1. Introduction
Within physical rehabilitation, remotely collating and aggregating data from patients has been suggested to have numerous benefits in terms of cost, clinical outcome and patient satisfaction [
1,
2]. Exercise biofeedback systems use a sensing platform to capture and interpret data to offer the user meaningful information about their performance [
3]. Many systems utilise one or more inertial measurement units (IMUs) to acquire biomechanical data [
4,
5,
6,
7,
8]. Some systems perform simple data processing tasks such as repetition counting, whilst others use more complex supervised machine learning (ML) models to offer greater granularity of feedback to the user such as joint angle measurement, repetition segmentation, or exercise technique biofeedback [
9,
10,
11].
The application of ML spans a variety of biomechanical contexts, with models developed to predict the effect of an intervention, perform activity recognition, predict disease progression or classify abnormal movement [
12]. Supervised learning is one of the main categories of ML and involves training a model which best maps input features to labelled outputs. This requires the developed algorithms to be provided with annotated training data, and the features analysed. The model is then trained using these data and the algorithms are tested with new unlabelled data to identify its accuracy [
9]. For example, supervised ML algorithms can be used with data collected from an IMU to classify exercise performance and technique, whereby the output of the model can give a binary prediction as to whether an exercise in a sequence (or set) was performed correctly or not [
10,
13,
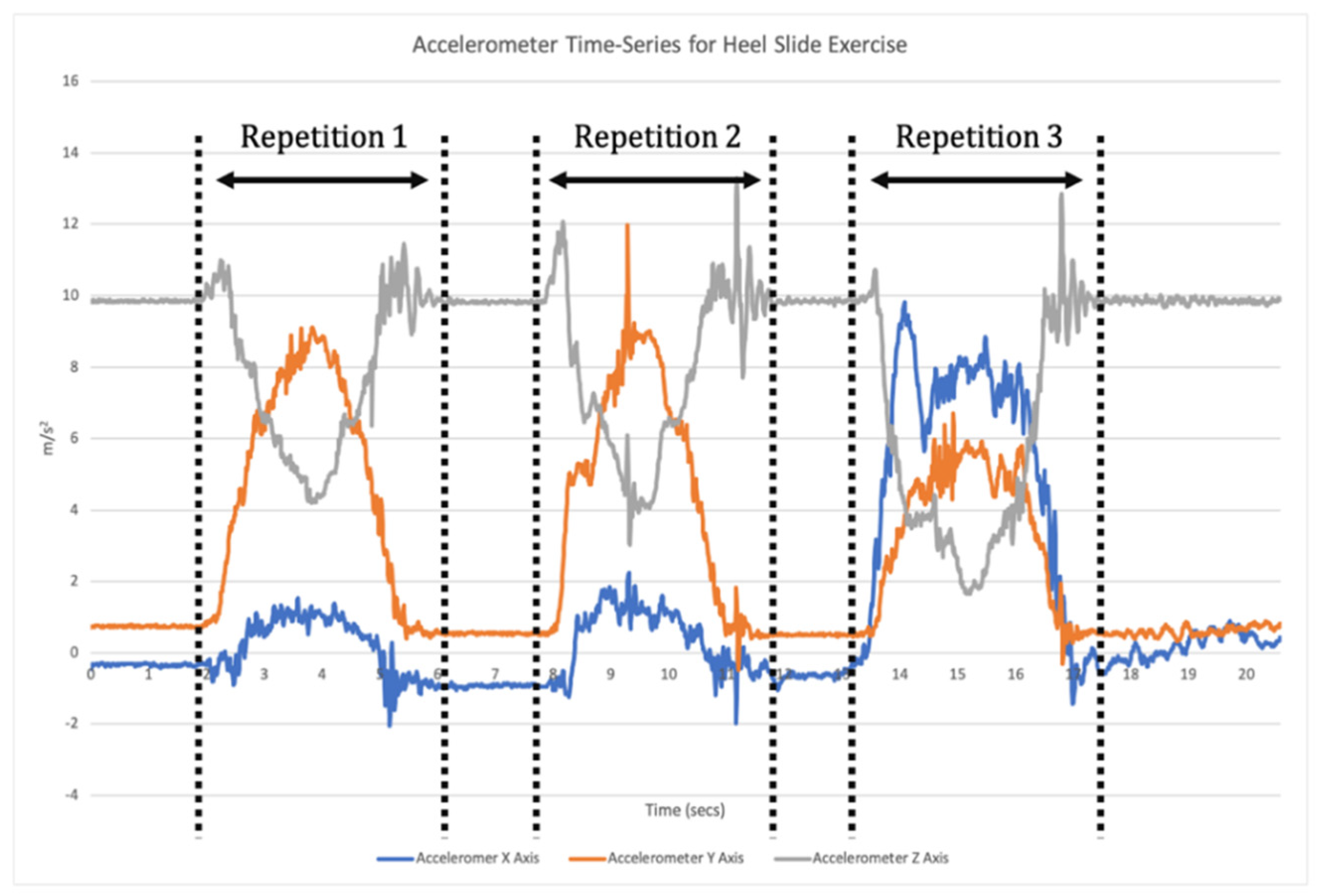
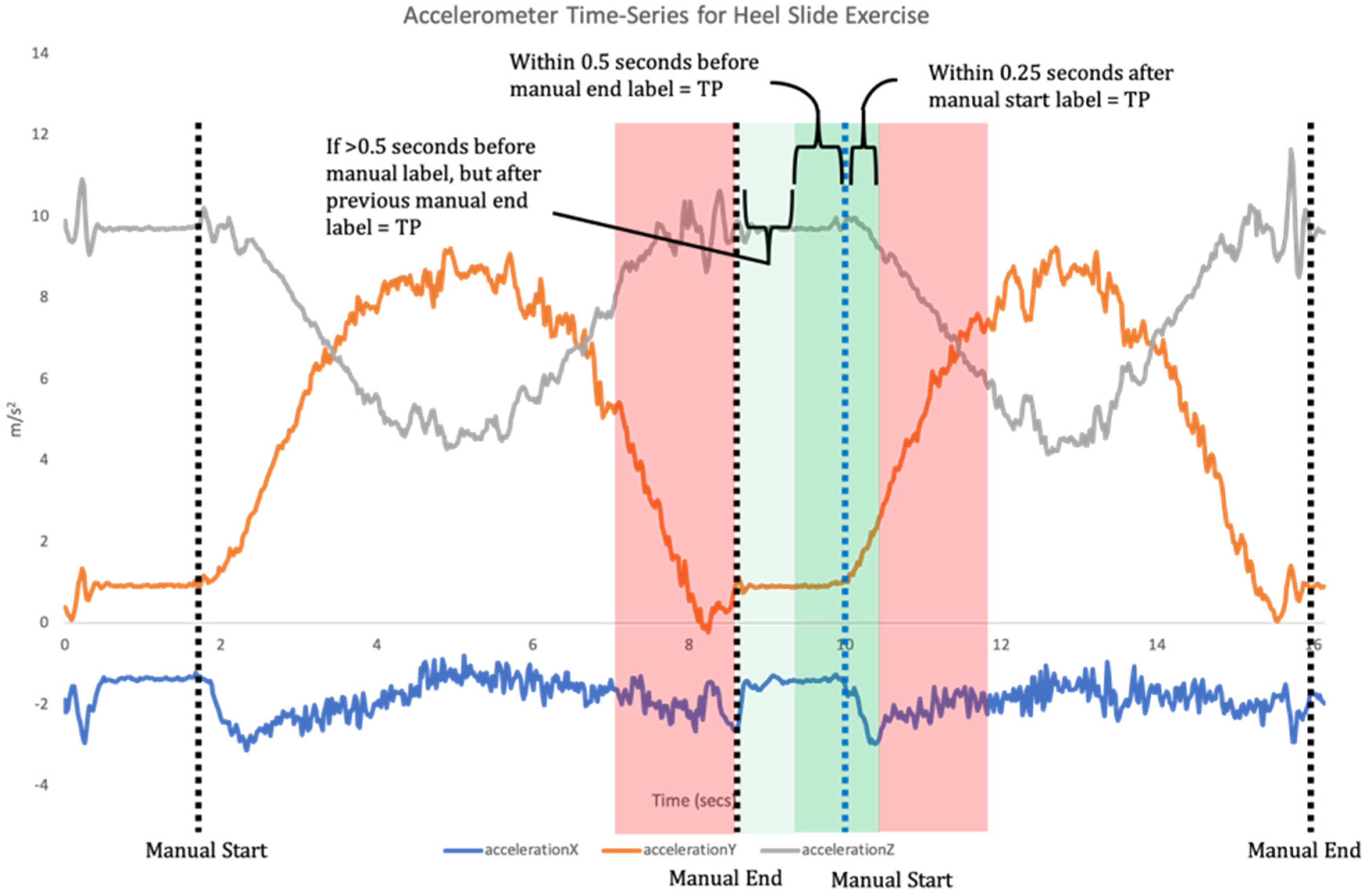
14]. This requires a two-step data analysis process. Firstly, when a time-series of sensor data is recorded the signal needs to be broken down into each individual repetition of the exercise (segmentation;
Figure 1). Segmentation goes beyond merely counting repetitions, but also isolates each individual repetition for the subsequent classification phase [
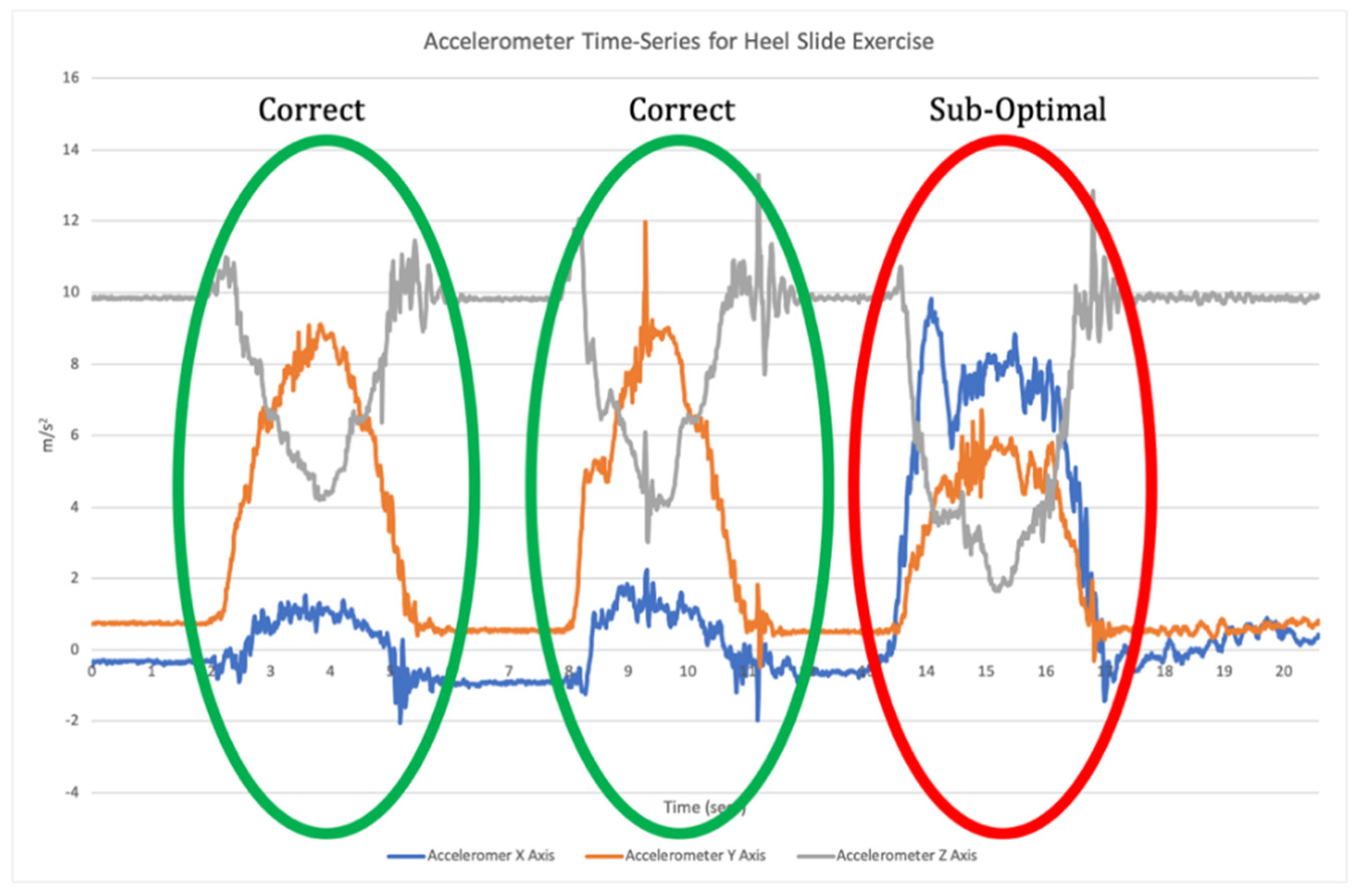
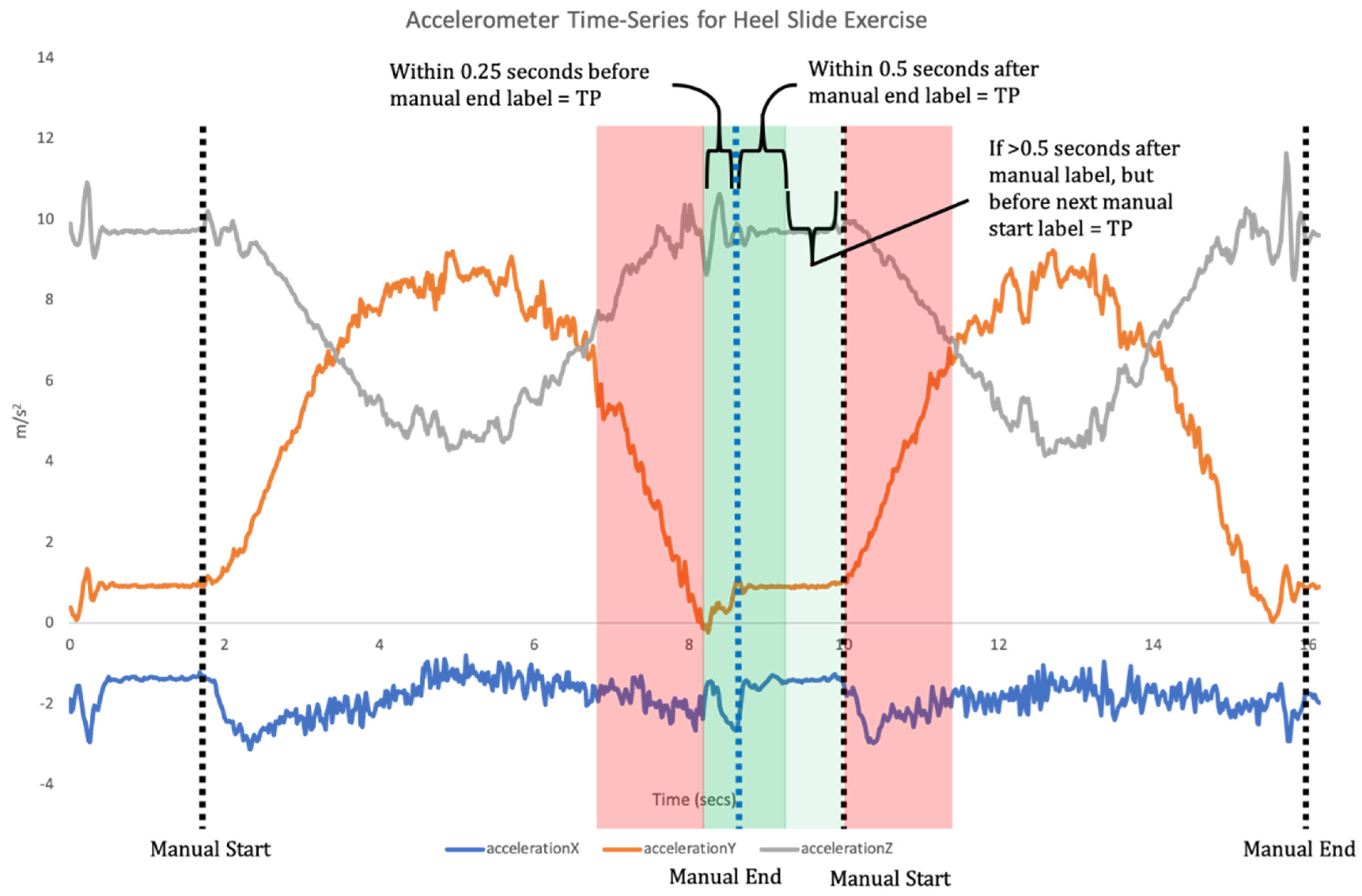
11]. Once individual repetitions have been segmented, classification is undertaken where the input data for each repetition are compared to the labelled training data to assess which label they match most closely to (
Figure 2).
It is common practice to test a number of different ML algorithms for early validation, with the best performing algorithm being used within the final model [
15,
16]; however, there are suggestions that the common cross-validation methods for evaluating ML models analysing IMU data do not provide a realistic reflection of system performance [
17].
The World Health Organisation has published guidelines for the evaluation of digital health interventions which include monitoring the functionality of a system, with functionality defined as
“the degree to which a product or system provides functions that meet stated and implied needs when used under specific conditions” [
18]. In the context of exercise biofeedback systems, we can assume that the ‘specific conditions’ relate to a real-world application that involves the patient performing exercise and receiving feedback outside a clinical or laboratory setting. However, there is a lack of research investigating the real-world validity of IMU based biofeedback systems with only one previous real-world validation of an exercise classification model identified in the literature [
19]. Thus, there is a clear need to assess these components in real-world environments in order to optimise their effectiveness. A framework for segmentation model validation has recently been proposed to assess segmentation accuracy using a staged approach, with testing in laboratory, pre-clinical and clinical settings [
20]. However, this does not consider the overall performance of an exercise biofeedback system incorporating classification models or continue to demonstrate an evaluation of a system in each of these settings.
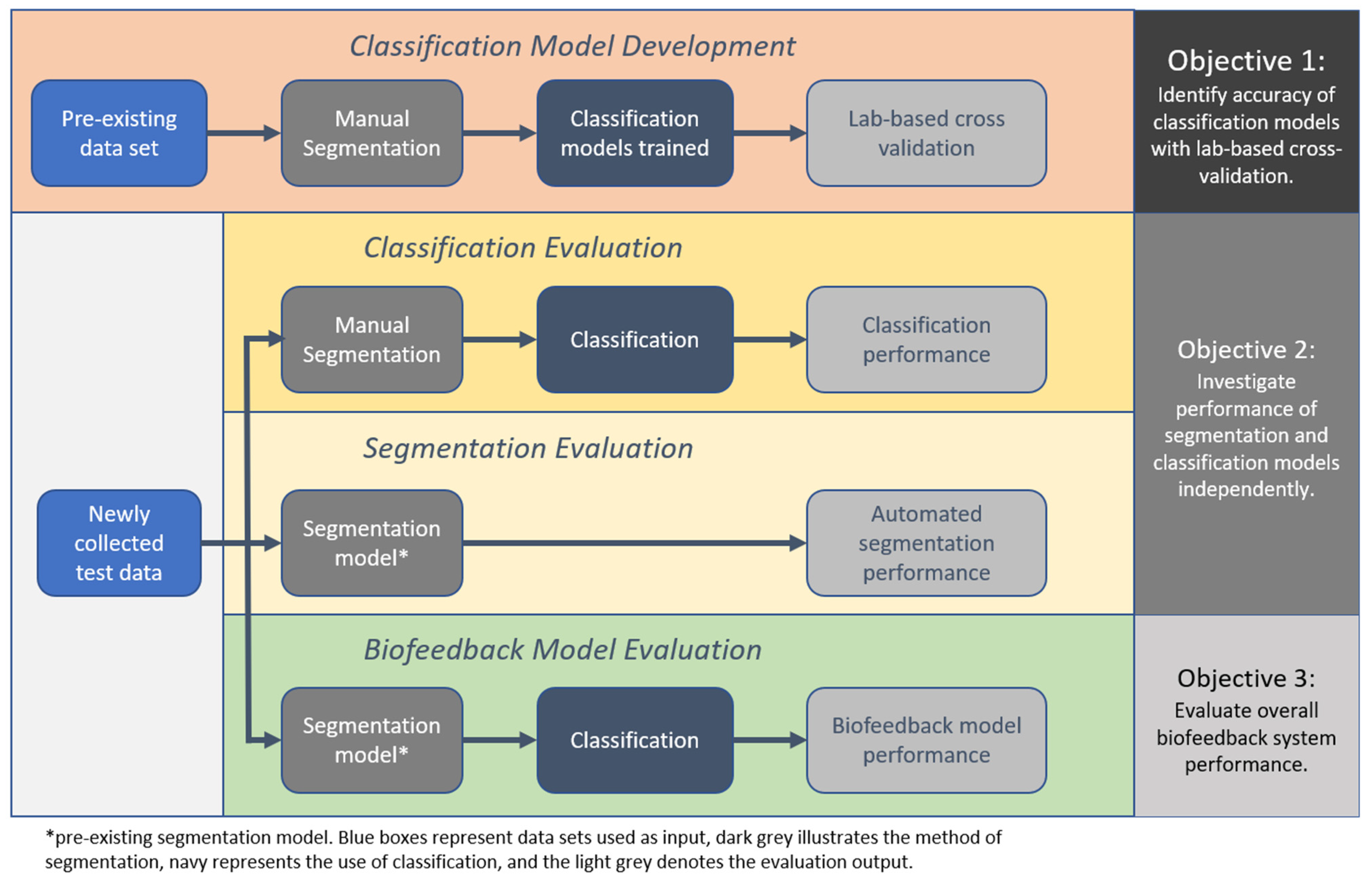
Therefore, the aim of this study was to conduct a thorough validation of an example IMU-based exercise biofeedback systems with testing in the laboratory through to clinical participants in the target-use conditions. Additionally, three objectives were identified; (1) to identify the accuracy of the classification models using the lab-based cross validation approach, (2) to investigate the performance of the segmentation and classification models independently with newly collected test data from a healthy and a clinical population, and (3) to evaluate overall biofeedback system performance with test data that have been algorithmically segmented and classified.
4. Discussion
This study has highlighted the importance of thorough validation of ML models in IMU based exercise biofeedback systems to ensure acceptable accuracy. The example system evaluated has illustrated the variations in performance across each phase of the process and demonstrates the shortcomings in accuracy of this system in the final clinical deployment, with the overall biofeedback model provided with a balanced clinical test set demonstrating ‘poor’ to ‘moderate’ levels of accuracy. Therefore, this particular model requires further development and refinement prior to wider scale implementation. This study highlights the importance of context when evaluating the performance of rehabilitation biofeedback systems. Making judgements on potential for performance in the real-world setting based on laboratory validation of underlying data models potentially reports an over-optimistic and unrealistic expectation of system accuracy, which in turn has implications for clinical applications.
Making direct comparisons between these results and the literature is difficult due to the lack of external validation that is conducted on ML models [
8,
17]. The majority of current research outlines LOSOCV and other similar lab-based methods such as k-fold cross-validation [
8,
10,
34,
35], rather than the evaluation of ML models with a newly collected and independent test-set. Shany et al. [
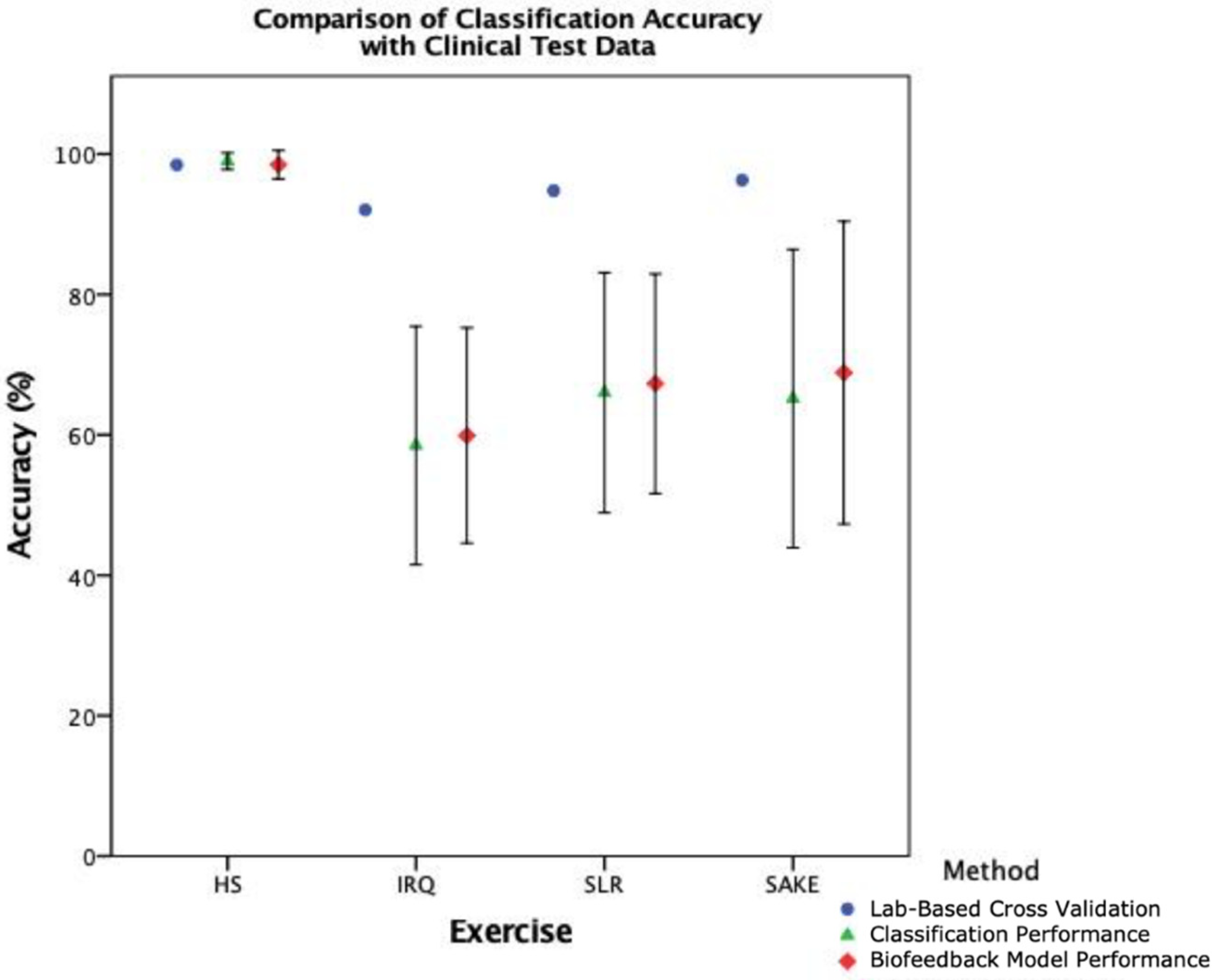
17] recommended that external validation is the most preferred option in the evaluation of ML models for fall risk prediction, and
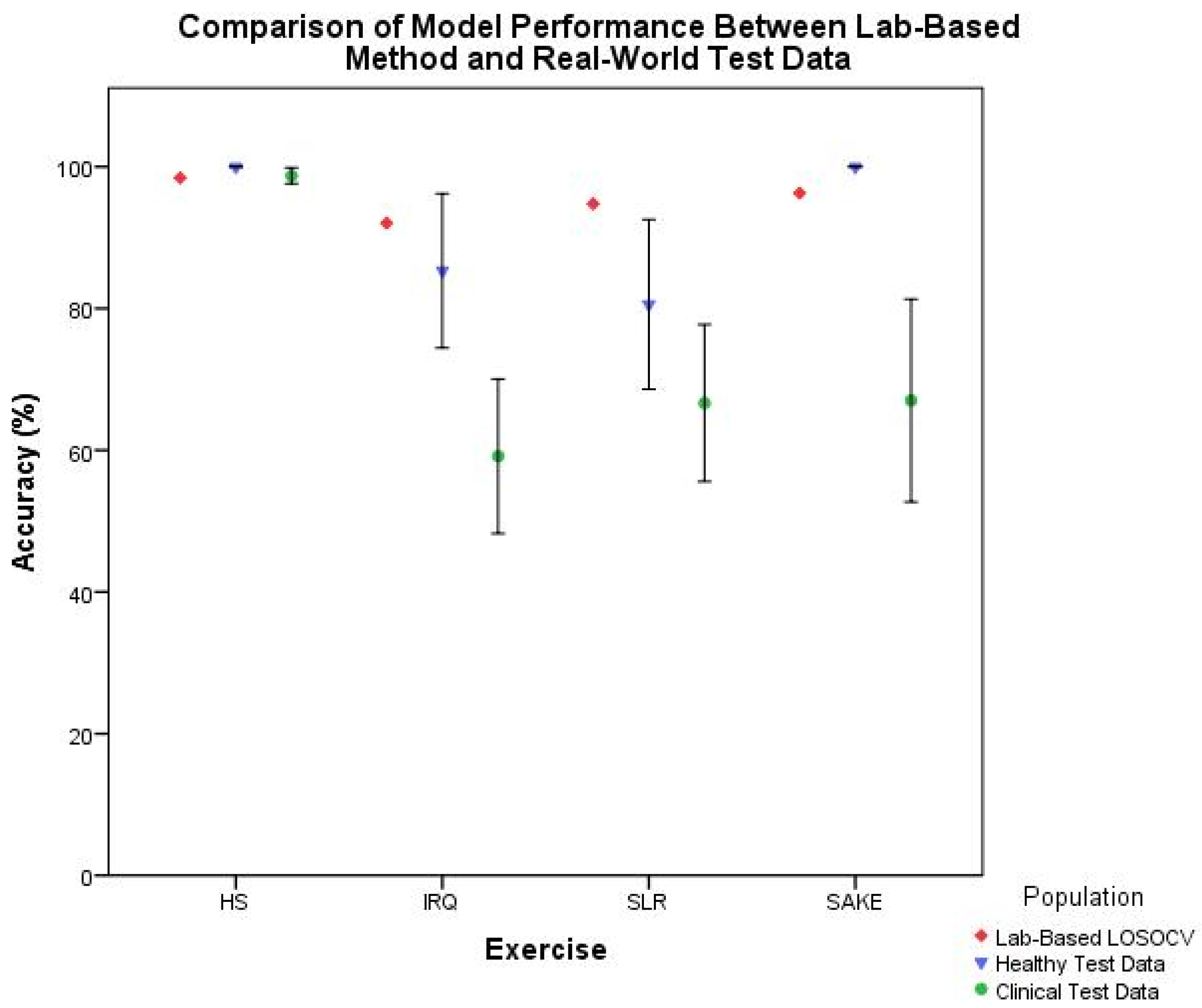
Figure 9 illustrates the trend in reducing accuracy with external test data in this clinical context. As such cross-validation results should arguably be seen as the best-case scenario rather than actual performance. This study has therefore taken the preferred validation option [
17], built on previous work [
11,
21], and moved past the lab-based validation methods into the real-world setting.
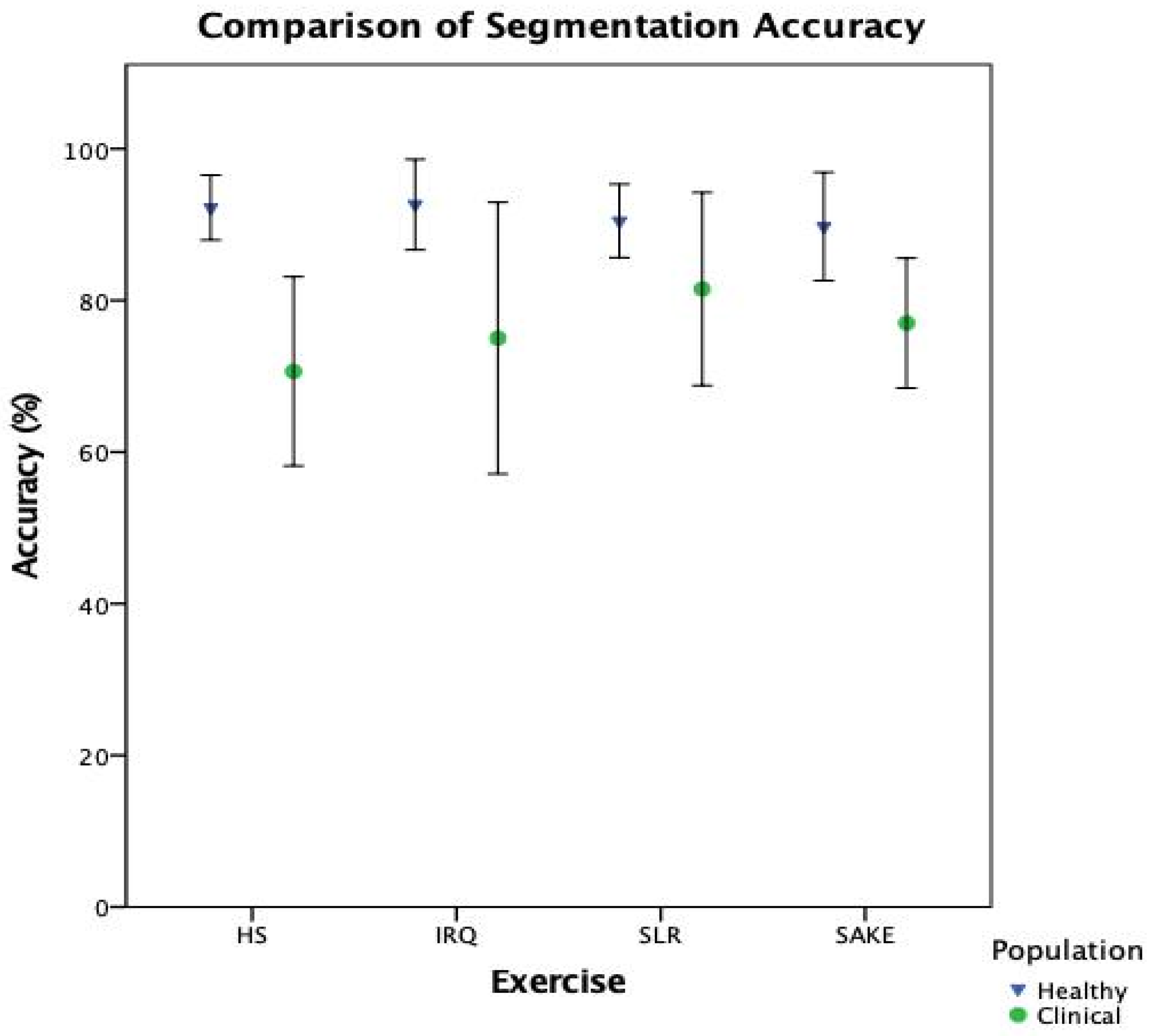
When breaking down the ML components of the system, it is clear that there is a notable reduction in performance in the clinical cohort across both classification (
Table 5) and segmentation (
Figure 7) when compared to healthy participants. However, interestingly, the slight improvement in overall biofeedback model performance, compared to classification performance in
Figure 8, would suggest that the reduction in segmentation accuracy has less of an effect on overall system functionality. This would indicate that the classification component, and the training data provided to the model, is the leading cause for the reduction in accuracy when deploying this system with clinical patients.
Therefore, this study has highlighted key criteria in the development of exercise classification models, particularly regarding the training data on which the models are built. Firstly, whilst the training data contained data from participants aged between 40 and 80 years of age undergoing lower limb rehabilitation [
21], none of the participants were undertaking rehabilitation following TKR or UKR. This could explain the reasons for the reduction in accuracy, as the classification models are being tested against training data which are too heterogenous, and must be closer matched to the population for which they are designed [
26]. During data collection using the example system, some participants preferred to complete their exercises on their bed, others on the sofa and some lying on the floor, thus demonstrating the heterogeneity of real-world use. However, the training data were collected in a controlled clinic environment. The presence of thick duvets, soft mattresses, and varying sizes of towels at home can all contribute to variance in the IMU data obtained, and therefore the outputs of the ML models. For pragmatic reasons, most training data sets for exercise biofeedback systems are collected in controlled environments; however, the results of this study would suggest that any ML model must be built on training data from those with the same clinical pathology and collected in the same manner as the target user to demonstrate acceptable levels of accuracy. Equally, it is arguable that given the greater balance between correct and sub-optimal repetitions in the clinical test data, these results are a better reflection of the actual performance of the models and that a balanced test set from healthy participants may have demonstrated a similar reduction in performance [
36]. Developers of these systems, and those reviewing their performance, must be cognisant of collecting a balanced test-set, whilst doing their best to avoid including deliberately produced examples of sub-optimal repetitions. Additionally, in this particular approach, further work in refining the classification models is required prior to clinical deployment, including exploring other feature selection approaches and hyperparameter tuning [
12].
This binary approach using supervised ML also has implications to both the technical and clinical feasibility of such systems. The need to collate large quantities of training data which contains both raw IMU data and video to label the ground truth is time consuming and requires access to large numbers of participants. There is subjectivity in labelling the exercise technique [
37], and the scalability of the method is impacted should it be sought to develop models for additional exercises. Finally, the clinical feasibility of using ML to classify exercise technique can be further questioned, as this method removes the human factors of therapy and the context of each individual patient. For example, it is possible that two patients performing the same exercises with the same clinician will receive markedly different feedback in clinical practice. This is due to the judgement the therapist makes on numerous factors including the rehabilitation timeline, patient’s personality, progress and goals. One patient may be ahead of schedule and moving quickly onto the next exercise, whereas another may be slow and lacking engagement, requiring a different message to be given by the therapist. Whilst in theory it is possible to train a machine to learn these variables, it is not necessarily feasible. As an alternative, during a user evaluation of this example system patients expressed a wish for a greater granularity in the feedback they receive such as joint angle or a quality score [
6].
There are a number of limitations to consider when reviewing the results of this study. It is important to highlight that this evaluation took place outside of the custom-built tablet application. This was done to ensure that there were no computational load or software programming issues; however, any such stability concerns must be considered when evaluating a system in its entirety. Secondly, the clinical test set contained multiple data sets from the same participant as they were tested over three different time points. Whilst it was outside the scope of the aims and objectives of this study, further analysis of exercise biofeedback performance within and between participants may be beneficial. Finally, as highlighted previously, the unbalanced test set for the HS exercise means that it is difficult to draw firm conclusions on the functionality of the models for this exercise.
Despite these limitations, this study has highlighted the deterioration in technical functionality of the ML models within an example biofeedback system when deployed with clinical patients compared to healthy controls. The results provide a clear illustration on the importance of not relying on the lab-based validation methods frequently reported in the literature and emphasises that this real-world validation is a crucial step in the development and implementation of IMU-based biofeedback ML systems.