroimage 2004, 23, S275–S288. [Google Scholar] [CrossRef]
- Sutoko, S.; Monden, Y.; Tokuda, T.; Ikeda, T.; Nagashima, M.; Funane, T.; Atsumori, H.; Kiguchi, M.; Maki, A.; Yamagata, T.; et al. Atypical dynamic-connectivity recruitment in attention-deficit/hyperactivity disorder children: An insight into task-based dynamic connectivity through an fNIRS study. Front. Hum. Neurosci. 2020, 14, 3. [Google Scholar] [CrossRef]
- Al-Shargie, F.; Kiguchi, M.; Badruddin, N.; Dass, S.C.; Hani AF, M.; Tang, T.B. Mental stress assessment using simultaneous measurement of EEG and fNIRS. Biomed. Opt. Express 2016, 7, 3882–3898. [Google Scholar] [CrossRef]
- Al-Shargie, F.; Tang, T.B.; Kiguchi, M. Assessment of mental stress effects on prefrontal cortical activities using canonical correlation analysis: An fNIRS-EEG study. Biomed. Opt. Express 2017, 8, 2583–2598. [Google Scholar] [CrossRef]
- Hoshi, Y. Functional near-infrared optical imaging: Utility and limitations in human brain mapping. Psychophysiology 2003, 40, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Hoshi, Y.; Kobayashi, N.; Tamura, M. Interpretation of near-infrared spectroscopy signals: A study with a newly developed perfused rat brain model. J. Appl. Physiol. 2001, 90, 1657–1662. [Google Scholar] [CrossRef] [PubMed]
- Latora, V.; Marchiori, M. Efficient behavior of small-world networks. Phys. Rev. Lett. 2001, 87, 198701. [Google Scholar] [CrossRef]
- Cui, X.; Bray, S.; Reiss, A.L. Speeded near infrared spectroscopy (NIRS) response detection. PLoS ONE 2010, 5, e15474. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, D.M.; Basak, S.C.; Mills, D. Assessing model fit by cross-validation. J. Chem. Inf. Comput. Sci. 2003, 43, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Tai, K.; Chau, T. Single-trial classification of NIRS signals during emotional induction tasks: Towards a corporeal machine interface. J. Neuroeng. Rehabil. 2009, 6, 39. [Google Scholar] [CrossRef]
- Sitaram, R.; Zhang, H.; Guan, C.; Thulasidas, M.; Hoshi, Y.; Ishikawa, A.; Shimizu, K.; Birbaumer, N. Temporal classification of multichannel near-infrared spectroscopy signals of motor imagery for developing a brain–computer interface. Neuroimage 2007, 34, 1416–1427. [Google Scholar] [CrossRef]
- Tanaka, H.; Katura, T. Classification of change detection and change blindness from near-infrared spectroscopy signals. J. Biomed. Opt. 2011, 16, 087001. [Google Scholar] [CrossRef]
- Misawa, T.; Takano, S.; Shimokawa, T.; Hirobayashi, S. A brain-computer interface for motor assist by the prefrontal cortex. Electron. Commun. Jpn. 2012, 95, 1–8. [Google Scholar] [CrossRef]
- Naseer, N.; Hong, K.-S. fNIRS-based brain-computer interfaces: A review. Front. Hum. Neurosci. 2015, 9, 3. [Google Scholar] [CrossRef]
- Naseer, N.; Qureshi, N.K.; Noori, F.M.; Hong, K.-S. Analysis of Different Classification Techniques for Two-Class Functional Near-Infrared Spectroscopy-Based Brain-Computer Interface. Comput. Intell. Neurosci. 2016, 2016, 5480760. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.A.; Naseer, N.; Qureshi, N.K.; Noori, F.M.; Nazeer, H.; Khan, M.U. fNIRS-based Neurorobotic Interface for gait rehabilitation. J. Neuroeng. Rehabil. 2018, 15, 7. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Jeong, J. Multiclass classification of hemodynamic responses for performance improvement of functional near-infrared spectroscopy-based brain-computer interface. J. Biomed. Opt. 2014, 19, 67009. [Google Scholar] [CrossRef] [PubMed]
- Shoushtarian, M.; Alizadehsani, R.; Khosravi, A.; Acevedo, N.; McKay, C.M.; Nahavandi, S.; Fallon, J.B. Objective measurement of tinnitus using functional near-infrared spectroscopy and machine learning. PLoS ONE 2020, 15, e0241695. [Google Scholar] [CrossRef]
- Cearns, M.; Hahn, T.; Baune, B.T. Recommendations and future directions for supervised machine learning in psychiatry. Transl. Psychiatry 2019, 9, 271. [Google Scholar] [CrossRef]
- Steardo, L.; Carbone, E.A.; De Filippis, R.; Pisanu, C.; Segura-Garcia, C.; Squassina, A.; De Fazio, P. Application of Support Vector Machine on fMRI Data as Biomarkers in Schizophrenia Diagnosis: A Systematic Review. Front. Psychiatry 2020, 11, 588. [Google Scholar] [CrossRef]
- Yu, Y.; Shen, H.; Zeng, L.-L.; Ma, Q.; Hu, D. Convergent and Divergent Functional Connectivity Patterns in Schizophrenia and Depression. PLoS ONE 2013, 8, e68250. [Google Scholar] [CrossRef]
- Sakai, K.; Yamada, K. Machine learning studies on major brain diseases: 5-year trends of 2014–2018. Jpn. J. Radiol. 2018, 37, 34–72. [Google Scholar] [CrossRef]
- Arbabshirani, M.R.; Plis, S.; Sui, J.; Calhoun, V.D. Single subject prediction of brain disorders in neuroimaging: Promises and pitfalls. Neuroimage 2016, 145, 137–165. [Google Scholar] [CrossRef]
- Stephan, K.E.; Schlagenhauf, F.; Huys, Q.; Raman, S.; Aponte, E.; Brodersen, K.; Rigoux, L.; Moran, R.; Daunizeau, J.; Dolan, R.; et al. Computational neuroimaging strategies for single patient predictions. Neuroimage 2016, 145, 180–199. [Google Scholar] [CrossRef]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software: An update SIGKDD. Explorations 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Keerthi, S.S.; Shevade, S.K.; Bhattacharyya, C.; Murthy, K.R.K. Improvements to Platt’s SMO algorithm for SVM classifier design. Neural Comput. 2001, 13, 637–649. [Google Scholar] [CrossRef]
- Üstün, B.; Melssen, W.J.; Buydens, L.M. Facilitating the application of support vector regression by using a universal Pearson VII function based kernel. Chemom. Intell. Lab. Syst. 2006, 81, 29–40. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, G.; Ou, Y.; Guo, W.; Peng, Y.; Hao, K.; Zhao, J.; Yang, Y.; Li, W.; Zhang, Y.; et al. Abnormal default-mode network homogeneity and its correlations with neurocognitive deficits in drug-naive first-episode adolescent-onset schizophrenia. Schizophr. Res. 2019, 215, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Yamamuro, K.; Kimoto, S.; Iida, J.; Kishimoto, N.; Tanaka, S.; Toritsuka, M.; Ikawa, D.; Yamashita, Y.; Ota, T.; Makinodan, M.; et al. Distinct patterns of blood oxygenation in the prefrontal cortex in clinical phenotypes of schizophrenia and bipolar disorder. J. Affect. Disord. 2018, 234, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, Y.; Zhang, Q.; Zhao, W.; Chen, X.; Zhai, J.; Chen, M.; Du, B.; Deng, X.; Ji, F.; et al. The effects of CACNA1C gene polymorphism on prefrontal cortex in both schizophrenia patients and healthy controls. Schizophr. Res. 2019, 204, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Okada, K.; Ota, T.; Iida, J.; Kishimoto, N.; Kishimoto, T. Lower prefrontal activity in adults with obsessive–compulsive disorder as measured by near-infrared spectroscopy. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2013, 43, 7–13. [Google Scholar] [CrossRef]
- Ota, T.; Iida, J.; Sawada, M.; Suehiro, Y.; Yamamuro, K.; Matsuura, H.; Tanaka, S.; Kishimoto, N.; Negoro, H.; Kishimoto, T. Reduced Prefrontal Hemodynamic Response in Pediatric Obsessive–Compulsive Disorder as Measured by Near-Infrared Spectroscopy. Child Psychiatry Hum. Dev. 2012, 44, 265–277. [Google Scholar] [CrossRef]
- Ahn, S.; Jun, S.C. Multi-Modal Integration of EEG-fNIRS for Brain-Computer Interfaces—Current Limitations and Future Directions. Front. Hum. Neurosci. 2017, 11, 503. [Google Scholar] [CrossRef]
- Batula, A.M.; Kim, Y.E.; Ayaz, H. Virtual and Actual Humanoid Robot Control with Four-Class Motor-Imagery-Based Optical Brain-Computer Interface. BioMed Res. Int. 2017, 2017, 1463512. [Google Scholar] [CrossRef]
- Orru, G.; Pettersson-Yeo, W.; Marquand, A.F.; Sartori, G.; Mechelli, A. Using support vector machine to identify imaging biomarkers of neurological and psychiatric disease: A critical review. Neurosci. Biobehav. Rev. 2012, 36, 1140–1152. [Google Scholar] [CrossRef] [PubMed]
- Sen, B.; Bernstein, G.A.; Xu, T.; Mueller, B.A.; Schreiner, M.W.; Cullen, K.R.; Parhi, K.K. Classification of obsessive-compulsive disorder from resting-state fMRI. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 3606–3609. [Google Scholar] [CrossRef]
- Chong, C.D.; Gaw, N.; Fu, Y.; Li, J.; Wu, T.; Schwedt, T.J. Migraine classification using magnetic resonance imaging resting-state functional connectivity data. Cephalalgia 2016, 37, 828–844. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, J.; Liu, Q.; Wang, Y. Multimodal MRI-based classification of migraine: Using deep learning convolutional neural network. Biomed. Eng. Online 2018, 17, 138. [Google Scholar] [CrossRef] [PubMed]
- Jorge-Hernandez, F.; Chimeno, Y.G.; Garcia-Zapirain, B.; Zubizarreta, A.C.; Beldarrain, M.A.G.; Fernandez-Ruanova, B. Graph theory for feature extraction and classification: A migraine pathology case study. Bio-Med. Mater. Eng. 2014, 24, 2979–2986. [Google Scholar] [CrossRef]
- Kawasaki, Y.; Suzuki, M.; Kherif, F.; Takahashi, T.; Zhou, S.-Y.; Nakamura, K.; Matsui, M.; Sumiyoshi, T.; Seto, H.; Kurachi, M. Multivariate voxel-based morphometry successfully differentiates schizophrenia patients from healthy controls. Neuroimage 2007, 34, 235–242. [Google Scholar] [CrossRef]
- Yassin, W.; Nakatani, H.; Zhu, Y.; Kojima, M.; Owada, K.; Kuwabara, H.; Gonoi, W.; Aoki, Y.; Takao, H.; Natsubori, T.; et al. Machine-learning classification using neuroimaging data in schizophrenia, autism, ultra-high risk and first-episode psychosis. Transl. Psychiatry 2020, 10, 278. [Google Scholar] [CrossRef]
- Pardo, P.J.; Georgopoulos, A.P.; Kenny, J.T.; Stuve, T.A.; Findling, R.L.; Schulz, S.C. Classification of adolescent psychotic disorders using linear discriminant analysis. Schizophr. Res. 2006, 87, 297–306. [Google Scholar] [CrossRef]
- D’souza, R.N.; Huang, P.Y.; Yeh, F.C. Structural Analysis and Optimization of Convolutional Neural Networks with a Small Sample Size. Sci. Rep. 2020, 10, 834. [Google Scholar] [CrossRef]
- Wickramaratne, S.D.; Mahmud, M.S. Conditional-GAN Based Data Augmentation for Deep Learning Task Classi-fier Improvement Using fNIRS Data. Front. Big Data 2021, 4, 659146. [Google Scholar] [CrossRef]
- Khalil, K.; Asgher, U.; Ayaz, Y. Novel fNIRS study on homogeneous symmetric feature-based transfer learning for brain-computer interface. Sci. Rep. 2022, 12, 3198. [Google Scholar] [CrossRef]
- Lyu, B.; Pham, T.; Blaney, G.; Haga, Z.; Sassaroli, A.; Fantini, S.; Aeron, S. Domain adaptation for robust workload level alignment between sessions and subjects using fNIRS. J. Biomed. Opt. 2021, 26, 022908. [Google Scholar] [CrossRef] [PubMed]
- Hennrich, J.; Herff, C.; Heger, D.; Schultz, T. Investigating Deep Learning for fNIRS Based BCI. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 2844–2847. [Google Scholar] [CrossRef]
- Chiarelli, A.M.; Croce, P.; Merla, A.; Zappasodi, F. Deep Learning for Hybrid EEG-fNIRS Brain-Computer In-terface: Application to Motor Imagery Classification. J. Neural Eng. 2018, 15, 036028. [Google Scholar] [CrossRef] [PubMed]
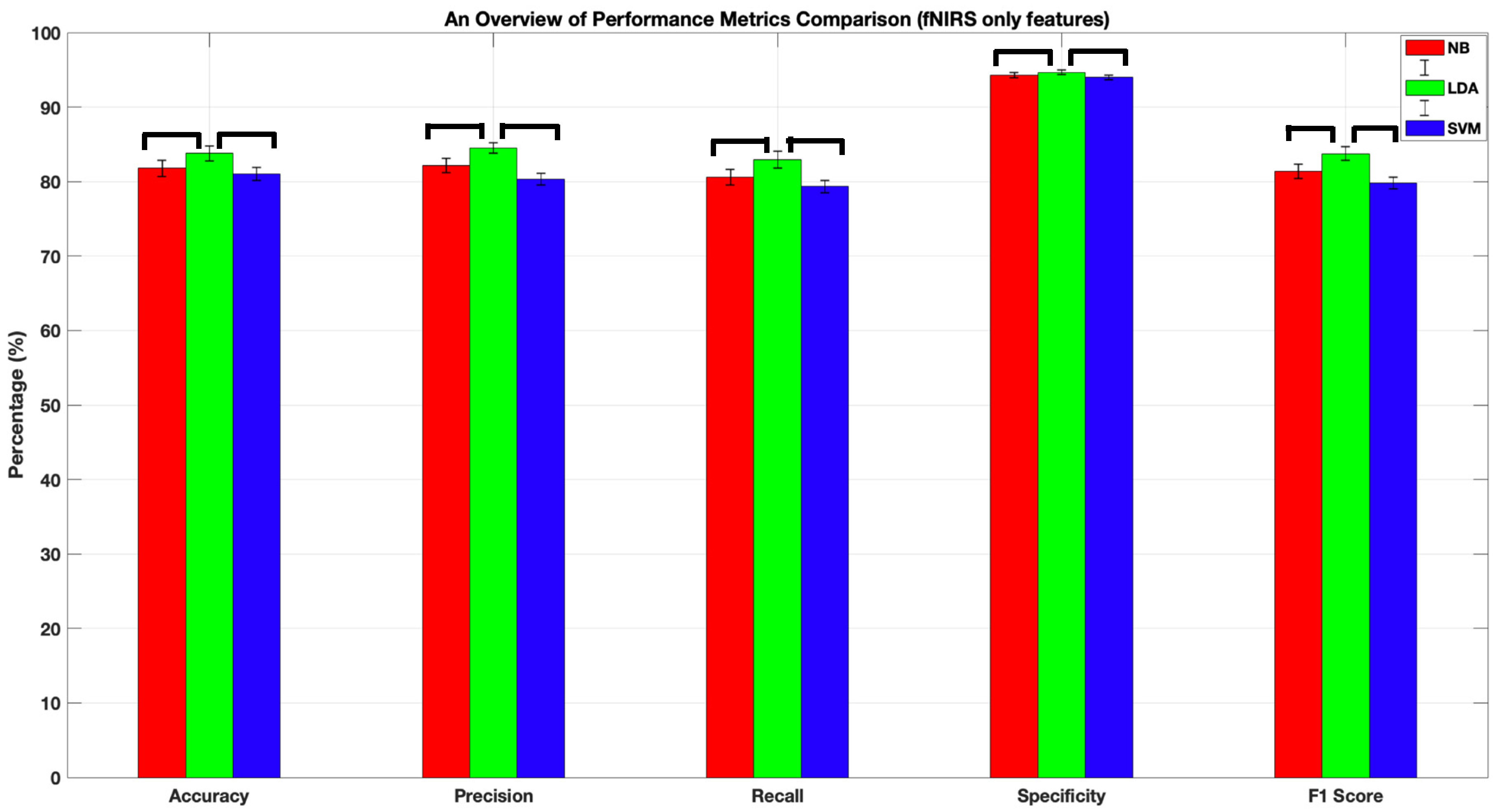
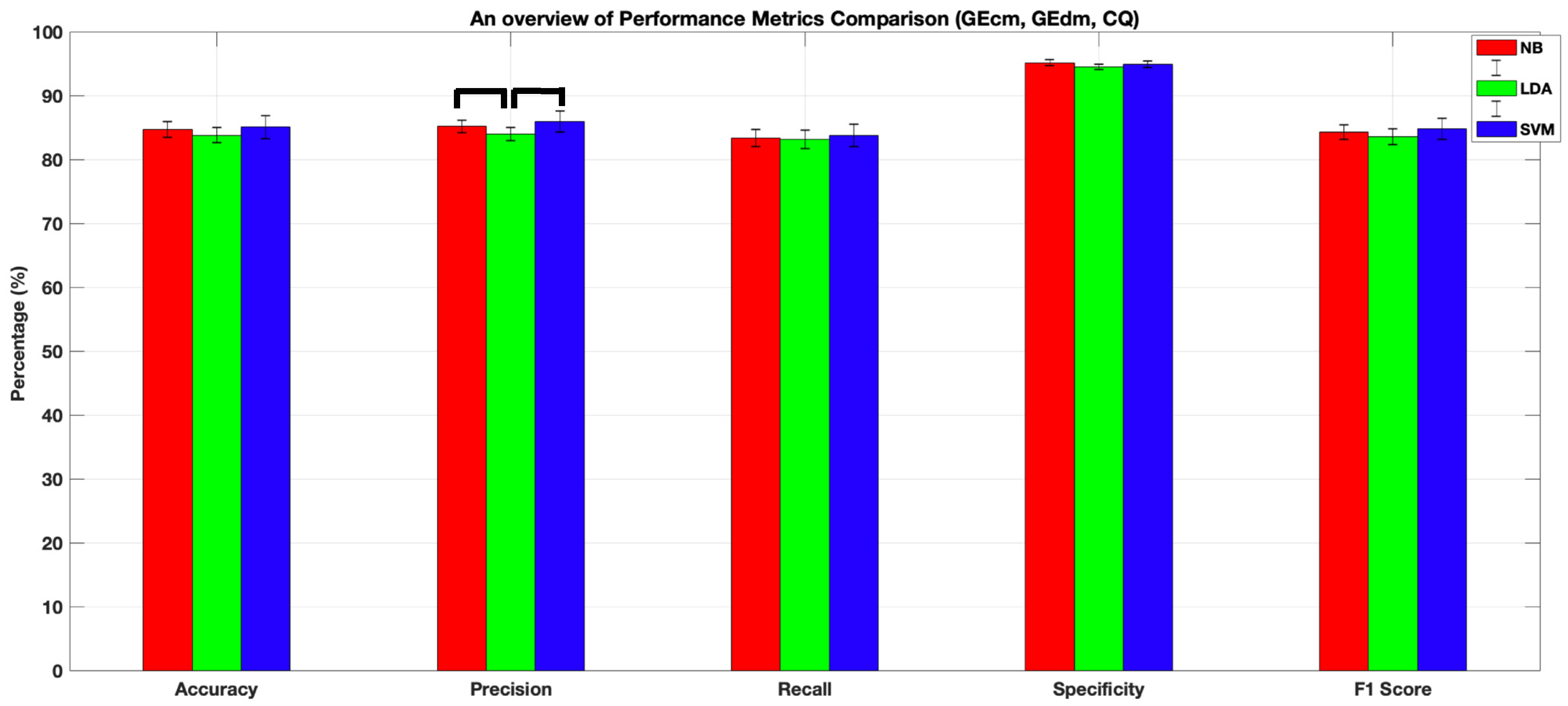
| Method | Accuracy | Precision | Recall | Specificity | F1-Score |
|---|---|---|---|---|---|
| NB | 81.77 ± 1.06 | 82.1 ± 1 | 81 ± 0.01 | 94 ± 0.004 | 81 ± 1 |
| LDA | 83.8 ± 1 | 85 ± 0.01 | 83 ± 0.01 | 95 ± 0.01 | 84 ± 0.01 |
| SVM | 81 ± 0.84 | 80 ± 0.01 | 79 ± 0.01 | 94 ± 0.003 | 80 ± 0.008 |
| Method | Accuracy | Precision | Recall | Specificity | F1-Score |
|---|---|---|---|---|---|
| NB | 84.68±1.3 | 85±0.01 | 83±0.01 | 95±0.01 | 84±0.01 |
| LDA | 83.8 ± 1.6 | 84 ± 1.1 | 83 ± 1.4 | 94 ± 0.04 | 84 ± 1.2 |
| SVM | 85±1.77 | 86±1.6 | 84±1.7 | 95±0.5 | 85±1.7 |
| Author/s (Year) | Sample Size | Classifier(s) | Number of Classes | Features | Mean Accuracy (%) |
|---|---|---|---|---|---|
| Sen et al. (2017) [74] | 16 OCD, 13 HC | SVM | 2 | Resting state network features derived from fMRI data | 80 |
| Chong et al. (2016) [75] | 58 MIG, 50 HC | Quadratic Discriminate Analysis | 2 | Resting state network features derived from fMRI data | 86 |
| Yang et al. (2018) [76] | 21 MIG without aura, 15 MIG with aura, 28 HC | Convolutional Neural Networks | 2 and 3 | Resting state network features derived from fMRI data | 85–99 (2 class), 87(3 class) |
| Hernandez et al. (2014) [77] | 15 HC, 20 MIG, 19 Medication Abuse | SVM | 2 | Graph theoretical features derived from fMRI data | 87 |
| Yu et al.(2013) [59] | 32 SCZ, 19 MDD, 38 HC | SVM | 3 | Resting state network features derived from fMRI data | 81 |
| Kawasaki et al.(2007) [78] | 30 SCZ, 30 HC | Multivariate Linear Model | 2 | Voxel based morphometry features extracted from MRI data | 80 |
| Yassin et al. (2020) [79] | 64 SCZ, 36 ASD, 106 HC | Logistic Regression | 3 | Cortical thickness and subcortical volume features derived from MRI data | 69 |
| Pardo et al. (2006) [80] | 10 SCZ, 10 BP, 8 HC | LDA | 3 | Neuropsychiatric test scores and structural metrics derived from MRI data | 96 |
| Present work | 20 MIG, 26 OCD, 21 SCZ, 13 HC | LDA, SVM, NB | 4 | Cognitive quotient and Global Efficiency metrics derived from fNIRS data | 84.7 (LDA), 83.8(NB), 85 (SVM) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erdoğan, S.B.; Yükselen, G. Four-Class Classification of Neuropsychiatric Disorders by Use of Functional Near-Infrared Spectroscopy Derived Biomarkers. Sensors 2022, 22, 5407. https://doi.org/10.3390/s22145407
Erdoğan SB, Yükselen G. Four-Class Classification of Neuropsychiatric Disorders by Use of Functional Near-Infrared Spectroscopy Derived Biomarkers. Sensors. 2022; 22(14):5407. https://doi.org/10.3390/s22145407
Chicago/Turabian StyleErdoğan, Sinem Burcu, and Gülnaz Yükselen. 2022. "Four-Class Classification of Neuropsychiatric Disorders by Use of Functional Near-Infrared Spectroscopy Derived Biomarkers" Sensors 22, no. 14: 5407. https://doi.org/10.3390/s22145407
APA StyleErdoğan, S. B., & Yükselen, G. (2022). Four-Class Classification of Neuropsychiatric Disorders by Use of Functional Near-Infrared Spectroscopy Derived Biomarkers. Sensors, 22(14), 5407. https://doi.org/10.3390/s22145407






