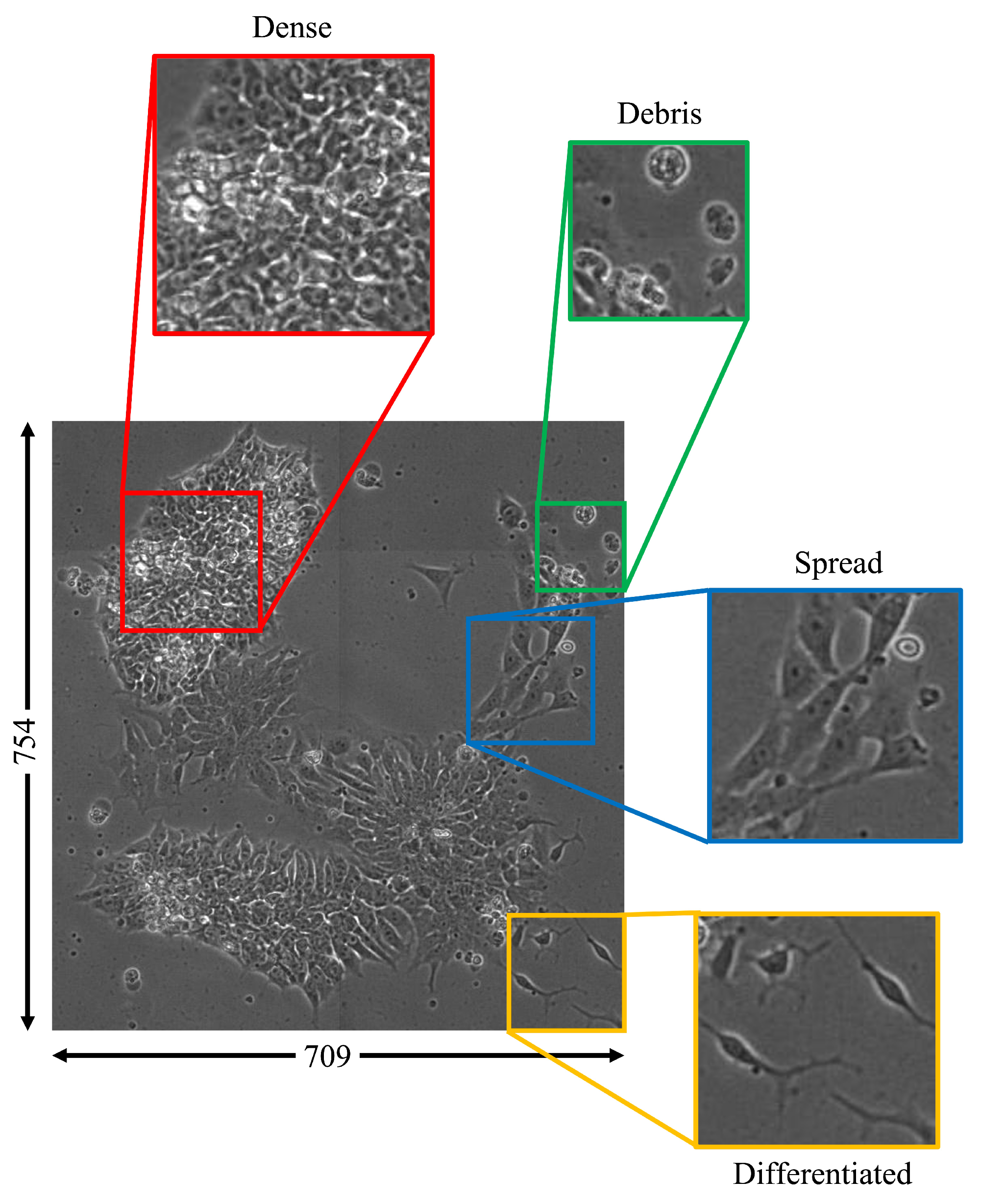
Figure 1.
Image examples for four morphological classes observable in a single cell colony (debris: green; dense: red; spread: blue; differentiated: yellow). Throughout the differentiation process, various proportions of each class can be found in cell colonies with contiguous cell boundaries. Classification of these multiclass images can be performed using image patches.
Figure 1.
Image examples for four morphological classes observable in a single cell colony (debris: green; dense: red; spread: blue; differentiated: yellow). Throughout the differentiation process, various proportions of each class can be found in cell colonies with contiguous cell boundaries. Classification of these multiclass images can be performed using image patches.
Figure 2.
Data preprocessing and classification schematic. The binary map of colony locations is used to segment colonies from the original image, which are then sorted by hand during ground-truth generation (left). Patches from the resulting dataset are used to train the GAN. Generated images are added to balance the dataset for the temporal CNN classification scheme (right), during which images are sorted into their individual classes through multiple hierarchical stages.
Figure 2.
Data preprocessing and classification schematic. The binary map of colony locations is used to segment colonies from the original image, which are then sorted by hand during ground-truth generation (left). Patches from the resulting dataset are used to train the GAN. Generated images are added to balance the dataset for the temporal CNN classification scheme (right), during which images are sorted into their individual classes through multiple hierarchical stages.
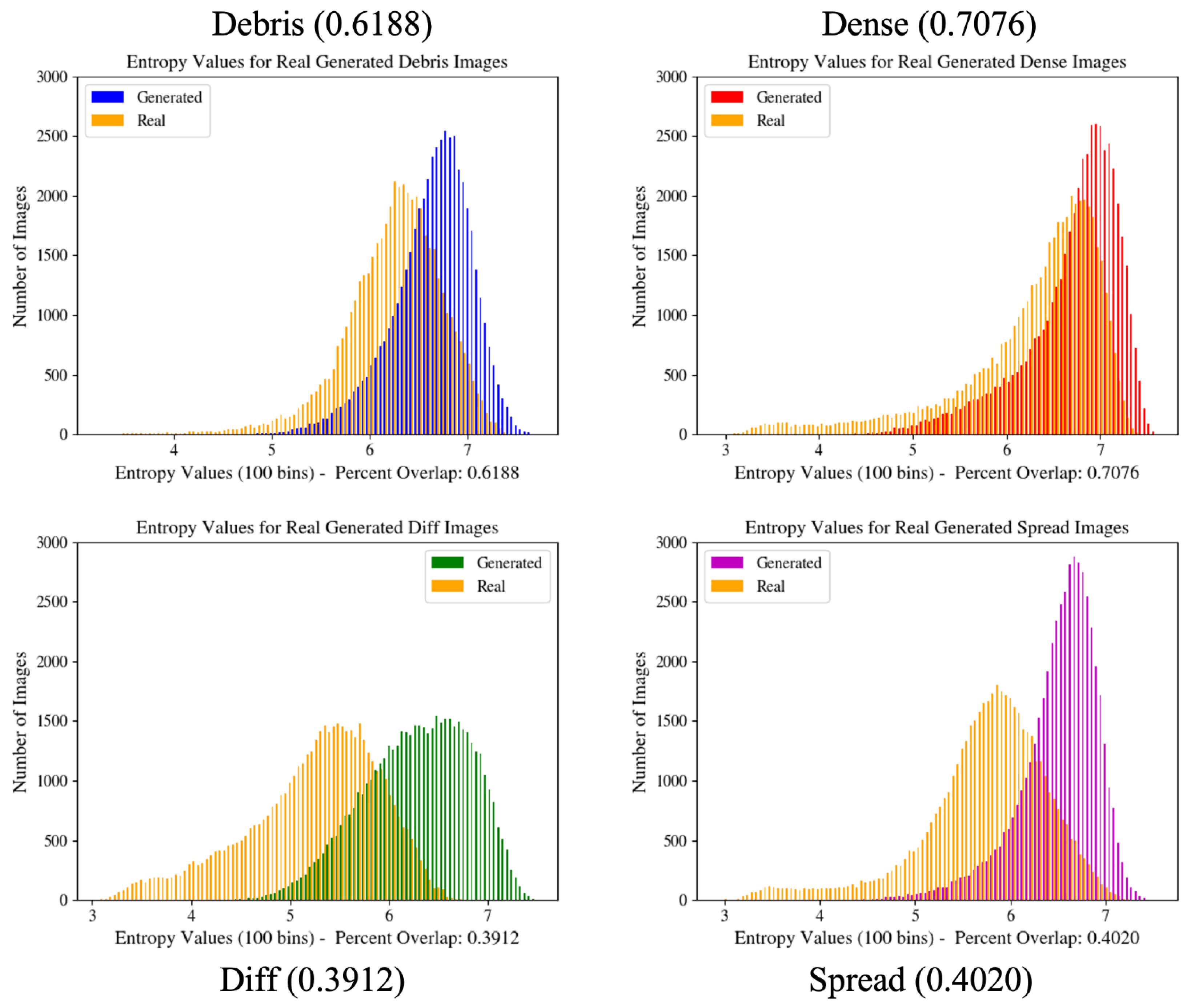
Figure 3.
Image entropy distribution histograms for GAN configurations. These graphs provide a quantitative measure of the overall generated image distribution in relation to the real image distribution and are used during GAN training to improve network learning. Values in parentheses indicate the percent overlap of the two graphs shown in the figure.
Figure 3.
Image entropy distribution histograms for GAN configurations. These graphs provide a quantitative measure of the overall generated image distribution in relation to the real image distribution and are used during GAN training to improve network learning. Values in parentheses indicate the percent overlap of the two graphs shown in the figure.
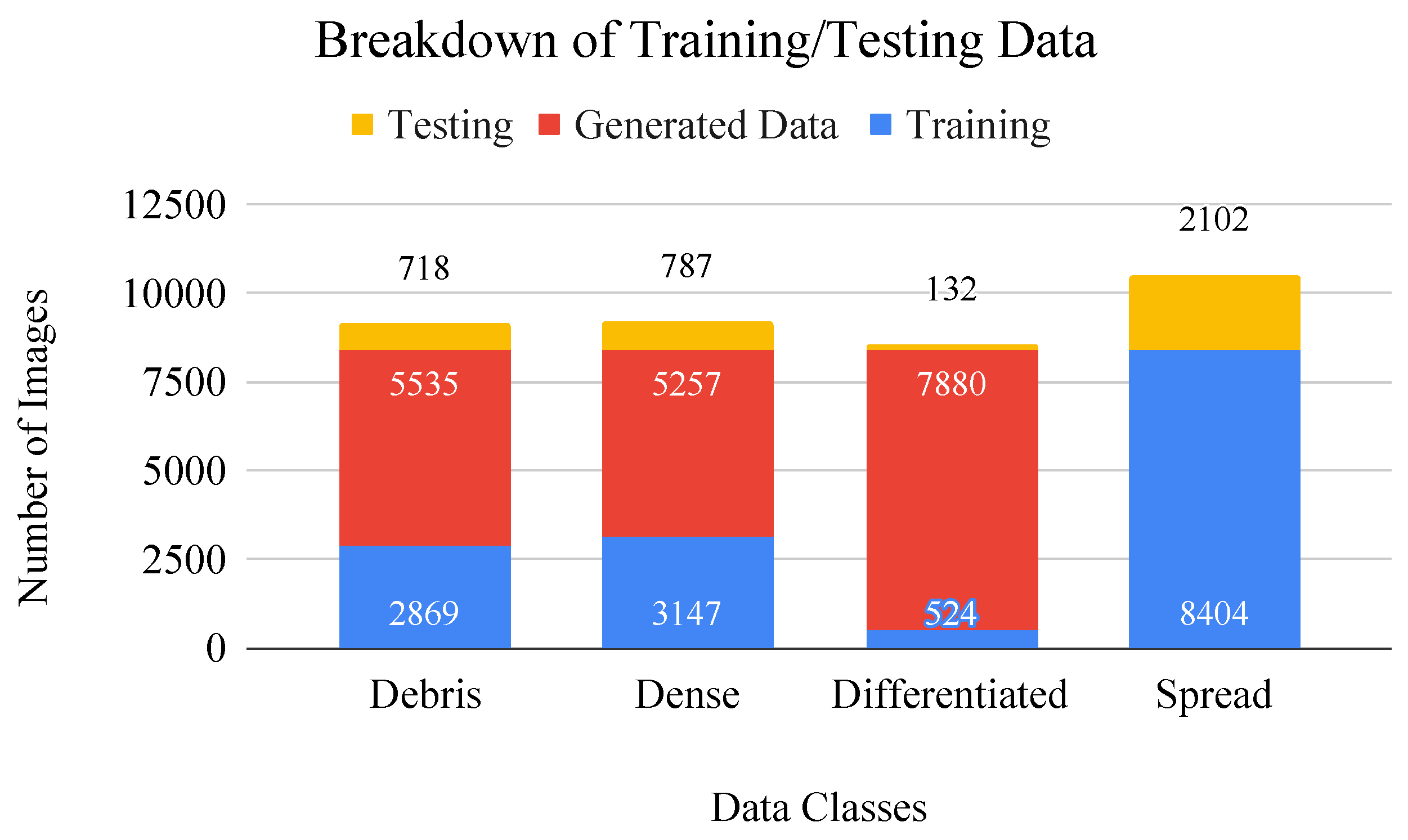
Figure 4.
Bar graph of data breakdown including values for training/testing (blue/yellow) split. Generated images (red) are added to the dataset to make up for class imbalances during CNN training.
Figure 4.
Bar graph of data breakdown including values for training/testing (blue/yellow) split. Generated images (red) are added to the dataset to make up for class imbalances during CNN training.
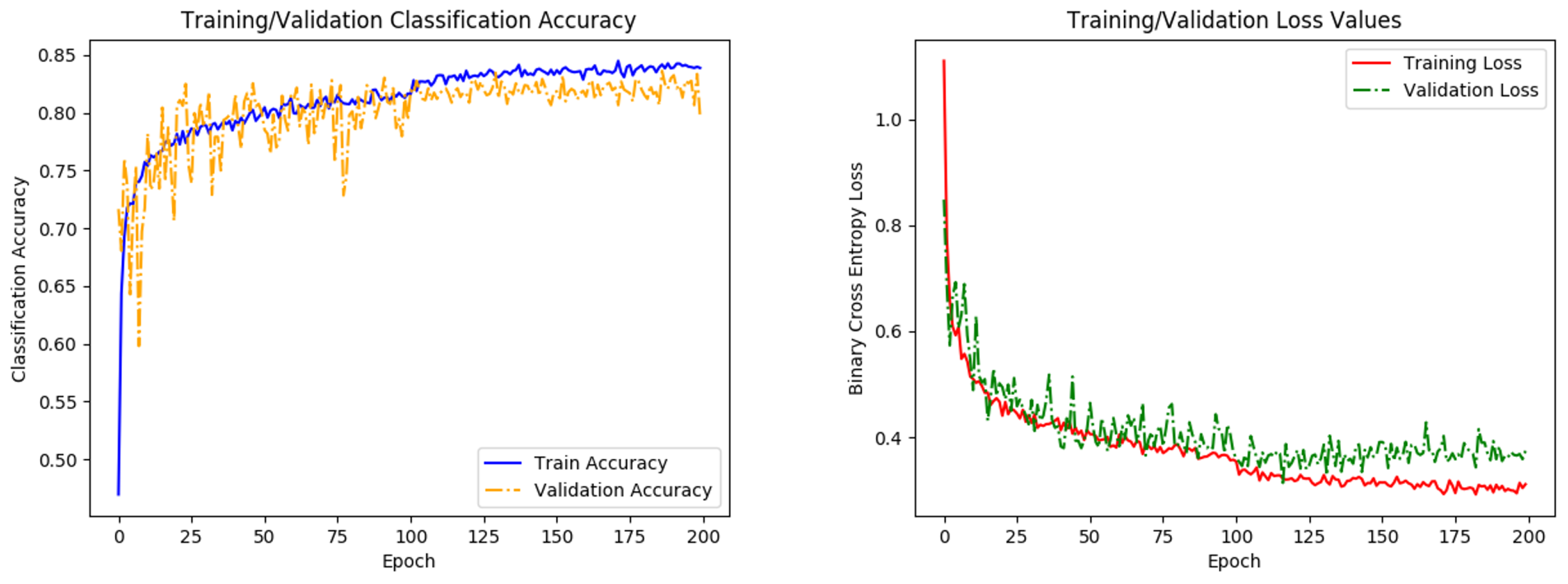
Figure 5.
Graphs of network accuracy (left) and cross-entropy loss (right) for training and validation datasets. A small respective bump/dip in accuracy/loss is observed at 100 epochs, where the learning rate parameter is reduced. Training levels out before the 200 epochs, indicating that the network has finished learning.
Figure 5.
Graphs of network accuracy (left) and cross-entropy loss (right) for training and validation datasets. A small respective bump/dip in accuracy/loss is observed at 100 epochs, where the learning rate parameter is reduced. Training levels out before the 200 epochs, indicating that the network has finished learning.
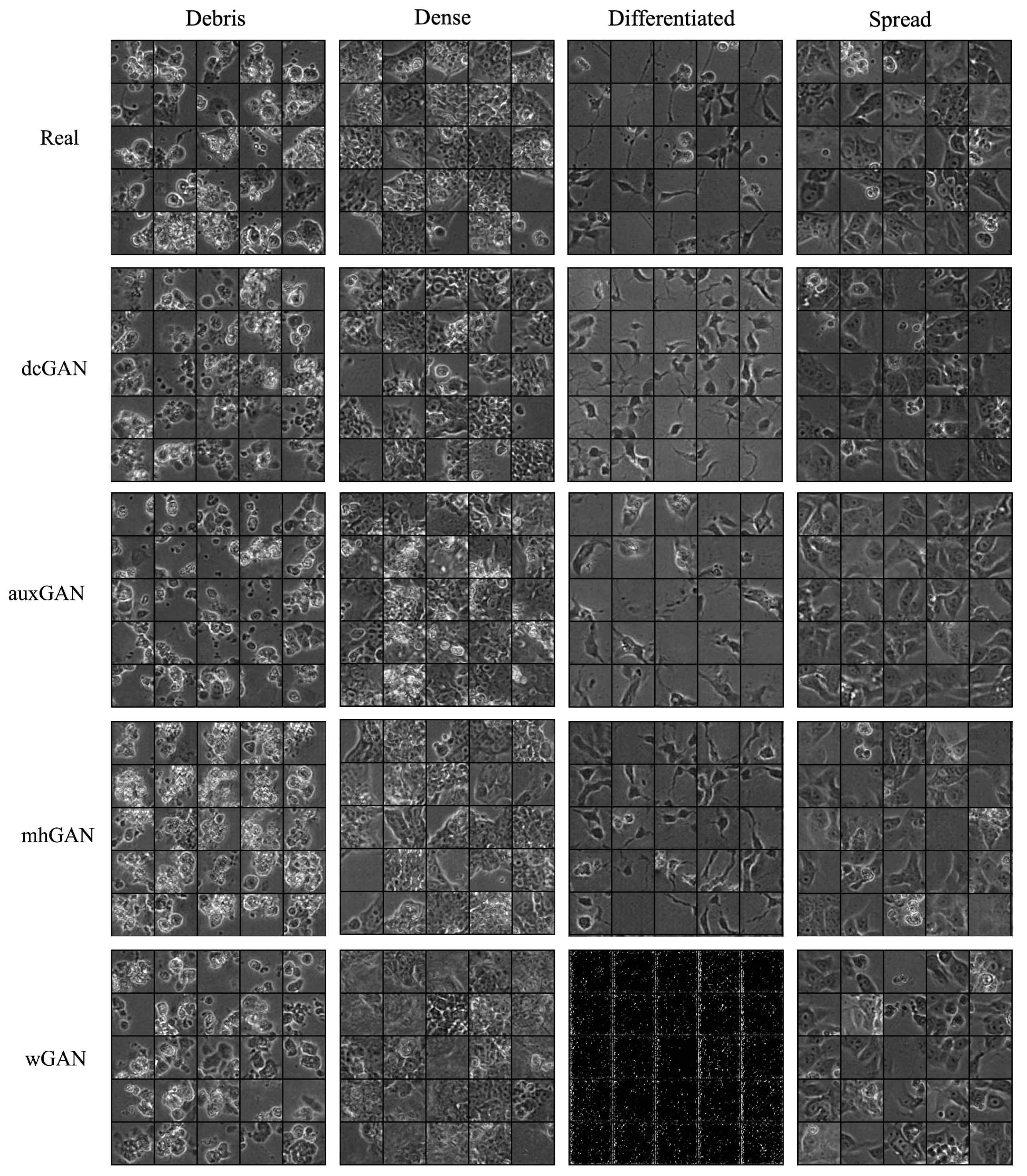
Figure 6.
Image patch samples for real and generated images. A comparison of classwise image features displays generally realistic image features indicative of morphological class. However, visual appearance of images provides only a qualitative measure of image quality, where quantitative metrics are necessary to determine image realness.
Figure 6.
Image patch samples for real and generated images. A comparison of classwise image features displays generally realistic image features indicative of morphological class. However, visual appearance of images provides only a qualitative measure of image quality, where quantitative metrics are necessary to determine image realness.
Figure 7.
Normalized generator inception score (red) and FID (blue) per training epoch with example images at various intervals for the spread class. Graphs include accompanying trend line. Training epoch numbers are marked by a white ’E’ in the bottom of each image. Agreement between inception score and FID can be seen in terms of their relative minimum and maximum values versus training epoch.
Figure 7.
Normalized generator inception score (red) and FID (blue) per training epoch with example images at various intervals for the spread class. Graphs include accompanying trend line. Training epoch numbers are marked by a white ’E’ in the bottom of each image. Agreement between inception score and FID can be seen in terms of their relative minimum and maximum values versus training epoch.
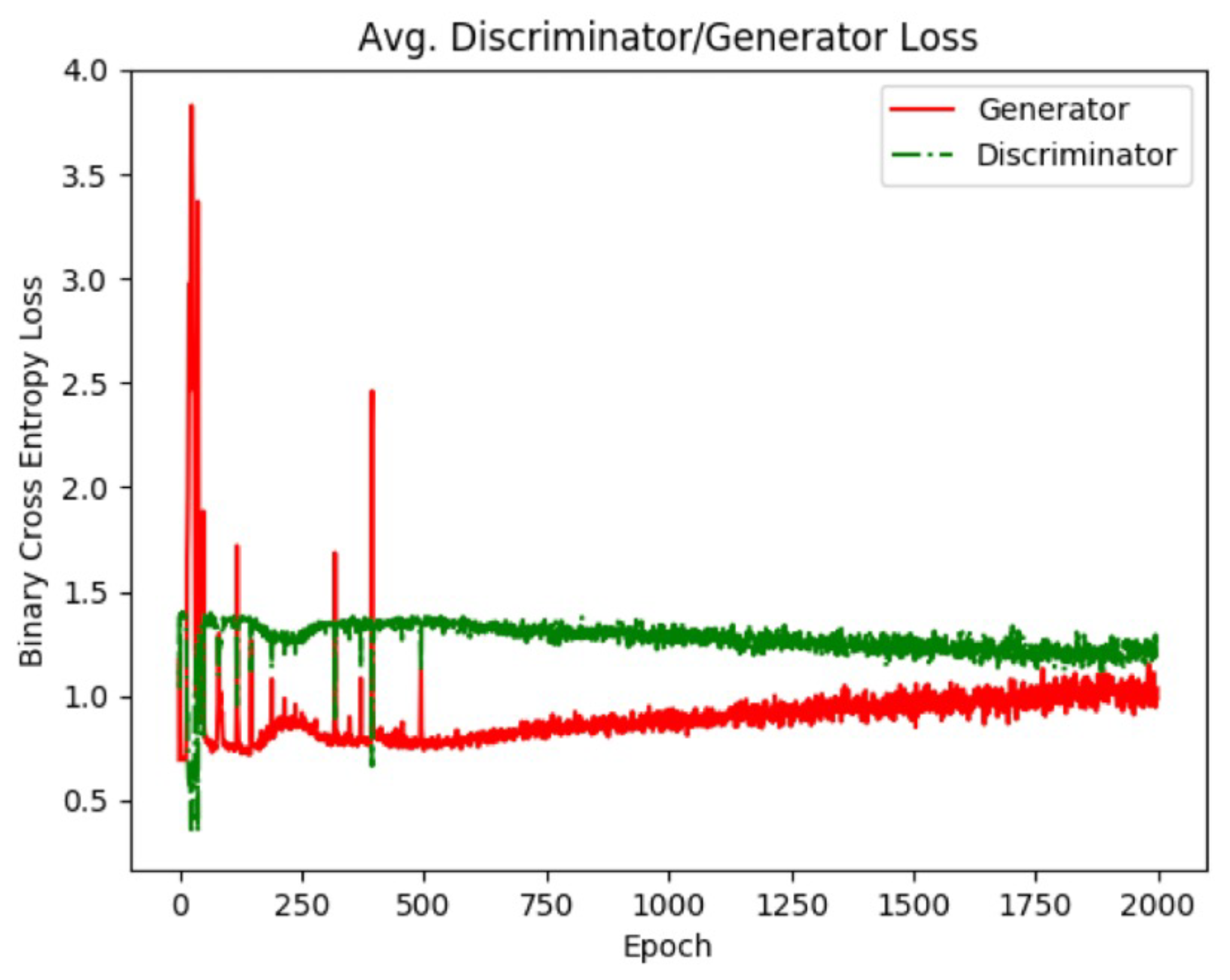
Figure 8.
Generator and discriminator loss values for the dense class. As training progresses, the GAN reaches an equilibrium which is when training is considered finished. Using individual GAN models for each image class allows the GAN to be trained for different amounts of time based on the image class.
Figure 8.
Generator and discriminator loss values for the dense class. As training progresses, the GAN reaches an equilibrium which is when training is considered finished. Using individual GAN models for each image class allows the GAN to be trained for different amounts of time based on the image class.
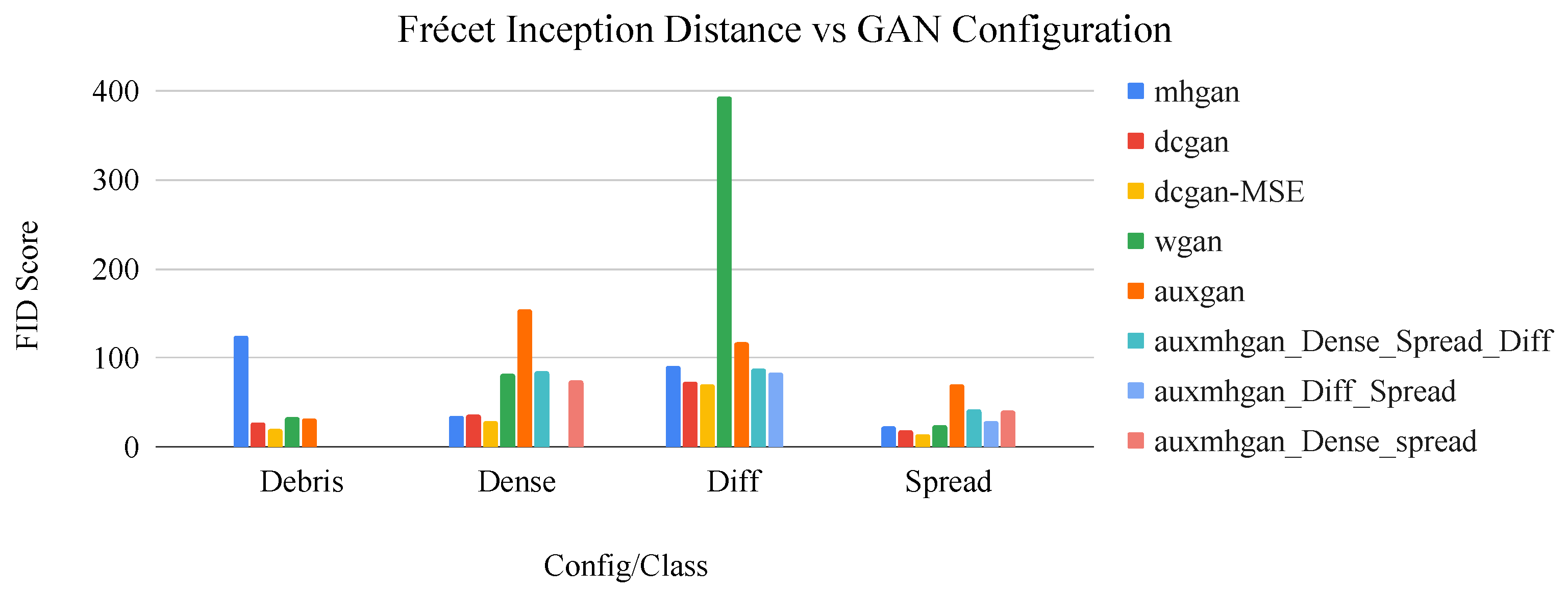
Figure 9.
Bar graph of network configuration vs. FID score by image class. The dcGAN+MSE configuration consistently displays the best performance in terms of this metric.
Figure 9.
Bar graph of network configuration vs. FID score by image class. The dcGAN+MSE configuration consistently displays the best performance in terms of this metric.
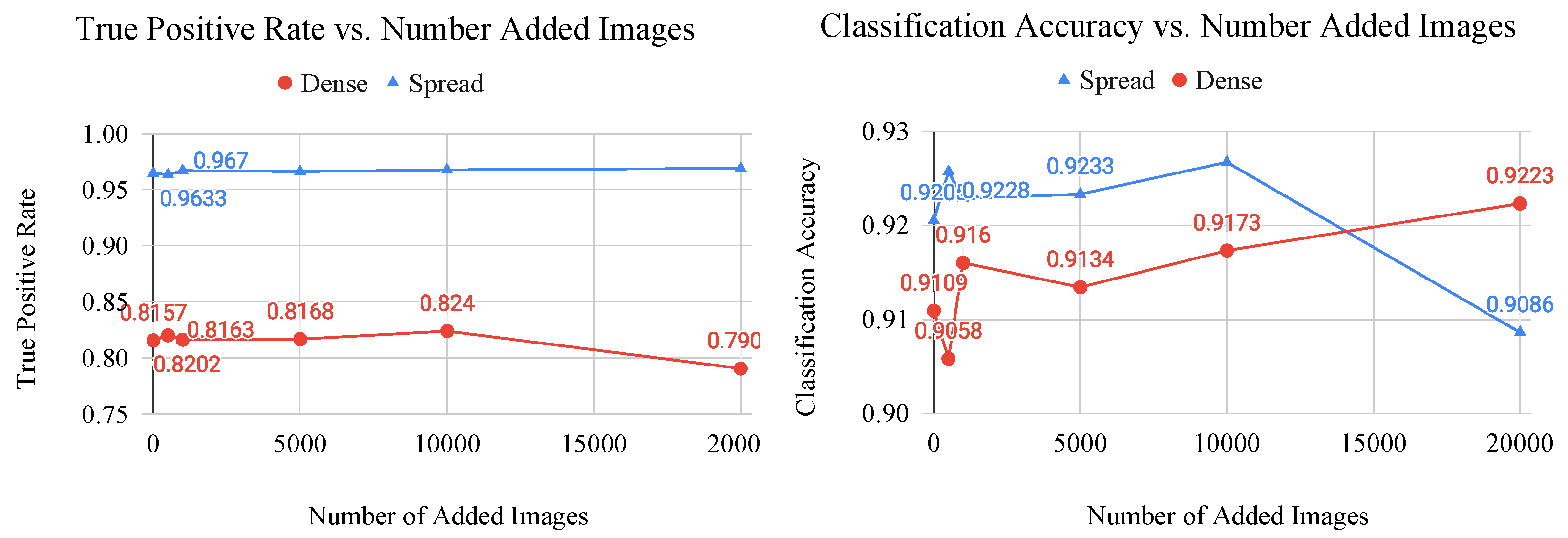
Figure 10.
Graphs of classification metric (left: true postitive rate, right: classification accuracy) vs. number of added generated images for the dense and spread classes. These graphs are used to determine the saturation point of a CNN, which is where the generated images no longer provide useful features to the model.
Figure 10.
Graphs of classification metric (left: true postitive rate, right: classification accuracy) vs. number of added generated images for the dense and spread classes. These graphs are used to determine the saturation point of a CNN, which is where the generated images no longer provide useful features to the model.
Table 1.
Morphological class descriptions and corresponding biological implications.
Table 1.
Morphological class descriptions and corresponding biological implications.
| Class | Morphological Description | Implication |
|---|
| Debris | Individual cells or aggregates cells showing circular morphology with high intensity white ‘halo’ marking distinct boundaries | Distressed, dead (apoptotic/necrotic) cells that float on top of colony indicating negative response to experimental conditions |
| Dense | Homogeneous aggregates of small cells with indiscernible cell boundaries, no clear nucleus | Induced pluripotent stem cell colonies that maintain undifferentiated status under current conditions |
| Spread | Homogeneous aggregates of large cells with discernible cell boundaries, clear nuclei, large protrusions | Down stream lineage intermediates or progenitor cells |
| Differentiated | Individual cells or spaced out aggregates of cells with distinct, dark cell bodies, high-intensity white boundaries, and dark axon like protrusions. | Differentiated neurons or neuronlike downstream lineages |
Table 2.
GAN Network Architecture: The input to the GAN generator (a) is a latent vector of length 100 multiplied which is processed through multiple convolutional (C2d) and upsampling (Up) layers. The output of the generator is a hyperbolic tangent (Tanh). The output of the discriminator (b) goes to a fully connected (FC) layer followed by a sigmoid function (Sig).
Table 2.
GAN Network Architecture: The input to the GAN generator (a) is a latent vector of length 100 multiplied which is processed through multiple convolutional (C2d) and upsampling (Up) layers. The output of the generator is a hyperbolic tangent (Tanh). The output of the discriminator (b) goes to a fully connected (FC) layer followed by a sigmoid function (Sig).
| Generator | Discriminator |
|---|
| Module | Size | Maps | Module | Size | Maps |
| Linear | | 1/512 | C2d | 64/32 | 1/64 |
| Up | 16/36 | 512/512 | C2d | 32/16 | 64/128 |
| C2d | 36/36 | 512/512 | C2d | 16/8 | 128/256 |
| Up | 36/64 | 512/512 | C2d | 8/4 | 256/512 |
| C2d | 64/64 | 512/256 | FC | 8192 | 512 |
| C2d | 64/64 | 256/1 | Sig(·) | 1/1 | -/- |
| Tanh(·) | 64/64 | 1/1 | | | |
Table 3.
GAN training hyperparameters were empirically determined to optimize network training efficiency.
Table 3.
GAN training hyperparameters were empirically determined to optimize network training efficiency.
| Training Hyperparameters |
|---|
| Parameter | Value |
| Learning Rate-Adam | 0.002 |
| 1—Adam | 0.5 |
| 2—Adam | 0.999 |
| Max feature maps—Discriminator | 512 |
| Max feature maps—Generator | 512 |
Table 4.
Overlap percentage for image entropy histograms across five trials, for which the entropy values of 50,000 random real and generated image patches each are plotted and the overlap is calculated. Variation in values is caused by randomly generated image patches that contain variability within the image. These values can be used to determine how well the generator has been able to model image features and can be correlated with the performance of downstream dataset augmentation tasks.
Table 4.
Overlap percentage for image entropy histograms across five trials, for which the entropy values of 50,000 random real and generated image patches each are plotted and the overlap is calculated. Variation in values is caused by randomly generated image patches that contain variability within the image. These values can be used to determine how well the generator has been able to model image features and can be correlated with the performance of downstream dataset augmentation tasks.
| Image Class | Overlap Percentage—Mean (std.) |
|---|
| Debris | 0.6182 (0.0026) |
| Dense | 0.7066 (0.0033) |
| Diff | 0.3936 (0.0018) |
| Spread | 0.3999 (0.0011) |
Table 5.
Data breakdown for four morphological classes. Class imbalances observed here are a factor of the natural growth and differentiation cycle of the cells.
Table 5.
Data breakdown for four morphological classes. Class imbalances observed here are a factor of the natural growth and differentiation cycle of the cells.
| Class | # Samples |
|---|
| Debris | 3587 |
| Dense | 3934 |
| Diff | 656 |
| Spread | 10,506 |
| Total | 18,683 |
Table 6.
Epoch values and corresponding inception scores at which the GAN generator is determined to be optimally trained, based on the plot of inception score vs. training epoch. Each GAN is trained on an individual class, and, therefore, requires a different level of training based on the number of images in each class, as well as complexity of features and other variables. The optimal GAN is used to generate images for each class to be used for dataset augmentation.
Table 6.
Epoch values and corresponding inception scores at which the GAN generator is determined to be optimally trained, based on the plot of inception score vs. training epoch. Each GAN is trained on an individual class, and, therefore, requires a different level of training based on the number of images in each class, as well as complexity of features and other variables. The optimal GAN is used to generate images for each class to be used for dataset augmentation.
| Image Class | Optimal Generator Epoch | Inception Score |
|---|
| Debris | 116 | 2.60 |
| Dense | 444 | 2.32 |
| Diff | 225 | 2.38 |
| Spread | 136 | 2.57 |
Table 7.
Frécet inception distance (FID) scores for each GAN configuration by image class. An x indicates where the GAN was not trained to generate the specific image class.
Table 7.
Frécet inception distance (FID) scores for each GAN configuration by image class. An x indicates where the GAN was not trained to generate the specific image class.
| Config./Class/FID | Debris | Dense | Diff. | Spread | Average |
|---|
| dcGAN | 27.73 | 36.32 | 72.77 | 18.51 | 38.83 |
| dcGAN + MSE | 19.5 | 29.5 | 70.7 | 13.67 | 33.34 |
| wGAN | 33.85 | 81.45 | 393.94 | 24.05 | 133.32 |
| auxGAN | 31.62 | 155.13 | 117.92 | 69.86 | 93.63 |
| mhGAN | 125.22 | 35.03 | 90.63 | 23.05 | 68.48 |
| aux-mhGAN (Dense, Diff, Spread) | x | 84.93 | 88.7 | 41.53 | 71.72 |
| aux-mhGAN (Dense, Spread) | x | x | 83.37 | 29.55 | 57.54 |
| aux-mhGAN (Diff, Spread) | x | 74.0 | x | 41.05 | 56.46 |
Table 8.
Classwise true positive rate for four-class CNN with and without dataset balancing. Several variations of balancing are used here, the most effective of which is supplementation using generated images in line with the temporal training configuration proposed in this paper. The p-value indicated by the * is calculated using Student’s t-test.
Table 8.
Classwise true positive rate for four-class CNN with and without dataset balancing. Several variations of balancing are used here, the most effective of which is supplementation using generated images in line with the temporal training configuration proposed in this paper. The p-value indicated by the * is calculated using Student’s t-test.
| Configuration/Class/TPR (Std.) | Debris | Dense | Diff. | Spread | Average |
|---|
| Unbalanced | 0.9141 (0.0144) | 0.8093 (0.0211) | 0.8807 (0.0342) | 0.9144 (0.0093) * | 0.8789 |
| Sampler Balanced | 0.8570 (0.0184) | 0.9300 (0.0189) | 0.9274 (0.0219) | 0.8410 (0.0073) | 0.8888 |
| Weight Balanced | 0.9030 (0.0312) | 0.8065 (0.0249) | 0.8439 (0.0715) | 0.9300 (0.0290) | 0.8708 |
| Generator Balanced | 0.9105 (0.0206) | 0.7940 (0.0116) | 0.8999 (0.0247) | 0.9172 (0.0124) | 0.8804 |
| Temporally Balanced | 0.9277 (0.0148) | 0.8157 (0.0142) | 0.8856 (0.0289) | 0.9646 (0.0040) * | 0.8984 |
Table 9.
Hierarchical tier 1: true positive rate for temporal combination of viable cell classes vs. debris cells. This stage acts as a filtration step to remove unviable and unhealthy colony areas.
Table 9.
Hierarchical tier 1: true positive rate for temporal combination of viable cell classes vs. debris cells. This stage acts as a filtration step to remove unviable and unhealthy colony areas.
| Configuration/Class/TPR (std.) | Debris | Dense/Diff./Spread | Average |
|---|
| Unbalanced | 0.9145 (0.0097) | 0.9570 (0.0058) | 0.9357 |
| Generator Balanced | 0.9277 (0.0148) | 0.9545 (0.0053) | 0.9411 |
Table 10.
Hierarchical tier 2: true positive rate for separation of dense/spread classes from differentiated. This tier serves to remove the mature cell colonies from the early and intermediate stage classes. The dense/spread classes have the highest level of misclassification, due to their relative proximity in terms of the downstream differentiation process, and subsequent similarity in texture features.
Table 10.
Hierarchical tier 2: true positive rate for separation of dense/spread classes from differentiated. This tier serves to remove the mature cell colonies from the early and intermediate stage classes. The dense/spread classes have the highest level of misclassification, due to their relative proximity in terms of the downstream differentiation process, and subsequent similarity in texture features.
| Configuration/Class/TPR (std.) | Diff. | Dense/Spread | Average |
|---|
| Unbalanced | 0.8792 (0.0255) | 0.9941 (0.0007) | 0.9367 |
| Generator Balanced | 0.8856 (0.0289) | 0.9935 (0.0007) | 0.9396 |
Table 11.
Hierarchical tier 3: true positive rate for classification of dense vs. spread. The balancing of the dense class, using generated images, in relation to the Spread class shows slight improvement over the unbalanced configuration, and marked improvement over the four-class configuration.
Table 11.
Hierarchical tier 3: true positive rate for classification of dense vs. spread. The balancing of the dense class, using generated images, in relation to the Spread class shows slight improvement over the unbalanced configuration, and marked improvement over the four-class configuration.
| Configuration/Class/TPR (std.) | Dense | Spread | Average |
|---|
| Unbalanced | 0.8187 (0.0140) | 0.9624 (0.0035) | 0.8906 |
| Generator Balanced | 0.8157 (0.0142) | 0.9646 (0.0040) | 0.8902 |
Table 12.
F1-score for unbalanced and generator-balanced temporal training configurations show an overall improvement for the balanced configuration on average, as well as statistically significant increases in the dense and spread classes. The p-value indicated by the * is calculated using Student’s t-test.
Table 12.
F1-score for unbalanced and generator-balanced temporal training configurations show an overall improvement for the balanced configuration on average, as well as statistically significant increases in the dense and spread classes. The p-value indicated by the * is calculated using Student’s t-test.
| Configuration/Class/F1 (std.) | Debris | Dense | Diff. | Spread | Average |
|---|
| Unbalanced | 0.8732 (0.0059) | 0.8430 (0.0082) * | 0.8580 (0.0164) | 0.9119 (0.0036) ** | 0.8715 |
| Generator-Balanced | 0.8599 (0.0099) | 0.8599 (0.0050) * | 0.8714 (0.0009) | 0.9433 (0.0030) ** | 0.8836 |