Feature Selection Using Enhanced Particle Swarm Optimisation for Classification Models
Abstract
1. Introduction
- Two new PSO variants for feature selection are proposed to overcome two major shortcomings of the original PSO algorithm, i.e., premature convergence and weak local exploitation capability around the near optimal solutions.
- The first proposed PSO model, i.e., PSOVA1 (PSO variant 1), comprises the following mechanisms: (1) a modified PSO operation with rectified global and personal best signals, (2) spiral search based local exploitation, (3) Gaussian distribution based swarm leader enhancement, as well as (4) mirroring and DE mutation operations for worst solution improvement.
- The second proposed PSO model, i.e., PSOVA2 (PSO variant 2), enhances PSOVA1 through four mechanisms: (1) an adaptive exemplar breeding mechanism incorporating multiple optimal signals, (2) search coefficient generation using sine, cosine, and hyperbolic tangent functions, (3) worst solution enhancement using a hybrid re-dispatching scheme, and (4) an exponential exploitation scheme for swarm leader improvement. Moreover, the search diversity and scopes in PSOVA2 are further elevated in comparison with those of PSOVA1. This is owing to the adoption of diverse exemplars to guide the search in each dimension, as well as the employment of versatile search trajectories to calibrate the particle positions.
- Evaluation using 13 datasets with a wide spectrum of dimensionalities: the empirical results indicate that both proposed models outperform five classical search methods and ten advanced PSO variants with significant advantages, evidenced by the statistical test outcomes.
2. Related Studies
2.1. Particle Swarm Optimisation
2.2. PSO Variants
2.3. PSO for Feature Selection
2.4. Research Motivations
3. The Proposed PSOVA1 Model
| Algorithm 1. The pseudo-code of the proposed PSOVA1 model. |
| 1 Start 2 Initialise a particle swarm using the Logistic chaotic map; 3 Evaluate each particle using the objective function f (x) and identify the pbest solution of each particle, and the global best solution, gbest; 4 Construct a Worst_memory, which stores the three weakest particles with the lowest fitness values, and identify the worst solution as gworst; 5 While (termination criteria are not met) 6 { 7 Conduct swarm leader enhancement using Gaussian distribution as defined in Equation (3); Use the new solution to replace gbest if it is fitter; 8 For (each particle i in the population) do 9 { 10 If (particle i belongs to Worst_memory) 11 { 12 If (particle i is gworst) 13 { 14 Construct an offspring solution by employing the local mutation operation based on gbest as defined in Equation (4), and use it to replace the global worst solution if the new offspring solution is fitter; 15 Else 16 Construct an offspring solution by employing the DE-based mutation operation based on three randomly selected pbest solutions as defined in Equations (5)–(6); 17 Evaluate the offspring solution and update the position of particle i in Worst_memory based on the annealing schedule as defined in Equation (7); 18 } End If 19 Update the pbest and gbest solutions; 20 } End If 21 } End For 22 For (each particle i in the population) do 23 { 24 If Rand < pswitch 25 { 26 Establish a memory of groupi which includes all neighboring pbest solutions with higher or equal fitness scores than that of the pbest solution of the current particle i, i.e., pbesti; 27 Identify the neighboring fitter pbest solution in groupi with the highest degree of dissimilarity to gbest, denoted as pbestD; 28 Calculate the ameliorated gbest solution, i.e., gbestM, by averaging the following two solutions, i.e., pbestD and gbest, as indicated in Equation (8); 29 Randomly select another neighboring fitter pbest solution from groupi, denoted as pbestR, 30 Calculate the ameliorated pbest solution, i.e., pbestM by averaging pbestR and the personal best solution of particle i, pbesti, as shown in Equation (9); 31 Conduct position updating using gbestM and pbestM for particle i as defined in Equation (10); 32 Else 33 Move particle i around gbest by following a logarithmic spiral search path as shown in Equation (11); 34 } End If 35 } End For 36 For (each particle i in the population) do 37 { 38 Evaluate each particle i using the objective function; 39 Update the pbest and gbest solutions; 40 } End For 41 } End While 42 Output gbest; 43 End |
3.1. A Swarm Leader Enhancing Mechanism
3.2. Mutation-Based Worst Solution Enhancement
3.3. Diversity-Enhanced PSO Evolving Strategy
3.4. An Intensified Spiral Exploitation Scheme
4. The Proposed PSOVA2 Model
| Algorithm 2. The pseudo-code of the proposed PSOVA2 model. |
| 1 Start 2 Initialise a particle swarm using the Logistic chaotic map; 3 Evaluate each particle using the objective function f (x) and identify the pbest solution of each particle, and the global best solution, gbest; 4 While (termination criteria are not met) 5 { 6 Conduct swarm leader enhancement as defined in Equations (26)–(27); 7 Implement the worse solution enhancement as defined in Equations (23)–(25); 8 For (each particle i in the population) do 9 { 10 Construct a breeding exemplar as defined in Equations (15)–(18); 11 Select a coefficient generation function from Equations (19)–(22) randomly; 12 For (each dimension j) do 13 { % Choose the target optimal signal to follow in each dimension 14 If Rand < 0.4 15 { 16 Choose the breeding exemplar as the target signal for position updating; 17 Else 18 Choose the gbest solution as the target signal for position updating; 19 } End If 20 Update the position of particle i on dimension j as defined in Equation (14); 21 } End For 22 } End For 23 For (each particle i in the population) do 24 { 25 Evaluate each particle i using the objective function; 26 Update pbest and gbest solutions; 27 } End For 28 } End While 29 Output gbest; 30 End |
4.1. A New Attraction Operation with Differentiated Search Trajectories
4.1.1. Exemplar Generation Using Adaptive Incorporation of Multiple Optimal Solutions
4.1.2. Nonlinear Search Coefficient Generation
4.2. A Hybrid Re-Dispatching Scheme for Enhancement of the Worst Solutions
4.3. Swarm Leader Enhancement Using an Adaptive Exponential Search Flight
| Algorithm 3. The pseudo-code of the hybrid PSOVA1/PSOVA2-KNN feature selection model. |
| 1 Start 2 Initialise a particle swarm using the Logistic chaotic map; 3 For (each particle i in the population) do 4 { 5 Convert particle i into a corresponding feature subset by selecting features on the dimensions where positive values are assigned; 6 Calculate classification performance of the feature subset encoded in particle i on the training data set using the KNN classifier; 7 Evaluate the fitness score of particle i based on its classification performance and number of selected features using the proposed objective function f (x) as shown in Equation (28); 8 Identify the pbest solution of each particle and the global best solution gbest; 9 } End For 10 While (termination criteria are not met) 11 { 12 Evolve swarm particles using the proposed mechanisms in PSOVA1 (i.e., line 7–35 in Algorithm 1) or PSOVA2 (i.e., line 6–22 in Algorithm 2); 13 For (each particle i in the population) do 14 { 15 Evaluate particle i using the objective function on the training set; 16 Update pbest and gbest solutions; 17 } End For 18 } End While 19 Output gbest; 20 Convert gbest into the identified optimal feature subset; 21 Calculate classification performance on the unseen test set based on the yielded optimal feature subset using the KNN classifier; 22 Output the test classification results & the selected features; 23 End |
5. Evaluation and Discussion
5.1. Data Sets
5.2. Parameter Settings
5.3. Results and Discussion
5.3.1. Classification Performance
5.3.2. Selected Feature Sizes
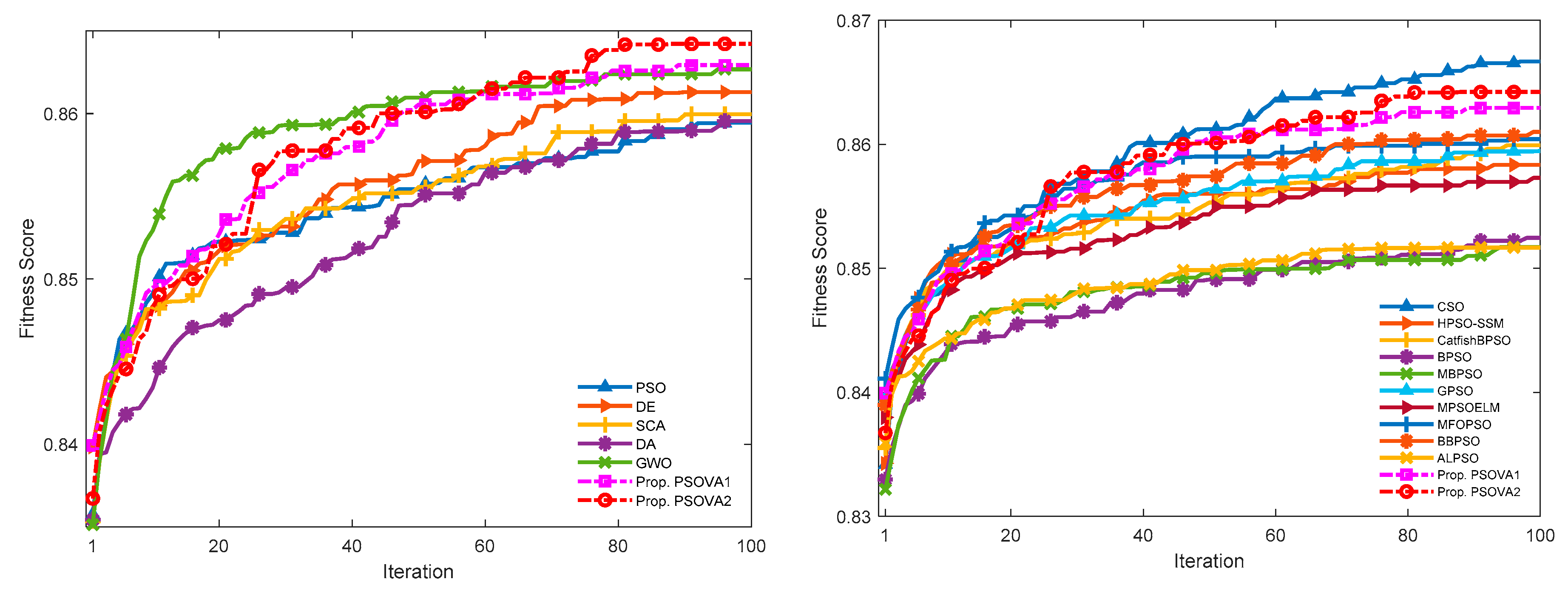
5.3.3. Convergence Rates and Computational Costs
5.3.4. Evaluation of The Proposed Mechanisms in PSOVA1 and PSOVA2
5.3.5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gheyas, I.A.; Smith, L.S. Feature subset selection in large dimensionality domains. Pattern Recognit. 2010, 43, 5–13. [Google Scholar] [CrossRef]
- Mafarja, M.; Heidari, A.A.; Faris, H.; Mirjalili, S.; Aljarah, I. Dragonfly Algorithm: Theory, Literature Review, and Application in Feature Selection. In Nature-Inspired Optimizers: Theories, Literature Reviews and Applications; Mirjalili, S., Dong, J.S., Lewis, A., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 47–67. [Google Scholar]
- Zhang, L.; Mistry, K.; Lim, C.P.; Neoh, S.C. Feature selection using firefly optimization for classification and regression models. Decis. Support Syst. 2018, 106, 64–85. [Google Scholar] [CrossRef]
- Xue, B.; Zhang, M.; Browne, W.N.; Yao, X. A Survey on Evolutionary Computation Approaches to Feature Selection. IEEE Trans. Evol. Comput. 2016, 20, 606–626. [Google Scholar] [CrossRef]
- Dash, M.; Liu, H. Feature selection for classification. Intell. Data Anal. 1997, 1, 131–156. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Zhou, T.; Lu, H.; Wang, W.; Yong, X. GA-SVM based feature selection and parameter optimization in hospitalization expense modeling. Appl. Soft Comput. 2019, 75, 323–332. [Google Scholar]
- Baig, M.Z.; Aslam, N.; Shum, H.P.; Zhang, L. Differential evolution algorithm as a tool for optimal feature subset selection in motor imagery EEG. Expert Syst. Appl. 2017, 90, 184–195. [Google Scholar] [CrossRef]
- Ghosh, A.; Datta, A.; Ghosh, S. Self-adaptive differential evolution for feature selection in hyperspectral image data. Appl. Soft Comput. 2013, 13, 1969–1977. [Google Scholar] [CrossRef]
- Xue, B.; Zhang, M.; Browne, W.N. Particle Swarm Optimization for Feature Selection in Classification: A Multi-Objective Ap-proach. IEEE Trans. Cybern. 2013, 43, 1656–1671. [Google Scholar] [CrossRef]
- Mirjalili, S. Moth-flame optimization algorithm: A novel nature-inspired heuristic paradigm. Knowl. Based Syst. 2015, 89, 228–249. [Google Scholar] [CrossRef]
- Jothi, G.; Hannah, H.I. Hybrid Tolerance Rough Set–Firefly based supervised feature selection for MRI brain tumor image classification. Appl. Soft Comput. 2016, 46, 639–651. [Google Scholar]
- Singh, U.; Singh, S.N. A new optimal feature selection scheme for classification of power quality disturbances based on ant colony framework. Appl. Soft Comput. 2019, 74, 216–225. [Google Scholar] [CrossRef]
- Abdel-Basset, M.; EI-Shahat, D.; EI-henawy, I.; Albuquerque, V.H.C.; Mirjalili, S. A new fusion of grey wolf optimizer algorithm with a two-phase mutation for feature selection. Expert Syst. Appl. 2020, 139, 112824. [Google Scholar] [CrossRef]
- Mafarja, M.; Mirjalili, S. Whale optimization approaches for wrapper feature selection. Appl. Soft Comput. 2018, 62, 441–453. [Google Scholar] [CrossRef]
- Sindhu, R.; Ngadiran, R.; Yacob, Y.M.; Zahri, N.A.H.; Hariharan, M. Sine–cosine algorithm for feature selection with elitism strategy and new updating mechanism. Neural Comput. Appl. 2017, 28, 2947–2958. [Google Scholar] [CrossRef]
- Hsieh, S.-T.; Sun, T.-Y.; Liu, C.-C.; Tsai, S.-J. Efficient Population Utilization Strategy for Particle Swarm Optimizer. IEEE Trans. Syst. Mancybern. Part B 2008, 39, 444–456. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Qin, A.; Suganthan, P.; Baskar, S. Comprehensive learning particle swarm optimizer for global optimization of multimodal functions. IEEE Trans. Evol. Comput. 2006, 10, 281–295. [Google Scholar] [CrossRef]
- Chen, K.; Zhou, F.-Y.; Yuan, X.-F. Hybrid particle swarm optimization with spiral-shaped mechanism for feature selection. Expert Syst. Appl. 2019, 128, 140–156. [Google Scholar] [CrossRef]
- Ahn, C.W.; An, J.; Yoo, J.-C. Estimation of particle swarm distribution algorithms: Combining the benefits of PSO and EDAs. Inf. Sci. 2012, 192, 109–119. [Google Scholar] [CrossRef]
- Iqbal, M.; de Oca, M.A.M. An Estimation of Distribution Particle Swarm Optimization Algorithm. In Ant Colony Optimization and Swarm Intelligence; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Kennedy, J.; Eberhart, R. Particle swarm optimization. In Proceedings of the ICNN’95-International Conference on Neural Networks, Perth, WA, Australia, 27 November–1 December 1995; Volume 4, pp. 1942–1948. [Google Scholar] [CrossRef]
- Pandit, D.; Zhang, L.; Chattopadhyay, S.; Lim, C.P.; Liu, C. A scattering and repulsive swarm intelligence algorithm for solving global optimization problems. Knowl. Based Syst. 2018, 156, 12–42. [Google Scholar] [CrossRef]
- Wang, H.; Sun, H.; Li, C.; Rahnamayan, S.; Pan, J.-S. Diversity enhanced particle swarm optimization with neighborhood search. Inf. Sci. 2013, 223, 119–135. [Google Scholar] [CrossRef]
- Tan, T.Y.; Zhang, L.; Neoh, S.C.; Lim, C.P. Intelligent skin cancer detection using enhanced particle swarm optimization. Knowl. Based Syst. 2018, 158, 118–135. [Google Scholar] [CrossRef]
- Tan, T.Y.; Zhang, L.; Lim, C.P. Intelligent skin cancer diagnosis using improved particle swarm optimization and deep learn-ing models. Appl. Soft Comput. 2019, 84, 105725. [Google Scholar] [CrossRef]
- Tan, T.Y.; Zhang, L.; Lim, C.P.; Fielding, B.; Yu, Y.; Anderson, E. Evolving Ensemble Models for Image Segmentation Using En-hanced Particle Swarm Optimization. IEEE Access 2019, 7, 34004–34019. [Google Scholar] [CrossRef]
- Chen, J.; Zheng, J.; Wu, P.; Zhang, L.; Wu, Q. Dynamic particle swarm optimizer with escaping prey for solving constrained non-convex and piecewise optimization problems. Expert Syst. Appl. 2017, 86, 208–223. [Google Scholar] [CrossRef]
- Li, M.; Chen, H.; Shi, X.; Liu, S.; Zhang, M.; Lu, S. A multi-information fusion “triple variables with iteration” inertia weight PSO algorithm and its application. Appl. Soft Comput. 2019, 84, 105677. [Google Scholar] [CrossRef]
- Xia, X.; Gui, L.; He, G.; Wei, B.; Zhang, Y.; Yu, F.; Wu, H.; Zhan, Z. An expanded particle swarm optimization based on mul-ti-exemplar and forgetting ability. Inf. Sci. 2020, 508, 105–120. [Google Scholar] [CrossRef]
- Chen, Q.; Sun, J.; Palade, V. Distributed Contribution-Based Quantum-Behaved Particle Swarm Optimization With Controlled Diversity for Large-Scale Global Optimization Problems. IEEE Access 2019, 7, 150093–150104. [Google Scholar] [CrossRef]
- Lin, A.; Sun, W.; Yu, H.; Wu, G.; Tang, H. Global genetic learning particle swarm optimization with diversity enhancement by ring topology. Swarm Evol. Comput. 2019, 44, 571–583. [Google Scholar] [CrossRef]
- Zhang, Z.; Yu, Y.; Zheng, S.; Todo, Y.; Gao, S. Exploitation Enhanced Sine Cosine Algorithm with Compromised Population Diversity for Optimization. In Proceedings of the 2018 IEEE International Conference on Progress in Informatics and Computing (PIC), Suzhou, China, 14–16 December 2018; pp. 1–7. [Google Scholar]
- Jordehi, A.R. Enhanced leader PSO (ELPSO): A new PSO variant for solving global optimisation problems. Appl. Soft Comput. 2015, 26, 401–417. [Google Scholar] [CrossRef]
- Kang, L.; Chen, R.-S.; Xiong, N.; Chen, Y.-C.; Hu, Y.-X.; Chen, C.-M. Selecting Hyper-Parameters of Gaussian Process Regression Based on Non-Inertial Particle Swarm Optimization in Internet of Things. IEEE Access 2019, 7, 59504–59513. [Google Scholar] [CrossRef]
- Yu, X.; Yu, X.; Lu, Y.; Yen, G.G.; Cai, M. Differential evolution mutation operators for constrained multi-objective optimization. Appl. Soft Comput. 2018, 67, 452–466. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, H.; Li, W.; Zhou, M.; Zhang, Y.; Chaovalitwongse, W.A. Comprehensive learning Particle Swarm Optimi-zation algorithm with local search for multimodal functions. IEEE Trans. Evol. Comput. 2019, 23, 718–731. [Google Scholar] [CrossRef]
- Xu, X.; Li, J.; Zhou, M.; Xu, J.; Cao, J. Accelerated Two-Stage Particle Swarm Optimization for Clustering Not-Well-Separated Data. IEEE Trans. Syst. Mancybern. Syst. 2020, 50, 4212–4223. [Google Scholar] [CrossRef]
- Elbaz, K.; Shen, S.-L.; Sun, W.-J.; Yin, Z.-Y.; Zhou, A. Prediction Model of Shield Performance During Tunneling via Incorporating Improved Particle Swarm Optimization Into ANFIS. IEEE Access 2020, 8, 39659–39671. [Google Scholar] [CrossRef]
- Elbaz, K.; Shen, S.-L.; Zhou, A.; Yin, Z.-Y.; Lyu, H.-M. Prediction of Disc Cutter Life During Shield Tunneling with AI via the Incorporation of a Genetic Algorithm into a GMDH-Type Neural Network. Engineering 2020. [Google Scholar] [CrossRef]
- Chen, Q.; Chen, Y.; Jiang, W. Genetic Particle Swarm Optimization-Based Feature Selection for Very-High-Resolution Re-motely Sensed Imagery Object Change Detection. Sensors 2016, 16, 1204. [Google Scholar] [CrossRef]
- Srisukkham, W.; Zhang, L.; Neoh, S.C.; Todryk, S.; Lim, C.P. Intelligent leukaemia diagnosis with bare-bones PSO based feature optimization. Appl. Soft Comput. 2017, 56, 405–419. [Google Scholar] [CrossRef]
- Mistry, K.; Zhang, L.; Neoh, S.C.; Lim, C.P.; Fielding, B. A Micro-GA Embedded PSO Feature Selection Approach to Intelligent Facial Emotion Recognition. IEEE Trans. Cybern. 2017, 47, 1496–1509. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.-D. A modified particle swarm optimization with multiple subpopulations for multimodal function optimization problems. Appl. Soft Comput. 2015, 33, 170–182. [Google Scholar] [CrossRef]
- Nayak, D.R.; Dash, R.; Majhi, B. Discrete ripplet-II transform and modified PSO based improved evolutionary extreme learning machine for pathological brain detection. Neurocomputing 2018, 282, 232–247. [Google Scholar] [CrossRef]
- Jin, X.; Xu, A.; Bie, R.; Guo, P. Machine learning techniques and chi-square feature selection for cancer classification using SAGE gene expression profiles. In Lecture Notes in Computer Science; Springer: Singapore, 2006; pp. 106–115. [Google Scholar]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Cheng, R.; Jin, Y. Feature selection for high-dimensional classification using a competitive swarm optimizer. Soft Com-Puting 2018, 22, 811–822. [Google Scholar] [CrossRef]
- Moradi, P.; Gholampour, M. A hybrid particle swarm optimization for feature subset selection by integrating a novel local search strategy. Appl. Soft Comput. 2016, 43, 117–130. [Google Scholar] [CrossRef]
- Tan, T.Y.; Zhang, L.; Lim, C.P. Adaptive melanoma diagnosis using evolving clustering, ensemble and deep neural networks. Knowl. Based Syst. 2020, 187, 104807. [Google Scholar] [CrossRef]
- Fielding, B.; Zhang, L. Evolving Image Classification Architectures with Enhanced Particle Swarm Optimisation. IEEE Access 2018, 6, 68560–68575. [Google Scholar] [CrossRef]
- Cheng, R.; Jin, Y. A Competitive Swarm Optimizer for Large Scale Optimization. IEEE Trans. Cybern. 2015, 45, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Vieira, S.M.; Mendonca, L.F.; Farinha, G.J.; Sousa, J.M.C. Modified binary PSO for feature selection using SVM applied to mor-tality prediction of septic patients. Appl. Soft Comput. 2013, 13, 3494–3504. [Google Scholar] [CrossRef]
- Chuang, L.-Y.; Tsai, S.-W.; Yang, C.-H. Improved binary particle swarm optimization using catfish effect for feature selection. Expert Syst. Appl. 2011, 38, 12699–12707. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, L.; Neoh, S.C.; Mistry, K.; Hossain, M.A. Intelligent affect regression for bodily expressions using hybrid particle swarm optimization and adaptive ensembles. Expert Syst. Appl. 2015, 42, 8678–8697. [Google Scholar] [CrossRef]
- Yang, X.-S. Nature-Inspired Optimization Algorithms; Yang, X.-S., Ed.; Elsevier: Oxford, UK, 2014; pp. 77–87. [Google Scholar]
- Verma, O.P.; Aggarwal, D.; Patodi, T. Opposition and dimensional based modified firefly algorithm. Expert Syst. Appl. 2016, 44, 168–176. [Google Scholar] [CrossRef]
- Emary, E.; Zawbaa, H.M.; Hassanien, A.E. Binary ant lion approaches for feature selection. Neurocomputing 2016, 213, 54–65. [Google Scholar] [CrossRef]
- Storn, R.; Price, K. Differential Evolution—A Simple and Efficient Heuristic for global Optimization over Continuous Spaces. J. Glob. Optim. 1997, 11, 341–359. [Google Scholar] [CrossRef]
- Mirjalili, S. SCA: A Sine Cosine Algorithm for solving optimization problems. Knowl. Based Syst. 2016, 96, 120–133. [Google Scholar] [CrossRef]
- Mirjalili, S. Dragonfly algorithm: A new meta-heuristic optimization technique for solving single-objective, discrete, and multi-objective problems. Neural Comput. Appl. 2016, 27, 1053–1073. [Google Scholar] [CrossRef]
- Mirjalili, S.; Mirjalili, S.M.; Lewis, A. Grey Wolf Optimizer. Adv. Eng. Softw. 2014, 69, 46–61. [Google Scholar] [CrossRef]
- Marinakis, Y.; Marinaki, M.; Dounias, G. Particle swarm optimization for pap-smear diagnosis. Expert Syst. Appl. 2008, 35, 1645–1656. [Google Scholar] [CrossRef]
- Labati, R.D.; Piuri, V.; Scotti, F. All-IDB: The acute lymphoblastic leukemia image database for image processing. In Proceedings of the 2011 18th IEEE International Conference on Image Processing, Brussels, Belgium, 11–14 September 2011; pp. 2045–2048. [Google Scholar]
- Blake, C.; Merz, C. Uci Repository of Machine Learning Databases; University of California: Irvine, CA, USA, 1998. [Google Scholar]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Re-pository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Neoh, S.C.; Srisukkham, W.; Zhang, L.; Todryk, S.; Greystoke, B.; Lim, C.P.; Hossain, M.A.; Aslam, N. An Intelligent Decision Support System for Leukaemia Diagnosis using Microscopic Blood Images. Sci. Rep. 2015, 5, 14938. [Google Scholar] [CrossRef]
- Mahé, P.; Arsac, M.; Chatellier, S.; Monnin, V.; Perrot, N.; Mailler, S.; Girard, V.; Ramjeet, M.; Surre, J.; Lacroix, B.; et al. Automatic identification of mixed bacterial species fingerprints in a MALDI-TOF mass-spectrum. Bioinformatics 2014, 30, 1280–1286. [Google Scholar] [CrossRef]
- Detrano, R.; Janosi, A.; Steinbrunn, W.; Pfisterer, M.; Schmid, J.-J.; Sandhu, S.; Guppy, K.H.; Lee, S.; Froelicher, V. International application of a new probability algorithm for the diagnosis of coronary artery disease. Am. J. Cardiol. 1989, 64, 304–310. [Google Scholar] [CrossRef]
- Latha, C.B.C.; Jeeva, S.C. Improving the accuracy of prediction of heart disease risk based on ensemble classification tech-niques. Inform. Med. Unlocked 2019, 16, 100203. [Google Scholar] [CrossRef]
- Zhang, L.; Srisukkham, W.; Neoh, S.C.; Lim, C.P.; Pandit, D. Classifier ensemble reduction using a modified firefly algorithm: An empirical evaluation. Expert Syst. Appl. 2018, 93, 395–422. [Google Scholar] [CrossRef]
- Long, W.; Jian, J.; Liang, X.; Tang, M. An exploration-enhanced grey wolf optimizer to solve high-dimensional numerical opti-mization. Eng. Appl. Artif. Intell. 2018, 68, 63–80. [Google Scholar] [CrossRef]
- Lawrence, T.; Zhang, L. IoTNet: An Efficient and Accurate Convolutional Neural Network for IoT Devices. Sensors 2019, 19, 5541. [Google Scholar] [CrossRef]
- Xie, H.; Zhang, L.; Lim, C.P.; Yu, Y.; Liu, C.; Liu, H.; Walters, J. Improving K-means clustering with enhanced Firefly Algo-rithms. Appl. Soft Comput. 2019, 84, 105763. [Google Scholar] [CrossRef]
- Zhang, L.; Mistry, K.; Neoh, S.C.; Lim, C.P. Intelligent facial emotion recognition using moth-firefly optimization. Knowl. Based Syst. 2016, 111, 248–267. [Google Scholar] [CrossRef]
- Kinghorn, P.; Zhang, L.; Shao, L. A region-based image caption generator with refined descriptions. Neurocomputing 2018, 272, 416–424. [Google Scholar] [CrossRef]
- Kinghorn, P.; Zhang, L.; Shao, L. A hierarchical and regional deep learning architecture for image description generation. Pattern Recognit. Lett. 2019, 119, 77–85. [Google Scholar] [CrossRef]


| Studies | Population Initialisation | Multiple Leaders | Exemplar Breeding Strategies | Modification of Existing Search Operations | Novel Search Mechanisms | Leader Enhancement | Other Diversity Enhancing Strategies |
|---|---|---|---|---|---|---|---|
| PSO [22] | Random | No (single leader) | No | No (the original PSO operation) | No | No | No |
| Wang et al. [24] | Random | No | No | No | Local and global neighborhood search based on the ring topology | No | Trial particle generation using a crossover factor & a DE operation |
| Lin et al. [32] | Random | No | Ring topology for exemplar generation | The updated PSO operation with the exemplar and the adaptive parameters | No | No | No |
| Chen et al. [31] | Random | No | No | Expansion-contraction coefficient and diversity measurement used in position updating | No | No | Genotype diversity measure and contribution-based fitness evaluation allocation |
| Chang [44] (MFOPSO) | Random | No | No | The search led by each sub-swarm leader | No | No | Multiple sub-swarms |
| Fielding et al. [51] | Random | No | No | Cosine-based adaptive search parameters | No | No | No |
| Srisukkham et al. [42] (BBPSOVA) | Random | The mean of all the personal bests | The average of the local and global best solutions | The average of the local and global optimal signals leading the attraction action | An evading action led by the mean of the worst indicators | No | Two sub-swarms |
| Tan et al. [27] (ALPSO) | Random | Two remote swarm leaders | The best leader and a remote second leader | Using helix search coefficients | Hybridization with SA and DE operations | No | No |
| Chen et al. [41] (GPSO) | Random | No | No | No | No | No | A crossover operator for population diversification |
| Nayak et al. [45] (MPSOELM) | Random | No | No | Using time-varying acceleration coefficients and an adaptive inertia weight | No | No | No |
| Jordehi [34] (ELPSO) | Random | No | No | No | No | 5-staged mutation | No |
| Kang et al. [35] | Random | No | No | A momentum element is used to replace the inertial component. | No | Mutation-based leader enhancement | No |
| Zhang et al. [33] | Random | No | No | No | Local search action using two randomly selected particles with a Gaussian search step | No | Distance-based population diversity estimation |
| Yu et al. [36] | Random | No | Solution selection based on domination relationships and density measurement | No | No | No | Infeasible solution enhancement using Gaussian mutation |
| Chen et al. [19] (HPSO-SSM) | Random | No | No | Using a logistic map to generate the inertia weight | Local exploitation using a spiral search operation | No | Nonlinear coefficients used for velocity updating |
| Cheng and Jin [52] (CSO) | Random | Winners from pairwise competition | No | Using a logarithmic linear regression relationship to generate the coefficient for the social component | Position updating by learning from the winner solution | No | No |
| Vieira et al. [53] (MBPSO) | Random | No | No | No | Resetting the swarm leader by deselecting features, and mutation on personal best solutions by flipping randomly | Using a mirroring operation when the maximum velocity is reached | |
| Chuang et al. [54] (CatfishBPSO) | Random | No | No | No | 10% worst solutions replaced by dimension-wise random assignment | No | No |
| Elbaz et al. [39] | Random | No | No | Using a time-varying adaptive inertia weight and a constriction factor for velocity updating | No | No | No |
| PSOVA1 (This research) | Logistic map | An enhanced hybrid global best signal | Enhancing local and global best solutions using neighboring personal best experiences | The updated PSO operation with enhanced local and global best signals. | Local exploitation using a spiral search operation | Swarm leader enhancement using Gaussian distributions | Mutation and DE-based worst solution enhancement |
| PSOVA2 (This research) | Logistic map | An adaptive exemplar incorporating multiple local and global best solutions | Exemplar generation using adaptive weightings between local and global optimal signals, as well as a dynamic number of local best solutions. | N/A | A new search operation using the exemplar or the swarm leader as the best signal, with search coefficients generated using sine, cosine and hyperbolic tangent functions. | Swarm leader enhancement using an adaptive exponential function | Worst solution enhancement using a hybrid re-dispatching scheme |
| Data Set | Number of Attributes | Number of Classes | Number of Instances |
|---|---|---|---|
| Crohn | 22,283 | 2 | 127 |
| Myeloma | 12,625 | 2 | 173 |
| Arcene | 10,000 | 2 | 200 |
| MicroMass | 1300 | 10 | 360 |
| Parkinsons | 753 | 2 | 756 |
| Activity | 561 | 6 | 1000 |
| Voice | 310 | 2 | 126 |
| Facial Expression | 301 | 2 | 1062 |
| Seizure | 178 | 2 | 4600 |
| ALL | 80 | 2 | 180 |
| Heart | 72 | 4 | 124 |
| Ionosphere | 33 | 2 | 253 |
| Wdbc | 30 | 2 | 569 |
| Algorithm | Parameters |
|---|---|
| PSO [22] | cognitive component = 2, social component = 2, inertial weight = , where and denote the current and maximum iteration numbers, respectively. |
| DE [59] | differential weight , crossover parameter = 0.4. |
| SCA [60] | , where . , , and , , and are four main search parameters. |
| DA [61] | separation factor = 0.1, alignment factor = 0.1, cohesion factor = 0.7, food factor = 1, enemy factor = 1, inertial weight = . |
| GWO [62] | , where is linearly decreasing from 2 to 0, and . , where . and are both coefficient vectors. |
| CSO [52] | , , , where , , and are search parameters randomly selected within [0, 1]. controlling parameter Φ = 0.1. |
| HPSO-SSM [19] | cognitive component = 2, social component = 2, inertial weight = Logistic map. , where is the particle position vector, while is the current iteration, and . . |
| BPSO [63] | cognitive component = 2, social component = 2, = 0.9, = 0.01, inertial weight = . |
| MBPSO [53] | cognitive component = 2, social component = 2, inertial weight = 1.4, mutation probability , where represents the dimensionality of the problem domain. |
| CatfishBPSO [54] | cognitive component = 2, social component = 2, inertial weight = 1, replacing rate = 0.1. |
| GPSO [41] | inertia weight = 0.9, cognitive component = social component = 1.5, crossover probability = 0.7, mutation probability = 0.3. |
| MPSOELM [45] | time-varying acceleration coefficients and an adaptive inertia weight. |
| MFOPSO [44] | inertia weight=0.9, cognitive component = social component = 2. |
| BBPSOVA [42] | search coefficients yielded by Logistic map. |
| ALPSO [27] | inertia weight=0.6, search parameters produced by helix functions. |
| Prop. PSOVA1 | cognitive component = 2, social component = 2, inertial weight = Logistic map, mutation probability threshold = 0.9, = Sinusoidal map. |
| Prop. PSOVA2 | switching probability for exemplar adoption = 0.4, initial weight for = 0.4, search coefficients implemented using exponential, sine, cosine, and hyperbolic tangent functions. |
| Data Sets | Metrics | PSO | DE | SCA | DA | GWO | CSO | HPSO-SSM | Catfish-BPSO | Prop. PSOVA1 | Prop. PSOVA2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Crohn | mean | 0.7556 | 0.7624 | 0.7479 | 0.7427 | 0.7786 | 0.7197 | 0.7675 | 0.7803 | 0.8128 | 0.8333 |
| std. | 6.74E-02 | 3.10E-02 | 3.18E-02 | 3.28E-02 | 3.07E-02 | 7.16E-02 | 3.10E-02 | 3.73E-02 | 2.90E-02 | 3.09E-02 | |
| Myeloma | mean | 0.7096 | 0.7288 | 0.7013 | 0.7032 | 0.7212 | 0.6917 | 0.7128 | 0.6910 | 0.7442 | 0.7545 |
| std. | 2.60E-02 | 2.29E-02 | 2.03E-02 | 2.42E-02 | 2.37E-02 | 6.01E-02 | 2.48E-02 | 1.56E-02 | 2.68E-02 | 2.66E-02 | |
| Arcene | mean | 0.7217 | 0.7244 | 0.7372 | 0.7183 | 0.7211 | 0.7372 | 0.7122 | 0.7100 | 0.7411 | 0.7694 |
| std. | 2.66E-02 | 2.78E-02 | 3.98E-02 | 3.71E-02 | 2.95E-02 | 3.79E-02 | 3.28E-02 | 3.77E-02 | 2.81E-02 | 3.58E-02 | |
| MicroMass | mean | 0.5897 | 0.6052 | 0.6061 | 0.5933 | 0.6124 | 0.5409 | 0.5903 | 0.5836 | 0.6455 | 0.6612 |
| std. | 4.34E-02 | 3.85E-02 | 5.13E-02 | 4.07E-02 | 4.38E-02 | 2.79E-02 | 4.12E-02 | 3.92E-02 | 4.59E-02 | 4.38E-02 | |
| Parkinsons | mean | 0.7949 | 0.7990 | 0.7922 | 0.7862 | 0.7940 | 0.7985 | 0.8000 | 0.7994 | 0.8115 | 0.8094 |
| std. | 1.74E-02 | 1.63E-02 | 2.48E-02 | 2.15E-02 | 1.91E-02 | 1.30E-02 | 1.77E-02 | 1.56E-02 | 1.88E-02 | 1.60E-02 | |
| Activity | mean | 0.8813 | 0.8919 | 0.8826 | 0.8785 | 0.8929 | 0.8876 | 0.8860 | 0.8785 | 0.9025 | 0.9117 |
| std. | 1.64E-02 | 1.55E-02 | 1.86E-02 | 2.23E-02 | 1.44E-02 | 1.60E-02 | 1.95E-02 | 1.42E-02 | 1.28E-02 | 1.53E-02 | |
| Voice | mean | 0.8237 | 0.8149 | 0.8202 | 0.8272 | 0.8219 | 0.7789 | 0.8237 | 0.8193 | 0.8526 | 0.8632 |
| std. | 5.00E-02 | 5.58E-02 | 4.66E-02 | 5.83E-02 | 5.42E-02 | 8.37E-02 | 5.09E-02 | 3.95E-02 | 4.28E-02 | 4.37E-02 | |
| Facial Expression | mean | 0.7187 | 0.6748 | 0.6891 | 0.6635 | 0.6844 | 0.6861 | 0.6914 | 0.6998 | 0.7351 | 0.7340 |
| std. | 4.64E-02 | 4.70E-02 | 4.05E-02 | 3.37E-02 | 4.68E-02 | 5.14E-02 | 3.86E-02 | 4.21E-02 | 4.60E-02 | 4.24E-02 | |
| Seizure | mean | 0.8459 | 0.8590 | 0.8543 | 0.8577 | 0.8655 | 0.8490 | 0.8461 | 0.8516 | 0.8698 | 0.8860 |
| std. | 5.08E-03 | 6.69E-03 | 1.12E-02 | 1.00E-02 | 2.01E-02 | 9.22E-03 | 5.28E-03 | 8.12E-03 | 5.13E-03 | 6.12E-03 | |
| ALL | mean | 0.8951 | 0.9167 | 0.9037 | 0.9025 | 0.8858 | 0.8728 | 0.8944 | 0.9123 | 0.9185 | 0.9241 |
| std. | 2.84E-02 | 2.69E-02 | 2.21E-02 | 1.91E-02 | 4.25E-02 | 5.59E-02 | 4.76E-02 | 3.28E-02 | 3.23E-02 | 3.26E-02 | |
| Heart | mean | 0.5963 | 0.6435 | 0.6620 | 0.5537 | 0.6398 | 0.5713 | 0.6444 | 0.5769 | 0.6731 | 0.7241 |
| std. | 8.33E-02 | 5.18E-02 | 5.56E-02 | 6.13E-02 | 6.35E-02 | 4.34E-02 | 4.83E-02 | 7.16E-02 | 4.63E-02 | 5.42E-02 | |
| Ionosphere | mean | 0.8171 | 0.8285 | 0.8320 | 0.8101 | 0.8197 | 0.8184 | 0.8189 | 0.8066 | 0.8351 | 0.8434 |
| std. | 2.70E-02 | 3.10E-02 | 2.94E-02 | 2.62E-02 | 2.28E-02 | 2.89E-02 | 2.60E-02 | 2.89E-02 | 2.49E-02 | 2.16E-02 | |
| Wdbc | mean | 0.9520 | 0.9534 | 0.9191 | 0.9458 | 0.9386 | 0.8828 | 0.9261 | 0.9497 | 0.9571 | 0.9585 |
| std. | 1.04E-02 | 1.60E-02 | 4.19E-02 | 2.36E-02 | 3.30E-02 | 3.33E-02 | 3.60E-02 | 1.67E-02 | 1.33E-02 | 9.59E-03 | |
| Data sets | Metrics | BPSO | MBPSO | GPSO | MPSO-ELM | MFO-PSO | BBPSO-VA | ALPSO | Prop. PSOVA1 | Prop. PSOVA2 | |
| Crohn | mean | 0.7427 | 0.7795 | 0.7504 | 0.7479 | 0.7726 | 0.7684 | 0.7889 | 0.8128 | 0.8333 | |
| std. | 3.00E-02 | 2.25E-02 | 1.86E-02 | 3.45E-02 | 3.67E-02 | 3.00E-02 | 3.08E-02 | 2.90E-02 | 3.09E-02 | ||
| Myeloma | mean | 0.6942 | 0.7051 | 0.7045 | 0.6917 | 0.7154 | 0.7128 | 0.7051 | 0.7442 | 0.7545 | |
| std. | 2.13E-02 | 1.94E-02 | 2.30E-02 | 2.56E-02 | 2.24E-02 | 2.81E-02 | 1.94E-02 | 2.68E-02 | 2.66E-02 | ||
| Arcene | mean | 0.7111 | 0.7117 | 0.7022 | 0.7106 | 0.7372 | 0.7200 | 0.7161 | 0.7411 | 0.7694 | |
| std. | 3.53E-02 | 2.79E-02 | 3.54E-02 | 3.18E-02 | 3.62E-02 | 4.23E-02 | 2.80E-02 | 2.81E-02 | 3.58E-02 | ||
| MicroMass | mean | 0.5758 | 0.5785 | 0.6052 | 0.5879 | 0.5915 | 0.6118 | 0.5994 | 0.6455 | 0.6612 | |
| std. | 3.58E-02 | 4.01E-02 | 3.46E-02 | 4.31E-02 | 4.77E-02 | 3.99E-02 | 5.30E-02 | 4.59E-02 | 4.38E-02 | ||
| Parkinsons | mean | 0.7988 | 0.7962 | 0.7953 | 0.7890 | 0.7822 | 0.7907 | 0.7950 | 0.8115 | 0.8094 | |
| std. | 1.97E-02 | 1.97E-02 | 1.85E-02 | 1.84E-02 | 2.49E-02 | 1.97E-02 | 2.00E-02 | 1.88E-02 | 1.60E-02 | ||
| Activity | mean | 0.8725 | 0.8775 | 0.8864 | 0.8785 | 0.8806 | 0.8848 | 0.8810 | 0.9025 | 0.9117 | |
| std. | 1.59E-02 | 1.15E-02 | 1.23E-02 | 1.54E-02 | 1.80E-02 | 1.52E-02 | 1.74E-02 | 1.28E-02 | 1.53E-02 | ||
| Voice | mean | 0.8263 | 0.8246 | 0.8526 | 0.8298 | 0.8377 | 0.8439 | 0.8175 | 0.8526 | 0.8632 | |
| std. | 4.43E-02 | 4.72E-02 | 5.07E-02 | 3.83E-02 | 7.03E-02 | 5.72E-02 | 6.19E-02 | 4.28E-02 | 4.37E-02 | ||
| Facial Expression | mean | 0.7170 | 0.7274 | 0.7177 | 0.7234 | 0.7031 | 0.7061 | 0.7032 | 0.7351 | 0.7340 | |
| std. | 3.56E-02 | 3.93E-02 | 4.33E-02 | 4.67E-02 | 5.89E-02 | 4.62E-02 | 4.40E-02 | 4.60E-02 | 4.24E-02 | ||
| Seizure | mean | 0.8370 | 0.8388 | 0.8492 | 0.8400 | 0.8519 | 0.8496 | 0.8430 | 0.8698 | 0.8860 | |
| std. | 4.74E-03 | 4.41E-03 | 5.62E-03 | 5.84E-03 | 5.14E-03 | 6.87E-03 | 7.06E-03 | 5.13E-03 | 6.12E-03 | ||
| ALL | mean | 0.8938 | 0.8988 | 0.9068 | 0.8981 | 0.9000 | 0.9019 | 0.9025 | 0.9185 | 0.9241 | |
| std. | 1.97E-02 | 3.32E-02 | 2.68E-02 | 2.78E-02 | 2.64E-02 | 2.25E-02 | 2.62E-02 | 3.23E-02 | 3.26E-02 | ||
| Heart | mean | 0.5815 | 0.5750 | 0.5944 | 0.5991 | 0.6426 | 0.6250 | 0.6333 | 0.6731 | 0.7241 | |
| std. | 5.91E-02 | 6.50E-02 | 5.67E-02 | 7.30E-02 | 8.86E-02 | 6.87E-02 | 7.29E-02 | 4.63E-02 | 5.42E-02 | ||
| Ionosphere | mean | 0.8276 | 0.8110 | 0.8189 | 0.8140 | 0.8171 | 0.8228 | 0.8197 | 0.8351 | 0.8434 | |
| std. | 2.60E-02 | 3.27E-02 | 2.03E-02 | 3.18E-02 | 2.83E-02 | 2.30E-02 | 2.26E-02 | 2.49E-02 | 2.16E-02 | ||
| Wdbc | mean | 0.9501 | 0.9454 | 0.9517 | 0.9481 | 0.9509 | 0.9540 | 0.9501 | 0.9571 | 0.9585 | |
| std. | 1.10E-02 | 2.12E-02 | 9.18E-03 | 1.63E-02 | 1.31E-02 | 1.06E-02 | 1.24E-02 | 1.33E-02 | 9.59E-03 | ||
| Data Sets | Metrics | PSO | DE | SCA | DA | GWO | CSO | HPSO-SSM | Catfish-BPSO | Prop. PSOVA1 | Prop. PSOVA2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Crohn | mean | 0.8202 | 0.8052 | 0.7943 | 0.7906 | 0.8236 | 0.7765 | 0.8137 | 0.8263 | 0.8550 | 0.8585 |
| std. | 3.69E-02 | 2.42E-02 | 2.44E-02 | 2.52E-02 | 2.42E-02 | 5.71E-02 | 2.39E-02 | 2.90E-02 | 2.24E-02 | 2.43E-02 | |
| Myeloma | mean | 0.8219 | 0.8411 | 0.8091 | 0.8105 | 0.8286 | 0.8026 | 0.8229 | 0.8034 | 0.8500 | 0.8551 |
| std. | 1.64E-02 | 1.45E-02 | 1.27E-02 | 1.47E-02 | 1.40E-02 | 4.68E-02 | 1.58E-02 | 1.01E-02 | 1.57E-02 | 1.63E-02 | |
| Arcene | mean | 0.6759 | 0.6757 | 0.6963 | 0.6780 | 0.6783 | 0.6959 | 0.6646 | 0.6574 | 0.6977 | 0.7130 |
| std. | 3.85E-02 | 3.87E-02 | 4.94E-02 | 4.89E-02 | 3.31E-02 | 5.40E-02 | 4.60E-02 | 5.14E-02 | 3.16E-02 | 4.18E-02 | |
| MicroMass | mean | 0.6349 | 0.6469 | 0.6428 | 0.6314 | 0.6445 | 0.5982 | 0.6350 | 0.6275 | 0.6759 | 0.6967 |
| std. | 4.26E-02 | 3.48E-02 | 4.79E-02 | 3.94E-02 | 4.36E-02 | 2.17E-02 | 4.21E-02 | 3.88E-02 | 4.19E-02 | 4.03E-02 | |
| Parkinsons | mean | 0.8691 | 0.8712 | 0.8670 | 0.8631 | 0.8686 | 0.8701 | 0.8720 | 0.8719 | 0.8798 | 0.8783 |
| std. | 1.15E-02 | 1.10E-02 | 1.73E-02 | 1.41E-02 | 1.33E-02 | 8.93E-03 | 1.13E-02 | 1.01E-02 | 1.32E-02 | 1.02E-02 | |
| Activity | mean | 0.8864 | 0.8962 | 0.8874 | 0.8833 | 0.8971 | 0.8930 | 0.8901 | 0.8838 | 0.9067 | 0.9131 |
| std. | 1.53E-02 | 1.49E-02 | 1.76E-02 | 2.16E-02 | 1.37E-02 | 1.65E-02 | 1.90E-02 | 1.34E-02 | 1.24E-02 | 1.44E-02 | |
| Voice | mean | 0.7180 | 0.7381 | 0.7265 | 0.7316 | 0.7208 | 0.6890 | 0.7339 | 0.7328 | 0.7764 | 0.7852 |
| std. | 9.23E-02 | 7.09E-02 | 8.03E-02 | 1.07E-01 | 9.79E-02 | 1.06E-01 | 8.13E-02 | 7.23E-02 | 6.94E-02 | 7.54E-02 | |
| Facial Expression | mean | 0.6458 | 0.6191 | 0.6288 | 0.6175 | 0.6287 | 0.5670 | 0.6292 | 0.6342 | 0.6572 | 0.6562 |
| std. | 3.18E-02 | 3.10E-02 | 2.51E-02 | 1.86E-02 | 3.14E-02 | 1.92E-01 | 2.54E-02 | 2.81E-02 | 3.02E-02 | 2.87E-02 | |
| Seizure | mean | 0.8197 | 0.8384 | 0.8359 | 0.8364 | 0.8486 | 0.8434 | 0.8199 | 0.8279 | 0.8526 | 0.8759 |
| std. | 7.33E-03 | 8.96E-03 | 1.50E-02 | 1.41E-02 | 2.90E-02 | 9.36E-03 | 8.21E-03 | 1.11E-02 | 8.08E-03 | 8.76E-03 | |
| ALL | mean | 0.9204 | 0.9345 | 0.9250 | 0.9266 | 0.9084 | 0.9037 | 0.9168 | 0.9331 | 0.9361 | 0.9408 |
| std. | 2.28E-02 | 2.17E-02 | 1.62E-02 | 1.37E-02 | 3.93E-02 | 4.34E-02 | 4.37E-02 | 2.51E-02 | 2.67E-02 | 2.60E-02 | |
| Heart | mean | 0.6039 | 0.6502 | 0.6661 | 0.5616 | 0.6436 | 0.5823 | 0.6513 | 0.5881 | 0.6783 | 0.7271 |
| std. | 8.59E-02 | 5.25E-02 | 5.68E-02 | 6.81E-02 | 6.72E-02 | 4.53E-02 | 4.88E-02 | 7.28E-02 | 4.63E-02 | 5.49E-02 | |
| Ionosphere | mean | 0.8439 | 0.8516 | 0.8550 | 0.8375 | 0.8427 | 0.8418 | 0.8452 | 0.8371 | 0.8562 | 0.8625 |
| std. | 2.06E-02 | 2.48E-02 | 2.33E-02 | 2.04E-02 | 2.23E-02 | 2.52E-02 | 2.05E-02 | 2.18E-02 | 2.05E-02 | 1.77E-02 | |
| Wdbc | mean | 0.9340 | 0.9355 | 0.8836 | 0.9246 | 0.9146 | 0.8286 | 0.8957 | 0.9308 | 0.9415 | 0.9432 |
| std. | 1.47E-02 | 2.34E-02 | 6.53E-02 | 3.57E-02 | 4.84E-02 | 5.04E-02 | 5.38E-02 | 2.47E-02 | 1.94E-02 | 1.31E-02 | |
| Data sets | Metrics | BPSO | MBPSO | GPSO | MPSO-ELM | MFO-PSO | BBPSO-VA | ALPSO | Prop. PSOVA1 | Prop. PSOVA2 | |
| Crohn | mean | 0.7889 | 0.8220 | 0.7945 | 0.7937 | 0.8188 | 0.8153 | 0.8306 | 0.8550 | 0.8585 | |
| std. | 2.19E-02 | 1.68E-02 | 1.35E-02 | 2.53E-02 | 2.89E-02 | 2.29E-02 | 2.40E-02 | 2.24E-02 | 2.43E-02 | ||
| Myeloma | mean | 0.8057 | 0.8189 | 0.8186 | 0.8031 | 0.8248 | 0.8234 | 0.8189 | 0.8500 | 0.8551 | |
| std. | 1.32E-02 | 1.23E-02 | 1.41E-02 | 1.58E-02 | 1.39E-02 | 1.74E-02 | 1.22E-02 | 1.57E-02 | 1.63E-02 | ||
| Arcene | mean | 0.6573 | 0.6590 | 0.6460 | 0.6602 | 0.6985 | 0.6732 | 0.6673 | 0.6977 | 0.7130 | |
| std. | 4.56E-02 | 3.58E-02 | 4.65E-02 | 4.13E-02 | 4.78E-02 | 5.20E-02 | 2.36E-02 | 3.16E-02 | 4.18E-02 | ||
| MicroMass | mean | 0.6219 | 0.6200 | 0.6451 | 0.6360 | 0.6308 | 0.6449 | 0.6444 | 0.6759 | 0.6967 | |
| std. | 4.05E-02 | 3.59E-02 | 3.05E-02 | 3.95E-02 | 4.30E-02 | 3.92E-02 | 5.00E-02 | 4.19E-02 | 4.03E-02 | ||
| Parkinsons | mean | 0.8716 | 0.8702 | 0.8695 | 0.8656 | 0.8612 | 0.8662 | 0.8688 | 0.8798 | 0.8783 | |
| std. | 1.30E-02 | 1.31E-02 | 1.26E-02 | 1.22E-02 | 1.70E-02 | 1.32E-02 | 5.02E-02 | 1.32E-02 | 1.02E-02 | ||
| Activity | mean | 0.8783 | 0.8824 | 0.8913 | 0.8842 | 0.8854 | 0.8895 | 0.8854 | 0.9067 | 0.9131 | |
| std. | 1.55E-02 | 1.14E-02 | 1.16E-02 | 1.47E-02 | 1.61E-02 | 1.45E-02 | 1.70E-02 | 1.24E-02 | 1.44E-02 | ||
| Voice | mean | 0.7368 | 0.7399 | 0.7804 | 0.7398 | 0.7598 | 0.7656 | 0.7272 | 0.7764 | 0.7852 | |
| std. | 7.58E-02 | 7.76E-02 | 7.78E-02 | 6.51E-02 | 1.06E-01 | 8.83E-02 | 5.07E-02 | 6.94E-02 | 7.54E-02 | ||
| Facial Expression | mean | 0.6527 | 0.6556 | 0.6372 | 0.6537 | 0.6404 | 0.6360 | 0.6371 | 0.6572 | 0.6562 | |
| std. | 2.63E-02 | 2.93E-02 | 3.04E-02 | 3.50E-02 | 4.15E-02 | 2.97E-02 | 4.81E-02 | 3.02E-02 | 2.87E-02 | ||
| Seizure | mean | 0.8066 | 0.8094 | 0.8243 | 0.8111 | 0.8282 | 0.8251 | 0.8155 | 0.8526 | 0.8759 | |
| std. | 6.61E-03 | 6.33E-03 | 7.72E-03 | 8.31E-03 | 6.98E-03 | 9.39E-03 | 9.87E-03 | 8.08E-03 | 8.76E-03 | ||
| ALL | mean | 0.9195 | 0.9241 | 0.9283 | 0.9237 | 0.9244 | 0.9215 | 0.9253 | 0.9361 | 0.9408 | |
| std. | 1.55E-02 | 2.44E-02 | 2.07E-02 | 2.02E-02 | 1.96E-02 | 1.94E-02 | 3.53E-02 | 2.67E-02 | 2.60E-02 | ||
| Heart | mean | 0.5904 | 0.5788 | 0.6006 | 0.6166 | 0.6442 | 0.6319 | 0.6381 | 0.6783 | 0.7271 | |
| std. | 6.62E-02 | 7.86E-02 | 6.33E-02 | 7.14E-02 | 8.75E-02 | 6.94E-02 | 7.52E-02 | 4.63E-02 | 5.49E-02 | ||
| Ionosphere | mean | 0.8521 | 0.8380 | 0.8452 | 0.8419 | 0.8426 | 0.8476 | 0.8453 | 0.8562 | 0.8625 | |
| std. | 1.82E-02 | 2.51E-02 | 1.50E-02 | 2.44E-02 | 2.45E-02 | 1.89E-02 | 3.31E-02 | 2.05E-02 | 1.77E-02 | ||
| Wdbc | mean | 0.9312 | 0.9239 | 0.9338 | 0.9286 | 0.9325 | 0.9366 | 0.9321 | 0.9415 | 0.9432 | |
| std. | 1.55E-02 | 3.10E-02 | 1.29E-02 | 2.33E-02 | 1.85E-02 | 1.53E-02 | 9.79E-03 | 1.94E-02 | 1.31E-02 | ||
| Data Sets | PSO | DE | SCA | DA | GWO | CSO | HPSO-SSM | Catfish-BPSO |
|---|---|---|---|---|---|---|---|---|
| Crohn | 2.25E-04 | 4.45E-07 | 5.04E-09 | 4.90E-10 | 9.82E-05 | 4.42E-08 | 2.14E-06 | 6.80E-04 |
| Myeloma | 1.35E-05 | 3.40E-02 | 1.25E-07 | 1.26E-06 | 9.24E-04 | 8.59E-05 | 7.27E-05 | 6.63E-10 |
| Arcene | 1.53E-02 | 3.53E-02 | 8.75E-01 | 2.44E-02 | 1.93E-02 | 6.16E-01 | 1.48E-03 | 4.41E-04 |
| MicroMass | 2.47E-04 | 7.55E-03 | 8.69E-03 | 3.50E-04 | 4.11E-02 | 1.05E-09 | 2.12E-04 | 2.90E-05 |
| Parkinsons | 1.65E-03 | 3.15E-02 | 6.60E-03 | 1.99E-05 | 2.38E-03 | 3.35E-02 | 4.69E-02 | 4.52E-02 |
| Activity | 3.93E-06 | 6.61E-03 | 1.27E-04 | 1.40E-05 | 1.19E-02 | 4.51E-05 | 1.05E-03 | 1.49E-07 |
| Voice | 3.21E-02 | 6.20E-03 | 9.98E-03 | 4.48E-02 | 2.78E-02 | 9.85E-04 | 3.35E-02 | 4.04E-03 |
| Facial Expression | 5.24E-01 | 8.72E-05 | 1.23E-03 | 1.75E-06 | 5.63E-04 | 4.14E-05 | 5.06E-04 | 4.69E-03 |
| Seizure | 3.07E-11 | 1.16E-03 | 3.33E-05 | 2.05E-04 | 5.49E-01 | 1.23E-08 | 7.52E-11 | 1.23E-07 |
| ALL | 7.85E-03 | 7.75E-01 | 4.79E-02 | 2.92E-02 | 3.45E-03 | 1.35E-03 | 3.82E-02 | 4.76E-01 |
| Heart | 1.44E-04 | 2.16E-02 | 2.94E-01 | 2.20E-09 | 3.15E-02 | 1.21E-09 | 3.84E-02 | 1.29E-06 |
| Ionosphere | 1.16E-02 | 6.10E-01 | 8.11E-01 | 1.15E-03 | 4.18E-02 | 3.82E-02 | 2.77E-02 | 2.06E-04 |
| Wdbc | 2.48E-02 | 5.23E-01 | 3.02E-05 | 1.30E-02 | 3.54E-02 | 5.44E-09 | 1.84E-04 | 1.84E-02 |
| Data sets | BPSO | MBPSO | GPSO | MPSOELM | MFOPSO | BBPSOVA | ALPSO | |
| Crohn | 1.45E-09 | 2.47E-05 | 2.25E-10 | 7.14E-09 | 2.94E-05 | 3.11E-06 | 4.13E-03 | |
| Myeloma | 1.44E-08 | 5.89E-07 | 1.00E-06 | 2.53E-08 | 1.14E-04 | 1.73E-04 | 4.49E-07 | |
| Arcene | 6.08E-04 | 6.28E-04 | 5.12E-05 | 6.38E-04 | 8.34E-01 | 2.15E-02 | 3.18E-03 | |
| MicroMass | 5.30E-06 | 1.13E-05 | 5.31E-03 | 1.38E-04 | 6.88E-04 | 1.99E-02 | 4.66E-03 | |
| Parkinsons | 3.93E-02 | 3.31E-02 | 4.74E-03 | 4.31E-04 | 4.41E-05 | 3.81E-04 | 6.72E-03 | |
| Activity | 1.07E-08 | 2.12E-08 | 3.99E-05 | 2.71E-07 | 6.55E-06 | 4.71E-05 | 9.65E-06 | |
| Voice | 2.91E-02 | 1.83E-02 | 8.87E-01 | 3.84E-02 | 4.52E-01 | 6.03E-01 | 1.61E-02 | |
| Facial Expression | 1.92E-02 | 3.40E-01 | 3.56E-02 | 3.28E-01 | 4.65E-02 | 1.45E-02 | 4.03E-02 | |
| Seizure | 2.92E-11 | 2.91E-11 | 4.85E-10 | 2.89E-11 | 3.40E-09 | 2.96E-09 | 4.62E-11 | |
| ALL | 1.98E-03 | 3.11E-02 | 1.29E-01 | 1.77E-02 | 2.38E-02 | 3.03E-02 | 4.75E-02 | |
| Heart | 2.87E-07 | 1.26E-07 | 1.56E-06 | 4.88E-05 | 4.85E-02 | 2.28E-03 | 9.35E-03 | |
| Ionosphere | 7.87E-01 | 4.58E-03 | 2.04E-02 | 3.37E-02 | 2.40E-02 | 1.22E-01 | 3.59E-02 | |
| Wdbc | 1.82E-02 | 4.16E-03 | 2.61E-02 | 4.01E-02 | 4.90E-02 | 2.13E-01 | 4.50E-02 | |
| Data Sets | PSO | DE | SCA | DA | GWO | CSO | HPSO-SSM | Catfish-BPSO |
|---|---|---|---|---|---|---|---|---|
| Crohn | 5.69E-05 | 2.58E-07 | 4.33E-09 | 8.51E-10 | 4.23E-05 | 2.57E-08 | 1.19E-06 | 2.44E-04 |
| Myeloma | 2.30E-07 | 5.30E-04 | 1.13E-09 | 1.12E-08 | 8.27E-06 | 2.13E-06 | 6.73E-07 | 5.56E-11 |
| Arcene | 1.83E-06 | 5.23E-06 | 1.85E-03 | 3.00E-06 | 2.25E-06 | 7.92E-04 | 6.24E-07 | 1.48E-06 |
| MicroMass | 1.43E-06 | 4.26E-05 | 2.06E-04 | 6.32E-06 | 6.27E-04 | 5.29E-11 | 2.36E-06 | 3.98E-07 |
| Parkinsons | 1.48E-03 | 2.70E-02 | 2.87E-03 | 6.40E-05 | 1.92E-03 | 1.06E-02 | 4.84E-02 | 3.30E-02 |
| Activity | 6.47E-08 | 3.65E-05 | 1.19E-06 | 1.79E-07 | 4.88E-05 | 1.24E-07 | 7.41E-06 | 2.49E-09 |
| Voice | 8.81E-03 | 2.39E-03 | 3.45E-03 | 2.01E-02 | 7.35E-03 | 1.86E-04 | 8.08E-03 | 1.67E-03 |
| Facial Expression | 4.15E-01 | 2.59E-05 | 1.04E-03 | 2.12E-07 | 3.30E-04 | 1.14E-04 | 2.71E-04 | 6.89E-03 |
| Seizure | 2.94E-11 | 7.60E-11 | 1.18E-10 | 1.21E-09 | 9.69E-04 | 2.97E-11 | 2.95E-11 | 3.98E-11 |
| ALL | 2.22E-03 | 5.08E-01 | 1.65E-02 | 1.03E-02 | 1.00E-03 | 3.80E-04 | 1.52E-02 | 2.86E-01 |
| Heart | 6.52E-07 | 8.33E-07 | 5.36E-05 | 1.30E-09 | 5.29E-06 | 1.30E-09 | 1.32E-07 | 1.09E-08 |
| Ionosphere | 2.53E-04 | 3.50E-02 | 1.00E-01 | 7.89E-06 | 3.26E-04 | 6.06E-04 | 5.16E-04 | 4.17E-06 |
| Wdbc | 1.33E-02 | 3.68E-01 | 1.84E-05 | 5.02E-03 | 1.93E-02 | 5.09E-09 | 1.22E-04 | 1.05E-02 |
| Data sets | BPSO | MBPSO | GPSO | MPSOELM | MFOPSO | BBPSOVA | ALPSO | |
| Crohn | 1.41E-09 | 1.23E-05 | 3.38E-10 | 6.63E-09 | 1.64E-05 | 1.36E-06 | 1.35E-03 | |
| Myeloma | 4.06E-10 | 3.70E-09 | 1.76E-08 | 1.21E-09 | 1.09E-06 | 3.52E-06 | 3.78E-09 | |
| Arcene | 7.05E-07 | 1.51E-07 | 1.40E-07 | 3.06E-07 | 6.70E-04 | 4.40E-05 | 4.58E-07 | |
| MicroMass | 2.43E-09 | 9.54E-08 | 1.13E-05 | 4.80E-07 | 9.00E-06 | 3.26E-04 | 9.12E-05 | |
| Parkinsons | 2.97E-02 | 1.20E-02 | 5.39E-03 | 5.00E-05 | 1.90E-05 | 2.32E-04 | 5.81E-03 | |
| Activity | 6.21E-10 | 8.34E-10 | 1.42E-07 | 4.88E-09 | 3.97E-08 | 2.17E-07 | 6.88E-08 | |
| Voice | 8.18E-03 | 7.50E-03 | 3.97E-01 | 1.16E-02 | 1.47E-01 | 2.22E-01 | 5.33E-03 | |
| Facial Expression | 6.16E-02 | 4.91E-01 | 4.89E-02 | 3.94E-01 | 4.64E-02 | 2.83E-02 | 3.61E-02 | |
| Seizure | 2.93E-11 | 2.92E-11 | 3.04E-11 | 2.90E-11 | 2.96E-11 | 2.95E-11 | 2.97E-11 | |
| ALL | 5.91E-04 | 1.09E-02 | 5.54E-02 | 5.70E-03 | 6.72E-03 | 1.44E-02 | 2.55E-02 | |
| Heart | 5.54E-09 | 1.76E-08 | 1.96E-08 | 1.02E-07 | 1.10E-04 | 1.18E-06 | 1.35E-05 | |
| Ionosphere | 2.88E-02 | 5.43E-05 | 1.26E-04 | 4.39E-04 | 2.69E-04 | 1.66E-03 | 3.35E-04 | |
| Wdbc | 4.84E-03 | 2.25E-03 | 8.69E-03 | 1.14E-02 | 2.00E-02 | 1.10E-01 | 1.07E-02 | |
| Data Sets | PSO | DE | SCA | DA | GWO | CSO | HPSO-SSM | Catfish-BPSO | Prop. PSOVA1 | Prop. PSOVA2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Crohn | 9468.8 | 8942.4 | 7594.7 | 8423.4 | 6292.6 | 1151.5 | 8846.2 | 9364.5 | 7026.6 | 7697.6 |
| Myeloma | 5654.6 | 5130.2 | 4462.9 | 4740.1 | 3680.3 | 1633.5 | 5236.9 | 5476.4 | 4059.0 | 4264.5 |
| Arcene | 3976.1 | 4046.1 | 3388.6 | 3695.4 | 2770.4 | 2545.3 | 3967.2 | 4424.8 | 3395.0 | 3412.4 |
| MicroMass | 548.6 | 527.2 | 439.8 | 485.9 | 330.6 | 1123.0 | 554.3 | 588.8 | 461.3 | 476.8 |
| Parkinsons | 323.3 | 310.2 | 266.3 | 283.2 | 209.8 | 492.0 | 323.6 | 361.6 | 273.1 | 274.8 |
| Activity | 237.6 | 222.9 | 184.0 | 208.2 | 146.3 | 394.4 | 232.7 | 255.7 | 194.0 | 185.5 |
| Voice | 128.0 | 121.4 | 108.3 | 118.1 | 86.7 | 65.0 | 122.0 | 140.2 | 108.6 | 109.5 |
| Facial Expression | 131.4 | 112.8 | 88.4 | 72.0 | 80.7 | 60.1 | 84.6 | 121.6 | 92.7 | 97.7 |
| Seizure | 61.0 | 38.4 | 25.3 | 33.4 | 19.7 | 5.1 | 58.0 | 39.7 | 19.4 | 12.2 |
| ALL | 26.5 | 23.0 | 18.4 | 29.5 | 12.8 | 9.5 | 25.4 | 28.8 | 19.0 | 15.8 |
| Heart | 28.8 | 23.9 | 20.9 | 27.8 | 17.8 | 56.7 | 26.4 | 31.9 | 21.8 | 24.6 |
| Ionosphere | 12.5 | 9.3 | 9.6 | 11.8 | 9.4 | 9.6 | 11.3 | 13.1 | 10.3 | 6.9 |
| Wdbc | 9.9 | 5.5 | 3.9 | 9.4 | 4.73 | 3.4 | 4.7 | 10.4 | 9.8 | 7.9 |
| Data sets | BPSO | MBPSO | GPSO | MPSO-ELM | MFO-PSO | BBPSO-VA | ALPSO | Prop. PSOVA1 | Prop. PSOVA2 | |
| Crohn | 11,134.8 | 11,106.7 | 10,030.2 | 10,188.4 | 6886.1 | 9093.0 | 9178.7 | 7026.6 | 7697.6 | |
| Myeloma | 6298.8 | 6299.0 | 5817.9 | 5924.5 | 4073.2 | 5299.6 | 5191.3 | 4059.0 | 4264.5 | |
| Arcene | 4977.2 | 4974.0 | 4484.6 | 4541.9 | 3014.3 | 4078.2 | 4051.2 | 3395.0 | 3412.4 | |
| MicroMass | 646.2 | 641.5 | 611.5 | 619.5 | 439.6 | 562.1 | 569.5 | 461.3 | 476.8 | |
| Parkinsons | 378.1 | 374.4 | 356.4 | 360.8 | 260.2 | 327.0 | 310.0 | 273.1 | 274.8 | |
| Activity | 277.2 | 277.8 | 261.4 | 272.9 | 195.1 | 237.8 | 241.7 | 194.0 | 185.5 | |
| Voice | 152.9 | 148.2 | 140.0 | 147.3 | 101.9 | 131.1 | 134.4 | 108.6 | 109.5 | |
| Facial Expression | 146.2 | 142.0 | 129.4 | 135.2 | 95.7 | 122.6 | 115.7 | 92.7 | 97.7 | |
| Seizure | 80.1 | 74.5 | 57.2 | 68.6 | 38.7 | 49.9 | 54.4 | 19.4 | 12.2 | |
| ALL | 35.4 | 33.3 | 27.9 | 33.6 | 23.1 | 23.7 | 31.6 | 19.0 | 15.8 | |
| Heart | 34.0 | 30.9 | 32.0 | 35.0 | 25.1 | 27.4 | 32.0 | 21.8 | 24.6 | |
| Ionosphere | 12.5 | 10.6 | 9.1 | 13.3 | 8.6 | 9.1 | 9.9 | 10.3 | 6.9 | |
| Wdbc | 10.8 | 6.8 | 9.1 | 11.8 | 7.6 | 8.6 | 9.6 | 9.8 | 7.9 | |
| Data Sets | PSO | DE | SCA | DA | GWO | CSO | HPSO-SSM | Catfish BPSO | Prop. PSOVA1 | Prop. PSOVA2 |
|---|---|---|---|---|---|---|---|---|---|---|
| Crohn | 3.60E-01 | 3.16E-01 | 3.57E-01 | 3.50E-01 | 2.96E-01 | 2.91E-01 | 3.18E-01 | 3.25E-01 | 3.47E-01 | 3.17E-01 |
| Myeloma | 3.00E-01 | 2.78E-01 | 3.10E-01 | 2.90E-01 | 2.48E-01 | 3.00E-01 | 2.80E-01 | 2.88E-01 | 2.90E-01 | 2.66E-01 |
| Seizure | 1.24E+01 | 1.24E+01 | 1.25E+01 | 1.25E+01 | 1.25E+01 | 1.25E+01 | 1.33E+01 | 1.24E+01 | 1.26E+01 | 1.16E+01 |
| Data sets | BPSO | MBPSO | GPSO | MPSO-ELM | MFO-PSO | BBPSO VA | ALPSO | Prop. PSOVA1 | Prop. PSOVA2 | |
| Crohn | 3.83E-01 | 3.55E-01 | 5.38E-01 | 3.80E-01 | 4.90E-01 | 4.30E-01 | 4.36E-01 | 3.47E-01 | 3.17E-01 | |
| Myeloma | 3.17E-01 | 3.09E-01 | 3.98E-01 | 3.20E-01 | 3.59E-01 | 3.47E-01 | 3.57E-01 | 2.90E-01 | 2.66E-01 | |
| Seizure | 1.26E+01 | 1.24E+01 | 1.27E+01 | 1.24E+01 | 1.26E+01 | 1.25E+01 | 1.25E+01 | 1.26E+01 | 1.16E+01 | |
| PSOVA1 | Mean Classification Accuracy Rate | PSOVA2 | Mean Classification Accuracy Rate | ||
|---|---|---|---|---|---|
| Seizure | Voice | Seizure | Voice | ||
| PSO | 0.8459 | 0.8237 | PSO | 0.8459 | 0.8237 |
| PSO + Leader enhancement | 0.8475 | 0.8281 | PSO + Leader enhancement | 0.8463 | 0.8254 |
| PSO + Leader & worse solution enhancement | 0.8510 | 0.8316 | PSO + Leader & worse solution enhancement | 0.8495 | 0.8298 |
| Leader & worse solution enhancement + ameliorated signals | 0.8672 | 0.8491 | Leader & worse solution enhancement + exemplar breeding | 0.8733 | 0.8535 |
| Leader & worse solution enhancement + ameliorated signals + spiral search | 0.8698 | 0.8526 | Leader & worse solution enhancement + exemplar breeding + coefficient generation | 0.8860 | 0.8632 |
| PSOVA1 | PSOVA2 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Standard PSO | PSO+ Mirroring | 1 (Leader Enhancement) | 1 + 2 (Worse Enhancement) | 1+2+3 (Diverse Signals) | 1+2+3+4 (Spiral) | 1 (Leader Enhancement) | 1 + 2 (Worse Enhancement) | 1+2+3 (Exemplar) | 1+2+3+4 (Coefficient) | ||
| Ackley | MEAN | 1.97E+01 | 1.76E+01 | 1.62E+01 | 7.19E+00 | 3.12E+00 | 1.69E+00 | 9.33E+00 | 6.79E+00 | 1.37E+00 | 9.07E-01 |
| MIN | 1.89E+01 | 1.46E+01 | 5.98E+00 | 5.15E+00 | 2.11E+00 | 4.92E-01 | 2.50E+00 | 3.67E+00 | 2.29E-01 | 1.36E-01 | |
| MAX | 1.98E+01 | 1.87E+01 | 1.99E+01 | 9.47E+00 | 4.48E+00 | 2.44E+00 | 1.44E+01 | 8.77E+00 | 2.43E+00 | 2.11E+00 | |
| STD | 1.68E-01 | 9.73E-01 | 4.81E+00 | 1.03E+00 | 4.77E-01 | 4.95E-01 | 3.05E+00 | 1.14E+00 | 5.90E-01 | 6.28E-01 | |
| Dixon | MEAN | 2.22E+05 | 1.17E+03 | 7.25E+02 | 6.81E+01 | 3.98E+01 | 9.40E+00 | 1.12E+02 | 5.09E+01 | 1.15E+01 | 6.49E+00 |
| MIN | 1.40E+01 | 1.58E+02 | 1.03E+02 | 4.82E+00 | 8.32E+00 | 1.66E+00 | 2.60E+00 | 4.67E+00 | 1.88E+00 | 1.39E+00 | |
| MAX | 9.77E+05 | 2.85E+03 | 2.91E+03 | 2.01E+02 | 1.56E+02 | 2.72E+01 | 3.40E+02 | 3.44E+02 | 5.45E+01 | 3.46E+01 | |
| STD | 2.45E+05 | 9.10E+02 | 5.37E+02 | 4.67E+01 | 3.29E+01 | 5.27E+00 | 1.38E+02 | 6.81E+01 | 1.17E+01 | 6.74E+00 | |
| Griewank | MEAN | 1.24E+02 | 1.52E+01 | 4.54E+00 | 9.28E-01 | 4.11E-01 | 1.76E-01 | 3.79E+00 | 9.86E-01 | 1.76E-02 | 6.28E-03 |
| MIN | 1.04E+00 | 3.47E+00 | 1.03E+00 | 5.99E-01 | 2.21E-02 | 2.09E-02 | 1.40E-01 | 2.61E-01 | 4.41E-03 | 2.21E-03 | |
| MAX | 2.71E+02 | 3.16E+01 | 1.75E+01 | 1.15E+00 | 8.34E-01 | 5.76E-01 | 9.10E+01 | 2.13E+00 | 4.55E-02 | 1.46E-02 | |
| STD | 6.48E+01 | 6.98E+00 | 3.70E+00 | 1.66E-01 | 2.47E-01 | 1.28E-01 | 1.65E+01 | 4.14E-01 | 1.07E-02 | 3.45E-03 | |
| Rastrigin | MEAN | 3.24E+02 | 2.43E+02 | 2.23E+02 | 1.14E+02 | 8.54E+01 | 5.79E+01 | 1.30E+02 | 1.07E+02 | 7.71E+01 | 6.43E+01 |
| MIN | 2.69E+02 | 1.85E+02 | 1.48E+02 | 2.28E+01 | 4.07E+01 | 2.73E+01 | 7.59E+01 | 5.65E+01 | 4.13E+01 | 3.45E+01 | |
| MAX | 3.96E+02 | 3.09E+02 | 3.08E+02 | 2.28E+02 | 1.31E+02 | 9.66E+01 | 1.78E+02 | 1.70E+02 | 1.14E+02 | 9.44E+01 | |
| STD | 3.64E+01 | 2.94E+01 | 4.05E+01 | 4.59E+01 | 2.17E+01 | 1.84E+01 | 2.63E+01 | 2.79E+01 | 1.86E+01 | 1.47E+01 | |
| Rothyp | MEAN | 1.02E+05 | 4.39E+04 | 1.63E+04 | 1.04E+04 | 7.69E+02 | 5.94E+00 | 1.30E+04 | 4.41E+03 | 5.47E+00 | 2.11E+00 |
| MIN | 1.70E+04 | 2.12E+04 | 4.23E+03 | 2.99E+03 | 2.52E+02 | 7.97E-01 | 3.15E+00 | 2.00E+01 | 1.29E+00 | 4.93E-01 | |
| MAX | 2.07E+05 | 8.25E+04 | 3.35E+04 | 2.48E+04 | 1.55E+03 | 1.98E+01 | 5.90E+04 | 2.56E+04 | 1.86E+01 | 4.78E+00 | |
| STD | 6.31E+04 | 1.51E+04 | 7.49E+03 | 4.72E+03 | 3.21E+02 | 5.14E+00 | 1.64E+04 | 6.36E+03 | 3.95E+00 | 1.08E+00 | |
| Rosenbrock | MEAN | 6.21E+05 | 2.63E+04 | 1.12E+04 | 9.80E+03 | 3.43E+02 | 7.43E+01 | 2.35E+04 | 2.94E+03 | 8.48E+01 | 6.56E+01 |
| MIN | 2.84E+05 | 9.42E+03 | 3.54E+03 | 2.76E+03 | 1.64E+02 | 2.52E+01 | 5.20E+01 | 8.18E+01 | 3.13E+01 | 3.10E+01 | |
| MAX | 1.47E+06 | 5.45E+04 | 3.95E+04 | 2.03E+04 | 7.57E+02 | 1.58E+02 | 8.17E+04 | 2.52E+04 | 1.90E+02 | 2.21E+02 | |
| STD | 2.32E+05 | 1.19E+04 | 8.35E+03 | 4.72E+03 | 1.50E+02 | 4.07E+01 | 2.90E+04 | 6.26E+03 | 4.91E+01 | 5.24E+01 | |
| Sphere | MEAN | 2.81E+01 | 1.42E+01 | 8.48E+00 | 4.04E+00 | 3.76E-01 | 9.16E-02 | 3.53E+00 | 8.80E-01 | 6.10E-02 | 4.00E-02 |
| MIN | 1.15E-02 | 5.75E+00 | 3.17E+00 | 1.96E+00 | 1.79E-01 | 2.90E-02 | 6.78E-03 | 3.18E-05 | 2.70E-02 | 2.24E-02 | |
| MAX | 7.87E+01 | 2.87E+01 | 1.81E+01 | 7.26E+00 | 7.48E-01 | 2.07E-01 | 2.63E+01 | 2.62E+01 | 1.08E-01 | 7.52E-02 | |
| STD | 2.47E+01 | 5.35E+00 | 3.33E+00 | 1.45E+00 | 1.39E-01 | 5.02E-02 | 9.06E+00 | 4.79E+00 | 2.26E-02 | 1.33E-02 | |
| Sumpow | MEAN | 7.07E-02 | 5.68E-02 | 1.28E-02 | 4.52E-03 | 9.81E-05 | 1.24E-06 | 3.55E-02 | 5.02E-03 | 2.87E-05 | 2.13E-05 |
| MIN | 9.19E-04 | 1.10E-03 | 6.47E-04 | 1.13E-04 | 2.22E-06 | 1.37E-09 | 3.54E-03 | 1.23E-04 | 2.27E-06 | 1.21E-06 | |
| MAX | 8.16E-01 | 1.82E-01 | 6.53E-02 | 1.36E-02 | 4.86E-04 | 1.50E-05 | 1.80E-01 | 3.68E-02 | 1.65E-04 | 7.90E-05 | |
| STD | 1.59E-01 | 4.33E-02 | 1.67E-02 | 4.38E-03 | 9.77E-05 | 2.85E-06 | 3.53E-02 | 8.04E-03 | 2.98E-05 | 1.78E-05 | |
| Zakharov | MEAN | 6.27E+02 | 4.11E+02 | 3.25E+02 | 1.70E+02 | 1.01E+02 | 8.27E+01 | 2.99E+02 | 1.56E+02 | 9.61E+01 | 7.39E+01 |
| MIN | 5.53E+02 | 3.38E+02 | 2.03E+02 | 7.22E+01 | 5.65E+01 | 5.07E+01 | 2.00E+02 | 6.55E+01 | 5.58E+01 | 4.81E+01 | |
| MAX | 7.63E+02 | 4.52E+02 | 4.32E+02 | 2.90E+02 | 1.52E+02 | 1.49E+02 | 3.84E+02 | 2.19E+02 | 1.34E+02 | 1.04E+02 | |
| STD | 5.56E+01 | 2.81E+01 | 6.42E+01 | 4.41E+01 | 2.20E+01 | 2.07E+01 | 4.57E+01 | 4.12E+01 | 1.81E+01 | 1.46E+01 | |
| Sumsqu | MEAN | 6.82E+02 | 4.26E+02 | 2.38E+02 | 6.48E+01 | 5.13E+00 | 2.95E+00 | 2.02E+02 | 4.40E+01 | 3.99E+00 | 2.07E+00 |
| MIN | 7.92E+01 | 2.26E+02 | 1.22E+02 | 2.56E+01 | 1.65E+00 | 6.65E-01 | 1.09E-02 | 4.40E-02 | 1.16E+00 | 6.20E-01 | |
| MAX | 1.34E+03 | 7.25E+02 | 3.63E+02 | 1.45E+02 | 1.04E+01 | 8.44E+00 | 4.98E+02 | 3.47E+02 | 1.12E+01 | 9.05E+00 | |
| STD | 3.35E+02 | 1.15E+02 | 7.35E+01 | 2.83E+01 | 2.28E+00 | 1.86E+00 | 1.39E+02 | 7.25E+01 | 2.90E+00 | 1.67E+00 | |
| Powell | MEAN | 4.91E+03 | 2.85E+03 | 5.03E+02 | 4.43E+02 | 3.72E+01 | 1.91E+01 | 3.08E+02 | 2.34E+02 | 2.63E+01 | 1.02E+01 |
| MIN | 5.46E+02 | 4.94E+02 | 4.01E+02 | 3.20E+02 | 1.89E+00 | 9.97E-01 | 2.87E-01 | 1.47E+00 | 7.82E+00 | 1.83E+00 | |
| MAX | 8.11E+03 | 6.87E+03 | 6.19E+02 | 5.62E+02 | 1.34E+02 | 9.65E+01 | 2.96E+03 | 1.88E+03 | 9.61E+01 | 3.90E+01 | |
| STD | 2.21E+03 | 1.96E+03 | 5.86E+01 | 6.24E+01 | 2.74E+01 | 1.89E+01 | 6.18E+02 | 4.20E+02 | 1.83E+01 | 7.72E+00 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, H.; Zhang, L.; Lim, C.P.; Yu, Y.; Liu, H. Feature Selection Using Enhanced Particle Swarm Optimisation for Classification Models. Sensors 2021, 21, 1816. https://doi.org/10.3390/s21051816
Xie H, Zhang L, Lim CP, Yu Y, Liu H. Feature Selection Using Enhanced Particle Swarm Optimisation for Classification Models. Sensors. 2021; 21(5):1816. https://doi.org/10.3390/s21051816
Chicago/Turabian StyleXie, Hailun, Li Zhang, Chee Peng Lim, Yonghong Yu, and Han Liu. 2021. "Feature Selection Using Enhanced Particle Swarm Optimisation for Classification Models" Sensors 21, no. 5: 1816. https://doi.org/10.3390/s21051816
APA StyleXie, H., Zhang, L., Lim, C. P., Yu, Y., & Liu, H. (2021). Feature Selection Using Enhanced Particle Swarm Optimisation for Classification Models. Sensors, 21(5), 1816. https://doi.org/10.3390/s21051816







