SDNN24 Estimation from Semi-Continuous HR Measures
Abstract
1. Introduction
2. Methods
2.1. Dataset and Feature Engineering
- nsr2db: Normal Sinus Rhythm RR Interval Database PhysioNet dataset [25]. This dataset contains beat annotations of 54 normal sinus rhythm subjects (30 men: 28–76 years; 24 women: 58–73 years) extracted from 23 h long ECG.
- chfdb: Congestive Heart Failure RR Interval Database PhysioNet dataset [26]. This dataset includes beat annotation files for 29 long-term ECG recordings of subjects aged 34 to 79, with congestive heart failure (New York Heart Association classes I, II, and III). Subjects included 8 men and 2 women; gender is not known for the remaining 21 subjects.
- mmash: Multilevel Monitoring of Activity and Sleep in Healthy people (MMASH) dataset [27,28] provides 24 h of continuous beat-to-beat heart data, triaxial accelerometer data, sleep quality, physical activity and psychological characteristics (i.e., anxiety status, stress events and emotions) for 22 healthy participants.
2.2. Data Preprocessing
2.3. PPG Error Estimation
2.4. SDNN24
2.4.1. Time Domain Analysis
- SDNN24: The standard deviation of NN intervals recorded during 24 h (Equation (1)).where and refer to each NN-interval value and the mean of 24 h NN-intervals, respectively. N is the number of NN-intervals recorded during 24 h. From a digital signal processing perspective, SDNN24, being the standard deviation of the 24 h long signal, is the power of the whole 24 h spectrum. As discussed in the introduction, we expect ULF and VLF to be the main contributors to the spectrum; therefore, we expect SDNN24 to correlate with VLF and ULF. This is the definition of SDNN24, and the ground truth we will try to estimate. To compute SDNN24 is necessary to have the continuous stream of inter-beats intervals signal, typically not available from wearable devices.
- SDNNi24: The mean of the standard deviations of the NN intervals calculated on segments with defined duration over 24 h (Equation (2)).where and reflect the number of segments and the number of NN-intervals in each segment, respectively. From a digital signal processing perspective, SDNNi24, being the standard deviation of short term measurements (usually 1 to 5 min), represents the average power of the short term spectrum. It will not be able to measure ULF and VLF (that comes from signals of periods longer than 5 min). To compute SDNN24 is necessary in order to have the inter-beats intervals data of each segment, typically not available from wearable devices.
- SDANN24: The standard deviation of the means of NN intervals calculated at segments of a defined duration over 24 h (Equation (3)).where refers to the mean of the NN-intervals in a segment. is the number of NN-intervals time windows recorded over 24 h. From a digital signal processing perspective SDANN24 can be considered similar to SDNN24 applied on a inter-beats intervals dataset after a low pass filter that dampened signals with a shorter period than the duration of the measured segments. To compute SDNN24, it is necessary to have the inter-beats intervals data of each segment, typically not available from wearable devices.
- SDANN24: The standard deviation of the Average NN intervals (ANN) derived from the HR, i.e., , calculated on segments with defined duration over 24 h (Equation (4)).where HR is computed as 60/. SDANN24 can be computed from data commonly collected by wrist-worn fitness wearable devices. In order to simulate error induced by wrist-worn devices, we randomly add a Gaussian error in each HR obtained during each NN-intervals time window. The bias for each time window’s length is provided in Table 1.
2.4.2. Frequency Domain Analysis for SDNN24
- SDANN24: The root square of the sum between NN intervals (ANN) variance derived from the average HR, i.e., ANN = ( is a random Gaussian bias of a specific time window length), calculated on segments with defined duration over 24 h (ANN24) and the mean of missing frequency variance (Equation (6)).where refers to the length of the segment in time-series (e.g., for 1, 5 and 10 min segments the are equal to 0.016, 0.0033 and 0.0016, respectively).With this approach, we attempt to remove the underestimation of SDNN24 by adding the portion of spectrum lost by using HR measures instead of inter-beats intervals data, simply adding the average power of the HRV spectrum above to the measured variance. The corrective factor is fixed for all subjects.
- SDANN24: The root square of the total power predicted in accordance with PSD (Equation (7)).where refers to the length of the segment in time-series (e.g., for 1, 5 and 10 min segments the are equal to 0.016, 0.0033 and 0.0016, respectively).With this approach we correct the underestimation by assuming that the missing high frequencies perfectly correlate with the measured low frequencies.
2.4.3. HR Circadian Rhythm
2.5. Validation
2.6. Healthy vs. Unhealthy Subjects
2.6.1. Statistical Analysis
2.6.2. Machine Learning Approach
- LR: Logistic Regression was performed using: only SDNN24 (); only SDNN24 (); all of the HR features as predictors, i.e., SDNN24, MESOR and Amplitude ();
- RF: Random Forest Classifiers (RF) were also performed using all the HR features as predictors;
- NN: Fully connected feed forward Neural Network, using all the HR features as predictors. We used Keras with the TensorFlow backend by using Python 3.8 programming language. We trained our neural networks on the Azure cloud, using bayesian sampling. The only explored topology was fully connected, with a single hidden layer, leaky relu activation function for the neurons of the hidden layer, a single output neuron with sigmoid activation function, and a dropout layer after the hidden layer. The tuned hyper-parameters were:
- The number of neurons in the hidden layer (between 1 and 8);
- Alpha value for the leaky relu activation function of the neurons in the hidden layer (between 0.0 and 1.0);
- The dropout rate (between 0% and 99%);
- The batch size (between 1 and 32).
The training set was split in train and validation using the train_test_split function from the sklearn python package, using a 80-20% split, ensuring stratification on the predicted class. The validation data were not used during hyper-parameter tuning. A total of 400 combinations of hyper-parameters were tested.
3. Results
3.1. SDNN24 Estimation
3.1.1. Time Domain Analysis
3.1.2. Frequency Domain Analysis
3.2. Healthy vs. Unhealthy Subjects
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shaffer, F.; Ginsberg, J. An Overview of Heart Rate Variability Metrics and Norms. Front. Public Health 2017, 5, 258. [Google Scholar] [CrossRef]
- Kleiger, R.E.; Miller, J.P.; Bigger, J.T., Jr.; Moss, A.J. Decreased heart rate variability and its association with increased mortality after acute myocardial infarction. Am. J. Cardiol. 1987, 59, 256–262. [Google Scholar] [CrossRef]
- Gao, L.; Chen, Y.; Shi, Y.; Xue, H.; Wang, J. Value of DC and DRs in prediction of cardiovascular events in acute myocardial infarction patients. Zhonghua Yi Xue Za Zhi 2016, 96. [Google Scholar] [CrossRef]
- Karcz, M.; Chojnowska, L.W.Z.R. Prognostic significance of heart rate variability in dilated cardiomyopathy. EP Eur. 2003, 87, 75–81. [Google Scholar] [CrossRef]
- Hillebrand, S.; Gast, K.; de Mutsert, R.; Swenne, C.; Jukema, J.; Middeldorp, S.; Rosendaal, F.; Dekkers, O. Heart rate variability and first cardiovascular event in populations without known cardiovascular disease: Meta-analysis and dose–response meta-regression. EP Eur. 2013, 15. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Shah, H.; Junior, C.; Sun, X.; Mitri, J.; Sambataro, M.; Sambado, L.; Gerstein, H.; Fonseca, V.; Doria, A.; et al. Intensive Risk Factor Management and Cardiovascular Autonomic Neuropathy in Type 2 Diabetes: The ACCORD Trial. Diabetes Care 2021, 44. [Google Scholar] [CrossRef] [PubMed]
- Hoshi, R.; Santos, I.; Dantas, E.; Andreão, R.; Mill, J.; Lotufo, P.; Bensenor, I. Reduced heart-rate variability and increased risk of hypertension—A prospective study of the ELSA-Brasil. J. Hum. Hypertens. 2021. [Google Scholar] [CrossRef] [PubMed]
- Ucak, S.; Dissanayake, H.; Sutherland, K.; de Chazal, P.; Cistulli, P. Heart rate variability and obstructive sleep apnea: Current perspectives and novel technologies. J. Sleep Res. 2021. [Google Scholar] [CrossRef]
- Hirten, R.; Danieletto, M.; Tomalin, L.; Choi, K.; Zweig, M.; Golden, E.; Kaur, S.; Helmus, D.; Biello, A.; Pyzik, R.; et al. Longitudinal Physiological Data fromaWearable Device Identifies SARS-CoV-2Infection andSymptoms and Predicts COVID-19 Diagnosis. MedWxiv 2020. [Google Scholar] [CrossRef]
- Buchhorn, R.; Baumann, C.; Willaschek, C. Heart Rate Variability in a Patient with Coronavirus Disease. Int. Cardiovasc. Forum J. 2020. [Google Scholar] [CrossRef]
- Josephine, M.; Lakshmanan, L.; Resmi, R.; Visu, P.; Ganesan, R.; Jothikumar, R. Monitoring and sensing COVID-19 symptoms as a precaution using electronic wearable devices. Int. J. Pervasive Comput. Commun. 2020, 16. [Google Scholar] [CrossRef]
- MacDonald, E.; Rose, R.; Quinn, T. Neurohumoral Control of Sinoatrial Node Activity and Heart Rate: Insight From Experimental Models and Findings From Humans. Front. Physiol. 2020, 11. [Google Scholar] [CrossRef]
- Yaniv, Y.; Lakatta, E.G. The end effector of circadian heart rate variation: The sinoatrial node pacemaker cell. BMB Rep. 2015, 48, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, A.A.; Weiser-Bitoun, I.; Billman, G.E.; Yaniv, Y. Signatures of the autonomic nervous system and the heart’s pacemaker cells in canine electrocardiograms and their applications to humans. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Haghi, M.; Thurow, K.; Stoll, R. Wearable Devices in Medical Internet of Things: Scientific Research and Commercially Available Devices. Healthc. Inform. Res. 2017, 59, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Li, K.H.C.; White, F.A.; Tipoe, T.; Liu, T.; Wong, M.C.; Jesuthasan, A.; Baranchuk, A.; Tse, G.; Yan, B.P. The current state of mobile phone apps for monitoring heart rate, heart rate variability, and atrial fibrillation: Narrative review. JMIR mHealth and uHealth 2019, 7, e11606. [Google Scholar] [CrossRef]
- Morelli, D.; Rossi, A.; Cairo, M.; Clifton, D. Analysis of the Impact of Interpolation Methods of Missing RR-Intervals Caused by Motion Artifacts on HRV Features Estimations. Sensors 2019, 19, 3163. [Google Scholar] [CrossRef]
- Morelli, D.; Bartoloni, L.; Colombo, M.; Clifton, D. Profiling the propagation of error from PPG to HRV features in a wearable physiological-monitoring device. Healthc. Technol. Lett. 2018, 5, 59–64. [Google Scholar] [CrossRef]
- Shcherbina, A.; Mattsson, C.; Waggott, D.; Salisbury, H.; Christle, J.; Hastie, T.; Wheeler, M.; Ashley, E. Accuracy in Wrist-Worn, Sensor-Based Measurements of Heart Rate and Energy Expenditure in a Diverse Cohort. J. Persersonalized Med. 2017, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Weiler, D.; Villajuan, S.; Edkins, L.; Cleary, S.; Saleem, J.J. Wearable Heart Rate Monitor Technology Accuracy in Research: A Comparative Study Between PPG and ECG Technology. Proc. Hum. Factors Ergon. Soc. Annu. Meet. 2017, 61. [Google Scholar] [CrossRef]
- Jo, E.; Lewis, K.; Directo, D.; Kim, M.; Dolezal, B. Validation of Biofeedback Wearables for Photoplethysmographic Heart Rate Tracking. J. Sport Med. 2016, 15, 540. [Google Scholar]
- Yaniv, Y.; Lyashkov, A.E.; Lakatta, E.G. The fractal-like complexity of heart rate variability beyond neurotransmitters and autonomic receptors: Signaling intrinsic to sinoatrial node pacemaker cells. Cardiovasc. Pharmacol. Open Access 2013, 2, 111. [Google Scholar] [CrossRef]
- Saul, J.; Albrecht, P.; Berger, R.; Cohen, R. Analysis of long term heart rate variability: Methods, 1/f scaling and implications. Comput. Cardiol. 1988, 14, 419–422. [Google Scholar]
- Bergfeldt, L.; Haga, Y. Power spectral and Poincaré plot characteristics in sinus node dysfunction. J. Appl. Physiol. 2003, 94. [Google Scholar] [CrossRef]
- Normal Sinus Rhythm RR Interval Database. PhysioNet. Available online: https://physionet.org/physiobank/database/nsr2db/ (accessed on 1 September 2020).
- Congestive Heart Failure RR Interval Database. PhysioNet. Available online: https://physionet.org/content/chf2db/1.0.0/ (accessed on 1 September 2020).
- Rossi, A.; Da Pozzo, E.; Menicagli, D.; Tremolanti, C.; Priami, C.; Sirbu, A.; Clifton, D.; Martini, C.; Morelli, D. Multilevel Monitoring of Activity and Sleep in Healthy People (Version 1.0.0). PhysioNet 2020. Available online: https://physionet.org/content/mmash/1.0.0/ (accessed on 1 September 2020).
- Rossi, A.; Da Pozzo, E.; Menicagli, D.; Tremolanti, C.; Priami, C.; Sirbu, A.; Clifton, D.; Martini, C.; Morelli, D. A Public Dataset of 24-h Multi-Levels Psycho-Physiological Responses in Young Healthy Adults. Data 2020, 5, 91. [Google Scholar] [CrossRef]
- Baek, H.; Cho, J. Novel heart rate variability index for wrist-worn wearable devices subject to motion artifacts that complicate measurement of the continuous pulse interval. Physiol. Meas. 2019, 40. [Google Scholar] [CrossRef]
- Rossi, A.; Pedreschi, D.; Clifton, D.; Morelli, D. Error Estimation of Ultra-Short Heart Rate Variability Parameters: Effect of Missing Data Caused by Motion Artifacts. Sensors 2020, 20, 7122. [Google Scholar] [CrossRef]
- Lomb, N. Least-squares frequency analysis of unequally spaced data. Astrophys. Space Sci. 1976, 39, 447–462. [Google Scholar] [CrossRef]
- Scargle, J.D. Studies in Astronomical Time Series Analysis. V. Bayesian Blocks, a New Method to Analyze Structure in Photon Counting Data. Astrophys. J. 1998, 504, 405. [Google Scholar] [CrossRef]
- Morelli, D.; Bartoloni, L.; Rossi, A.; DA, C. A computationally efficient algorithm to obtain an accurate and interpretable model of the effect of circadian rhythm on resting heart rate. Pysiolog. Meas. 2019, 40. [Google Scholar] [CrossRef] [PubMed]
- Camm, A.J.; Malik, M.; Bigger, J.T.; Breithardt, G.; Cerutti, S.; Cohen, R.J.; Coumel, P.; Fallen, E.L.; Kennedy, H.L.; Kleiger, R.E.; et al. Heart rate variability: Standards of measurement, physiological interpretation and clinical use. Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Circulation 1996, 927–934. [Google Scholar] [CrossRef]
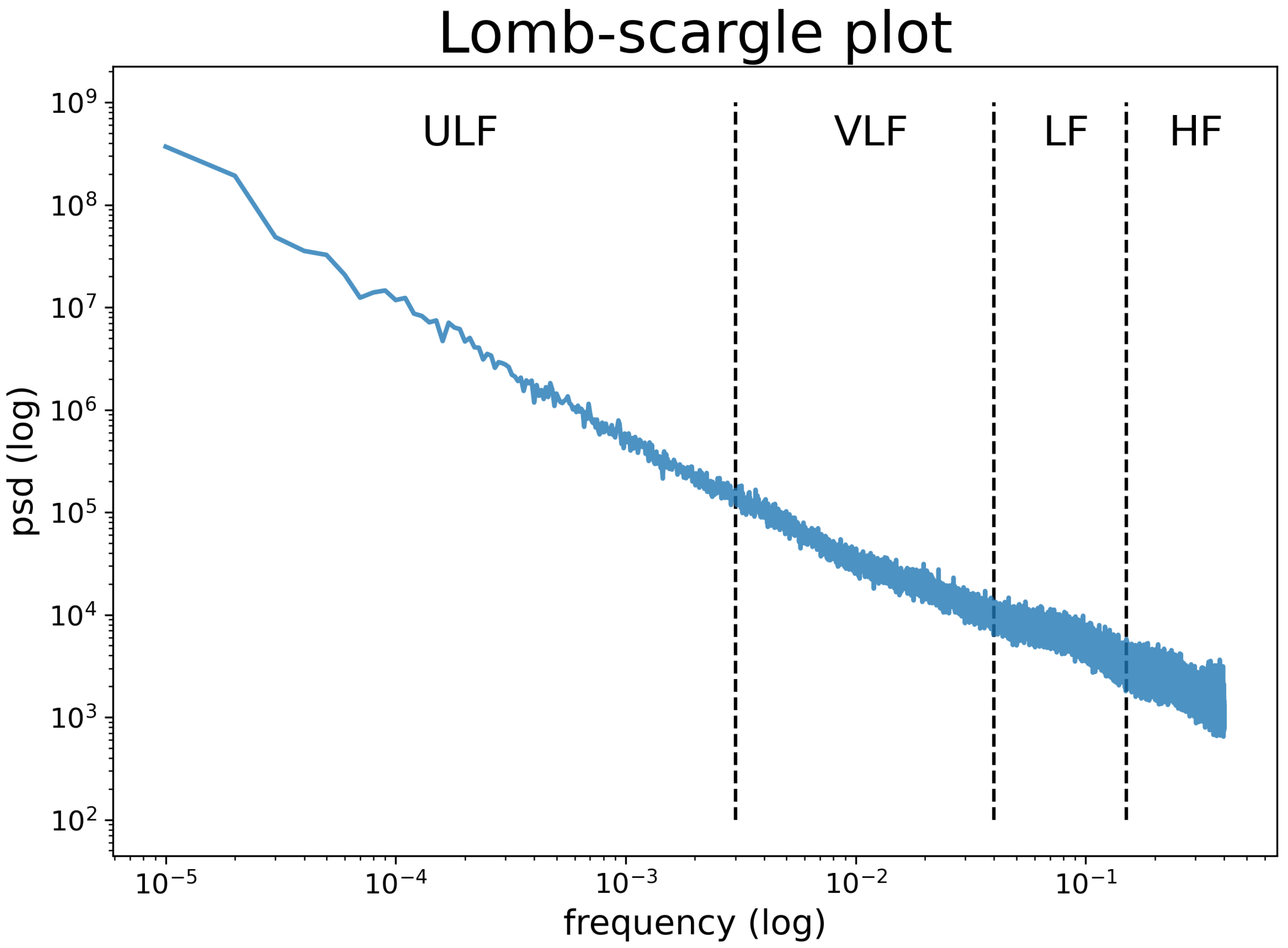
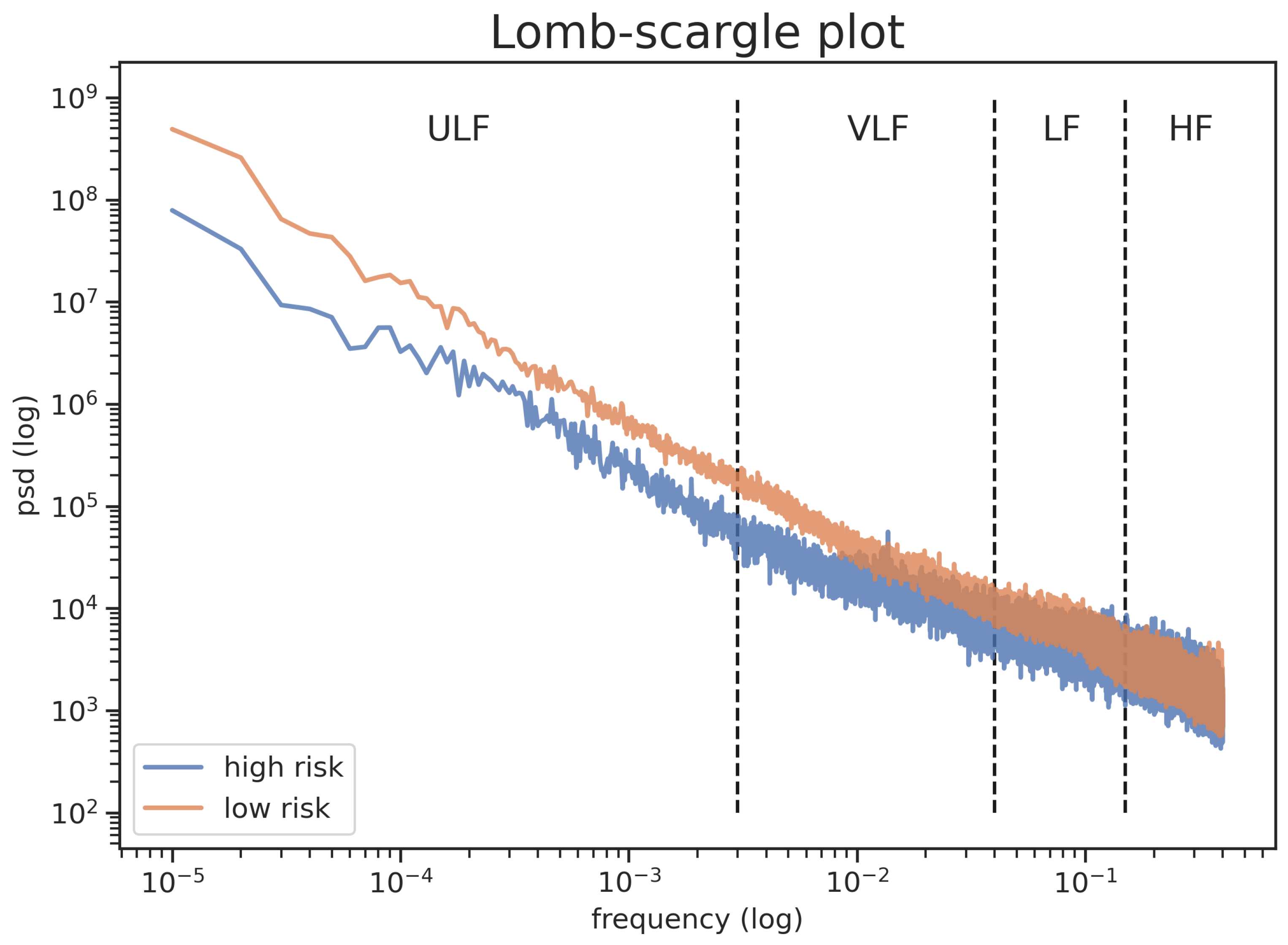
| Time Window | Error |
|---|---|
| 1 min | 0.03 ± 5.91 |
| 5 min | 0.33 ± 5.09 |
| 10 min | −0.16 ± 4.71 |
| 30 min | −0.60 ± 4.01 |
| 60 min | −0.35 ± 3.95 |
| Segment | SDNN24 | SDNNi24 | SDANN24 | SDANN24 |
|---|---|---|---|---|
| 1 min | 133.73 ± 45.98 | 56.28 ± 18.51 * | 119.38 ± 49.51 * | 120.96 ± 48.91 * |
| 5 min | 59.47 ± 21.52 * | 117.33 ± 48.03 * | 118.62 ± 47.00 * | |
| 10 min | 60.77 ± 23.02 * | 116.72 ± 47.37 * | 117.94 ± 46.30 * | |
| 30 min | 63.11 ± 26.26 * | 115.75 ± 46.69 * | 117.12 ± 45.96 * | |
| 60 min | 64.61 ± 28.52 * | 114.87 ± 46.23 * | 116.29 ± 45.85 * |
| Segment | Lower Frequency | Higher Frequency | Total Power |
|---|---|---|---|
| 1 min max frequency = 1.67 | 1.17± 8.68 (20.44%) | 4.57± 2.86 (79.56%) | 5.74± 3.66 |
| 5 min max frequency = 3.33 Hz | 1.12± 8.47 (19.41%) | 4.62± 2.86 (80.59%) | |
| 10 min max frequency = 1.67 Hz | 1.09± 8.35 (18.88%) | 4.66± 2.87 (81.22%) | |
| 30 min max frequency = 5.55 Hz | 1.03± 8.09 (17.85%) | 4.72± 2.90 (82.25%) | |
| 60 min max frequency = 2.78 Hz | 9.77± 7.90 (17.01%) | 4.77± 2.92 (82.98%) |
| Segment | SDNN24 | SDNN24 | SDANN24 | SDANN24 |
|---|---|---|---|---|
| 1 min | 173.53 ± 25.76 | 164.00 ± 32.52 | 213.63 ± 20.81 * | 175.65 ± 25.65 |
| 5 min | 153.02 ± 32.54 * | 209.23 ± 19.79 * | 165.64 ± 31.59 | |
| 10 min | 149.84 ± 31.00 * | 198.92 ± 23.90 * | 161.51 ± 30.96 | |
| 30 min | 146.02 ± 32.47 * | 197.60 ± 24.55 * | 164.21 ± 35.98 | |
| 60 min | 142.11 ± 35.02 * | 198.38 ± 31.10 * | 155.23 ± 40.00 |
| Segment | Features | High Risk | Low Risk | t-score |
|---|---|---|---|---|
| — | SDNN24 (ms) | 86.54 ± 43.29 | 142.14 ± 31.05 | 6.66 * |
| 1 min | SDNN24 (ms) | 67.86 ± 37.23 | 132.61 ± 30.79 | 8.28 * |
| 5 min | SDNN24 (ms) | 64.46 ± 37.23 | 128.31 ± 30.63 | 8.27 * |
| 10 min | SDNN24 (ms) | 62.55 ± 36.67 | 126.02 ± 30.52 | 8.29 * |
| 30 min | SDNN24 (ms) | 58.37 ± 35.45 | 121.96 ± 30.44 | 8.44 * |
| 60 min | SDNN24 (ms) | 55.75 ± 34.60 | 118.81 ± 30.26 | 8.49 * |
| Model | Class | Precision | Recall | F1-score |
|---|---|---|---|---|
| Low | 0.84 | 1.00 | 0.91 | |
| High | 1.00 | 0.50 | 0.67 | |
| Low | 0.76 | 1.00 | 0.86 | |
| High | 1.00 | 0.17 | 0.29 | |
| Low | 0.80 | 1.00 | 0.89 | |
| High | 1.00 | 0.33 | 0.50 | |
| Low | 0.80 | 1.00 | 0.89 | |
| High | 1.00 | 0.33 | 0.50 | |
| * | Low | 0.94 | 1.00 | 0.97 |
| High | 1.00 | 0.83 | 0.91 | |
| Low | 0.80 | 1.00 | 0.89 | |
| High | 1.00 | 0.33 | 0.50 | |
| NN | Low | 0.94 | 1.00 | 0.97 |
| High | 0.80 | 1.00 | 0.89 | |
| Low | 0.72 | 0.81 | 0.76 | |
| High | 0.25 | 0.17 | 0.20 | |
| Low | 0.73 | 1.00 | 0.84 | |
| High | 0.00 | 0.00 | 0.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morelli, D.; Rossi, A.; Bartoloni, L.; Cairo, M.; Clifton, D.A. SDNN24 Estimation from Semi-Continuous HR Measures. Sensors 2021, 21, 1463. https://doi.org/10.3390/s21041463
Morelli D, Rossi A, Bartoloni L, Cairo M, Clifton DA. SDNN24 Estimation from Semi-Continuous HR Measures. Sensors. 2021; 21(4):1463. https://doi.org/10.3390/s21041463
Chicago/Turabian StyleMorelli, Davide, Alessio Rossi, Leonardo Bartoloni, Massimo Cairo, and David A. Clifton. 2021. "SDNN24 Estimation from Semi-Continuous HR Measures" Sensors 21, no. 4: 1463. https://doi.org/10.3390/s21041463
APA StyleMorelli, D., Rossi, A., Bartoloni, L., Cairo, M., & Clifton, D. A. (2021). SDNN24 Estimation from Semi-Continuous HR Measures. Sensors, 21(4), 1463. https://doi.org/10.3390/s21041463







