Electrochemical DNA Biosensor That Detects Early Celiac Disease Autoantibodies
Abstract
1. Introduction
2. Materials and Methods
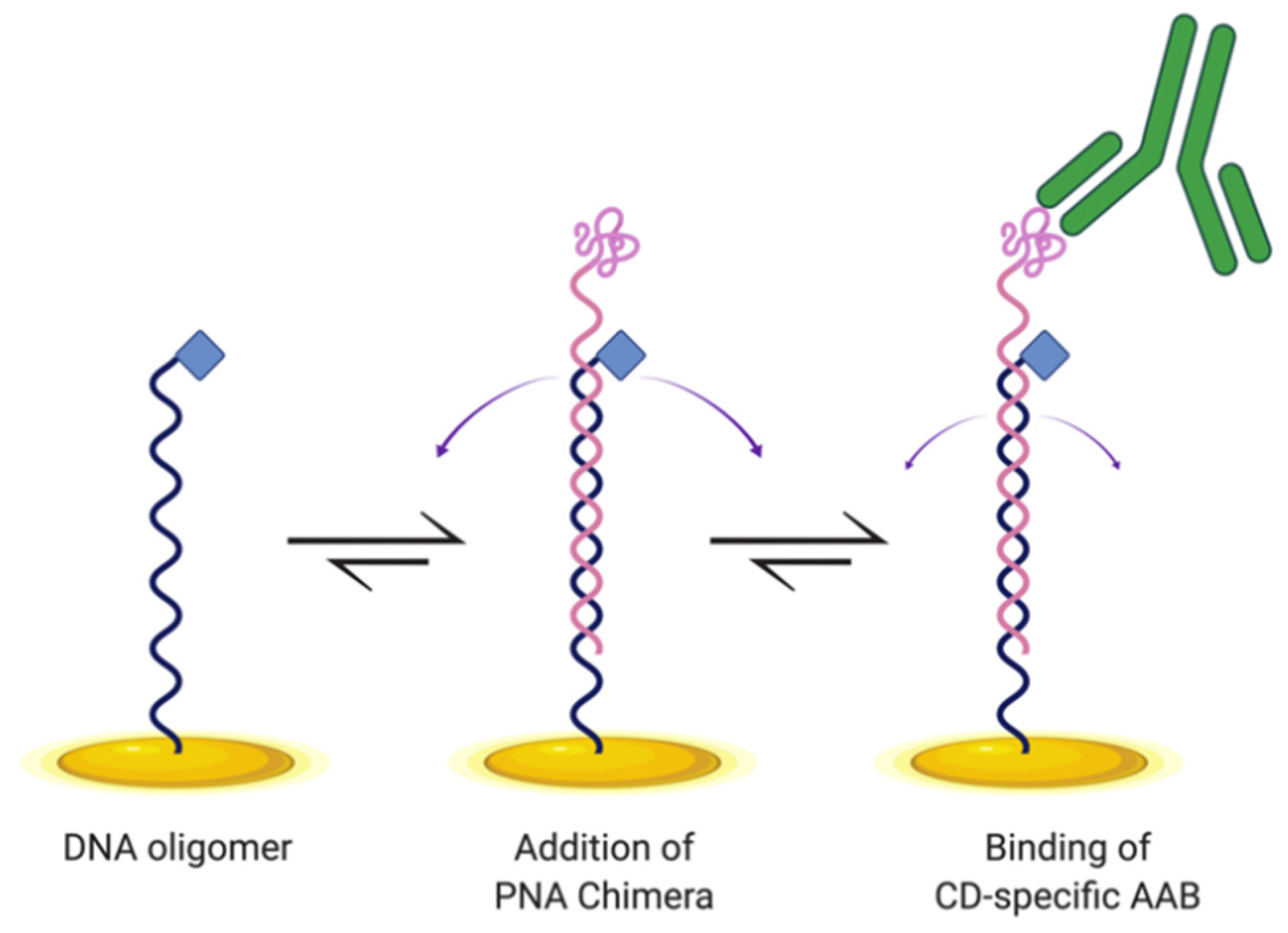
2.1. Biosensor Design and Preparation
2.2. Electrochemical and Control Parameters
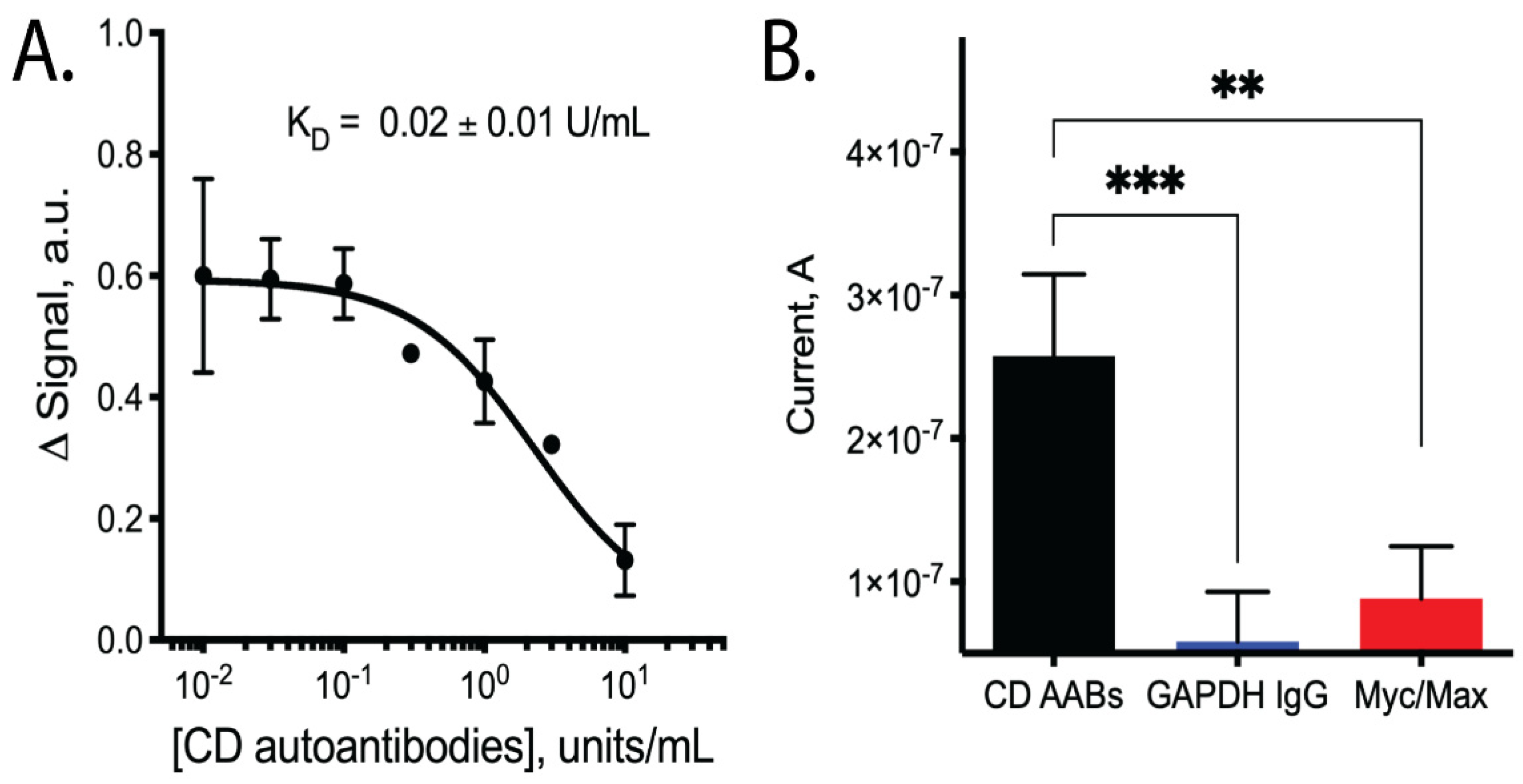
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rubio-Tapia, A.; Kyle, R.A.; Kaplan, E.L.; Johnson, D.R.; Page, W.; Erdtmann, F.; Brantner, T.L.; Kim, W.R.; Phelps, T.K.; Lahr, B.D.; et al. Increased prevalence and mortality in undiagnosed celiac disease. Gastroenterology 2009, 137, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Choung, R.S.; Unalp-Arida, A.; Ruhl, C.E.; Brantner, T.L.; Everhart, J.E.; Murray, J.A. Less hidden celiac disease but increased gluten avoidance without a diagnosis in the united states: Findings from the national health and nutrition examination surveys from 2009 to 2014. Mayo Clin. Proc. 2017, 92, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Caio, G.; Volta, U.; Sapone, A.; Leffler, D.A.; De Giorgio, R.; Catassi, C.; Fasano, A. Celiac disease: A comprehensive current review. BMC Med. 2019, 17, 142. [Google Scholar] [CrossRef]
- Oxentenko, A.S.; Rubio-Tapia, A. Celiac disease. Mayo Clin. Proc. 2019, 94, 2556–2571. [Google Scholar] [CrossRef]
- Cichewicz, A.B.; Mearns, E.S.; Taylor, A.; Boulanger, T.; Gerber, M.; Leffler, D.A.; Drahos, J.; Sanders, D.S.; Craig, K.J.T.; Lebwohl, B. Diagnosis and treatment patterns in celiac disease. Dig. Dis. Sci. 2019, 64, 2095–2106. [Google Scholar] [CrossRef] [PubMed]
- Matthias, T.; Neidhöfer, S.; Pfeiffer, S.; Prager, K.; Reuter, S.; Gershwin, M.E. Novel trends in celiac disease. Cell. Mol. Immunol. 2011, 868, 121–125. [Google Scholar] [CrossRef]
- Rashtak, S.; Ettore, M.W.; Homburger, H.A.; Murray, J.A. Comparative usefulness of deamidated gliadin antibodies in the diagnosis of celiac disease. Clin. Gastroenterol. Hepatol. 2008, 6, 426–432. [Google Scholar] [CrossRef]
- Di Pisa, M.; Pascarella, S.; Scrima, M.; Sabatino, G.; Real-Fernández, F.; Chelli, M.; Renzi, D.; Calabrò, A.; D’Ursi, A.M.; Papini, A.M.; et al. Synthetic peptides reproducing tissue transglutaminase–gliadin complex neo-epitopes as probes for antibody detection in celiac disease patients’ sera. J. Med. Chem. 2015, 58, 1390–1399. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Electrochemical biosensors: Towards point-of-care cancer diagnostics. Biosens. Bioelectron. 2006, 21, 1887–1892. [Google Scholar] [CrossRef]
- Yin, Y.; Zhao, X.S. Kinetics and dynamics of DNA hybridization. Acc. Chem. Res. 2011, 44, 1172–1181. [Google Scholar] [CrossRef]
- Bonham, A.J.; Hsieh, K.; Ferguson, B.S.; Valle-Belisle, A.; Ricci, F.; Soh, H.T.; Plaxco, K.W. Quantification of transcription factor binding in cell extracts using an electrochemical, structure-switching biosensor. J. Am. Chem. Soc. 2012, 134, 3346–3348. [Google Scholar] [CrossRef]
- Lubin, A.A.; Plaxco, K.W. Folding-based electrochemical biosensors: The case for responsive nucleic acid architectures. Acc. Chem. Res. 2010, 43, 496–505. [Google Scholar] [CrossRef] [PubMed]
- Santos-Cancel, M.; Simpson, L.W.; Leach, J.B.; White, R.J. Direct, real-time detection of adenosine triphosphate release from astrocytes in three-dimensional culture using an integrated electrochemical aptamer-based sensor. ACS Chem. Neurosci. 2019, 10, 2070–2079. [Google Scholar] [CrossRef] [PubMed]
- White, R.J.; Kallewaard, H.M.; Hsieh, K.; Patterson, A.S.; Kasehagen, J.B.; Cash, K.J.; Uzawa, T.; Soh, H.T.; Plaxco, K.W. A wash-free, electrochemical platform for the quantitative, multiplexed detection of specific antibodies. Anal. Chem. 2011, 84, 1098–1103. [Google Scholar] [CrossRef][Green Version]
- Bonham, A.J.; Paden, N.G.; Ricci, F.; Plaxco, K.W. Detection of IP-10 protein marker in undiluted blood serum via an electrochemical E-DNA scaffold sensor. Analyst 2013, 138, 5580–5583. [Google Scholar] [CrossRef] [PubMed]
- Fetter, L.; Richards, J.; Daniel, J.; Roon, L.; Rowland, T.J.; Bonham, A.J. Electrochemical aptamer scaffold biosensors for detection of botulism and ricin toxins. Chem. Commun. 2015, 8–11. [Google Scholar] [CrossRef]
- Arroyo-Currás, N.; Somerson, J.; Vieira, P.A.; Ploense, K.L.; Kippin, T.E.; Plaxco, K.W. Real-time measurement of small molecules directly in awake, ambulatory animals. Proc. Natl. Acad. Sci. USA 2017, 114, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Rowe, A.A.; White, R.J.; Bonham, A.J.; Plaxco, K.W. Fabrication of electrochemical-dna biosensors for the reagentless detection of nucleic acids, proteins and small molecules. J. Vis. Exp. 2011, 52, e2922. [Google Scholar] [CrossRef]
- Ferguson, B.S.; Hoggarth, D.A.; Maliniak, D.; Ploense, K.; White, R.J.; Woodward, N.; Hsieh, K.; Bonham, A.J.; Eisenstein, M.; Kippin, T.E.; et al. Real-time, aptamer-based tracking of circulating therapeutic agents in living animals. Sci. Transl. Med. 2013, 5, 213ra165. [Google Scholar] [CrossRef]
- Vallée-Bélisle, A.; Ricci, F.; Plaxco, K.W. Engineering biosensors with extended, narrowed, or arbitrarily edited dynamic range. J. Am. Chem. Soc. 2012, 134, 2876–2879. [Google Scholar] [CrossRef]
- Meneghello, A.; Tartaggia, S.; Alvau, M.D.; Polo, F.; Toffoli, G. Biosensing technologies for therapeutic drug monitoring. Curr. Med. Chem. 2017, 25. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Lai, R.Y. Effects of DNA probe and target flexibility on the performance of a “signal-on” electrochemical DNA sensor. Anal. Chem. 2014, 86, 17. [Google Scholar] [CrossRef] [PubMed]
- González-Fernández, E.; Staderini, M.; Avlonitis, N.; Murray, A.F.; Mount, A.R.; Bradley, M. Effect of spacer length on the performance of peptide-based electrochemical biosensors for protease detection. Sens. Actuators B 2018, 255, 3040–3046. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, A.B.N.; Maldonado, M.; Poch, D.; Sodia, T.; Smith, A.; Rowland, T.J.; Bonham, A.J. Electrochemical DNA Biosensor That Detects Early Celiac Disease Autoantibodies. Sensors 2021, 21, 2671. https://doi.org/10.3390/s21082671
Nguyen ABN, Maldonado M, Poch D, Sodia T, Smith A, Rowland TJ, Bonham AJ. Electrochemical DNA Biosensor That Detects Early Celiac Disease Autoantibodies. Sensors. 2021; 21(8):2671. https://doi.org/10.3390/s21082671
Chicago/Turabian StyleNguyen, Anna B. N., Marcos Maldonado, Dylan Poch, Tyler Sodia, Andrew Smith, Teisha J. Rowland, and Andrew J. Bonham. 2021. "Electrochemical DNA Biosensor That Detects Early Celiac Disease Autoantibodies" Sensors 21, no. 8: 2671. https://doi.org/10.3390/s21082671
APA StyleNguyen, A. B. N., Maldonado, M., Poch, D., Sodia, T., Smith, A., Rowland, T. J., & Bonham, A. J. (2021). Electrochemical DNA Biosensor That Detects Early Celiac Disease Autoantibodies. Sensors, 21(8), 2671. https://doi.org/10.3390/s21082671








