Performance Evaluation of Deep Learning-Based Prostate Cancer Screening Methods in Histopathological Images: Measuring the Impact of the Model’s Complexity on Its Processing Speed
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset
2.2. CNN Models
2.3. Benchmark
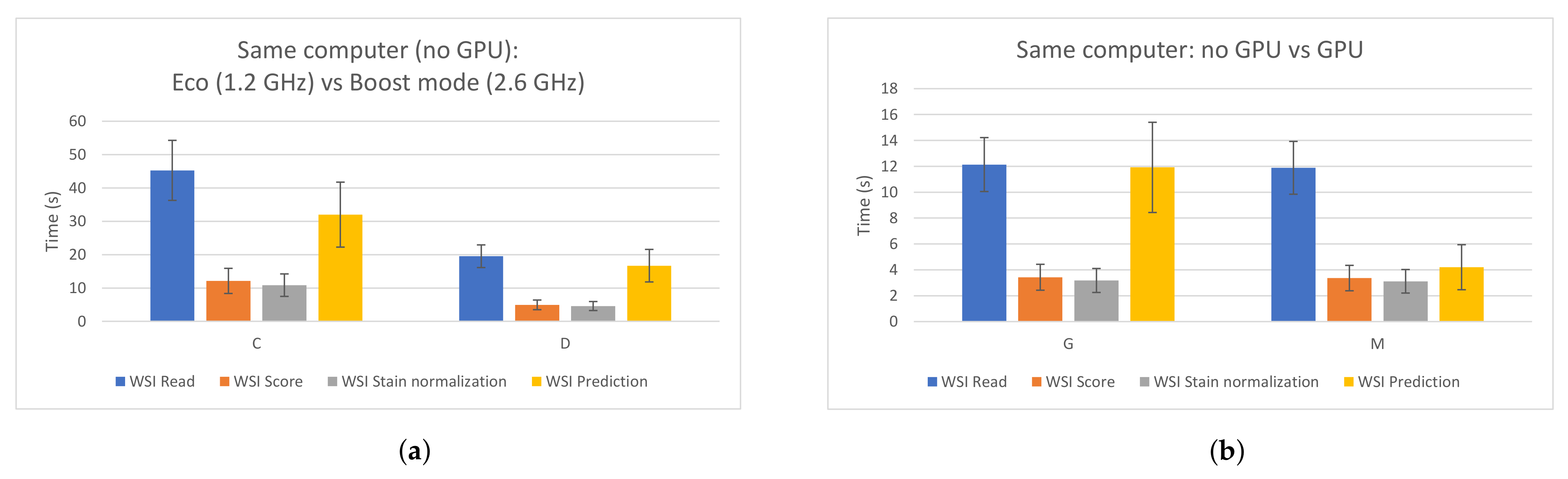
3. Results
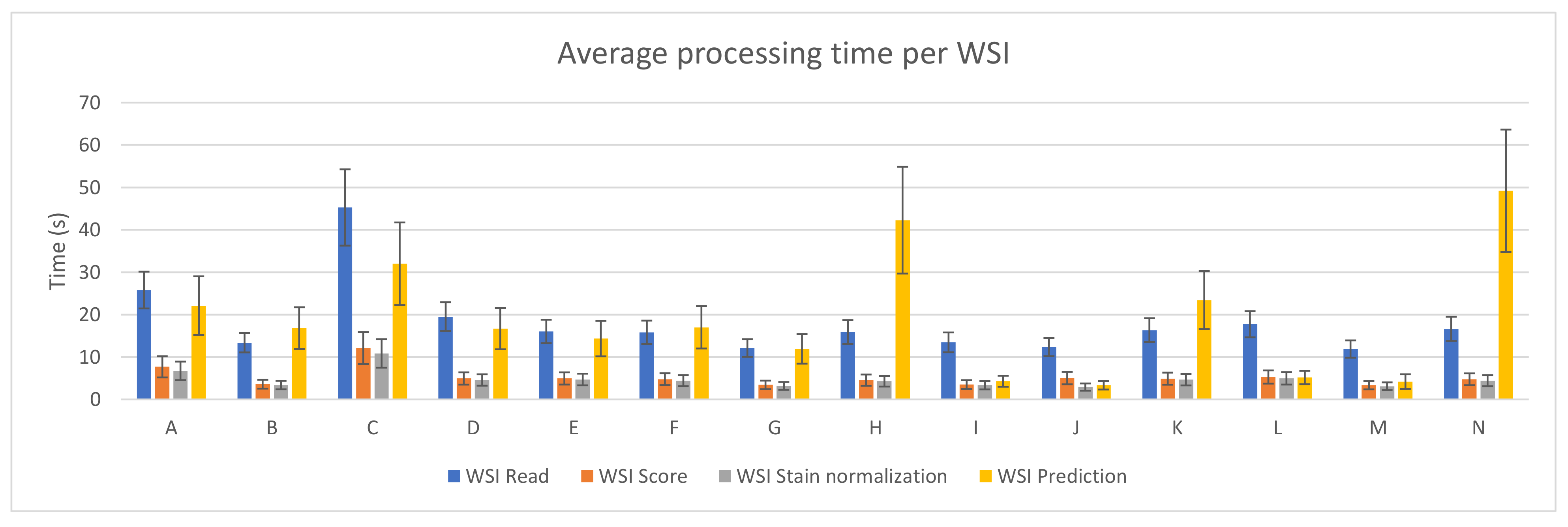
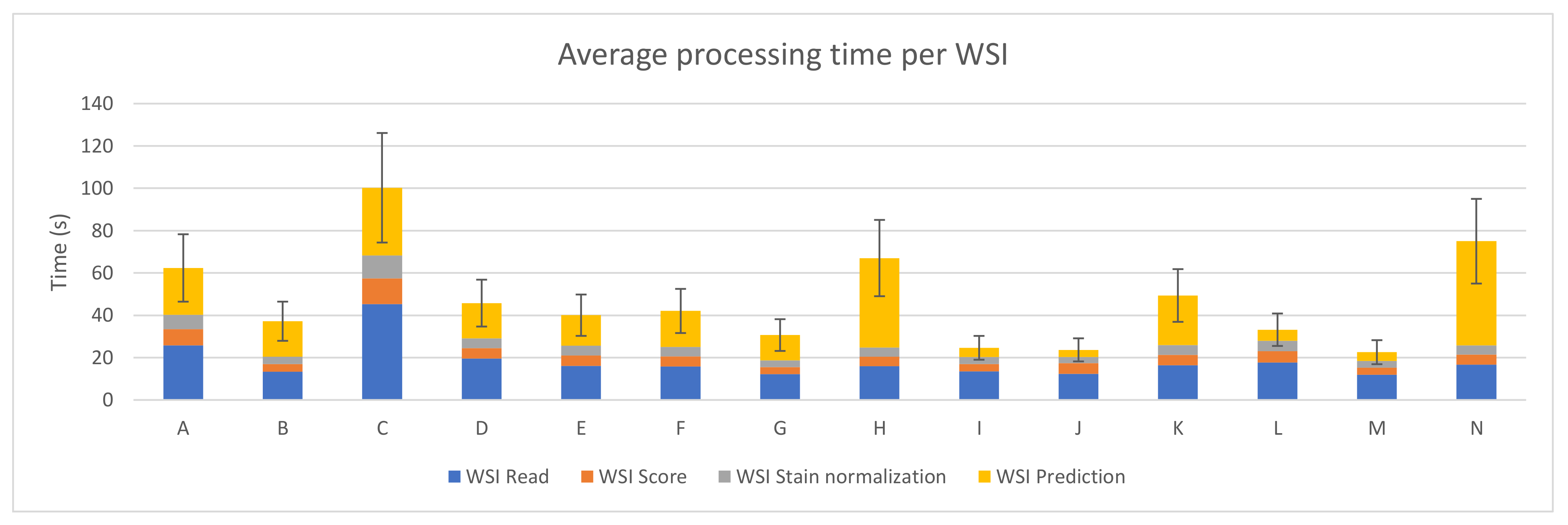
3.1. PROMETEO Evaluation
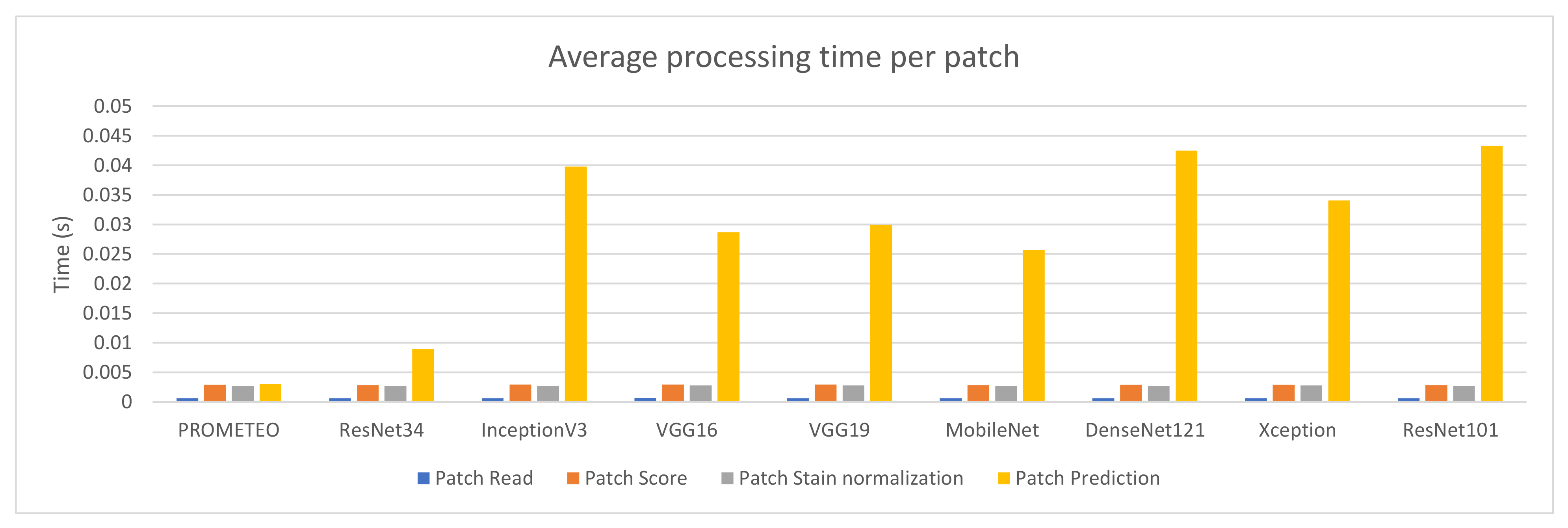
3.2. Performance Comparison for Different State-of-the-Art Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| AUC | Area Under Curve |
| CAD | Computer-Aided Diagnosis |
| CNN | Convolutional Neural Network |
| CPU | Central Processing Unit |
| DL | Deep Learning |
| DRE | Digital Rectal Examination |
| GB | Gigabyte |
| GGS | Gleason Grading System |
| GPU | Graphic Processing Unit |
| H&E | Hematoxylin and Eosin |
| PC | Personal Computer |
| PCa | Prostate Cancer |
| PSA | Prostate-Specific Antigen |
| RNN | Recurrent Neural Network |
| TPU | Tensor Processing Unit |
| WHO | World Health Organization |
| WSI | Whole-Slide Image |
Appendix A. PROMETEO Evaluation
| Device | CPU | GPU |
|---|---|---|
| A | Intel® Core™ i7-8850U @ 1.80 GHz 4 cores, 8 threads | - |
| B | Intel® Core™ i9-7900X @ 3.30 GHz 10 cores, 20 threads | - |
| C | Intel® Core™ i7-6700HQ @ 1.20 GHz 4 cores, 8 threads | - |
| D | Intel® Core™ i7-6700HQ @ 2.60 GHz 4 cores, 8 threads | - |
| E | Intel® Core™ i5-6500 @ 3.20 GHz 4 cores, 4 threads | - |
| F | Intel® Core™ i7-4770K @ 3.50 GHz 4 cores, 8 threads | - |
| G | Intel® Core™ i7-8700K @ 3.70 GHz 6 cores, 12 threads | - |
| H | Intel® Core™ i7-4970 @ 3.60 GHz 4 cores, 8 threads | - |
| I | Intel® Core™ i9-7900X @ 3.30 GHz 10 cores, 20 threads | NVIDIA® GeForce™ GTX 1080 Ti 11 GB GDDR5X |
| J | AMD® Ryzen™ 9 3900X @ 4.20 GHz 12 cores, 24 threads | NVIDIA® GeForce™ GTX 1080 Ti 11 GB GDDR5X |
| K | Intel® Core™ i5-6500 @ 3.20 GHz 4 cores, 4 threads | NVIDIA® GeForce™ GT 730 2 GB GDDR5 |
| L | Intel® Core™ i7-4770K @ 3.50 GHz 4 cores, 8 threads | NVIDIA® GeForce™ GTX 1080 Ti 11 GB GDDR5X |
| M | Intel® Core™ i7-8700K @ 3.70 GHz 6 cores, 12 threads | NVIDIA® GeForce™ GTX 1080 Ti 11 GB GDDR5X |
| N | Intel® Core™ i7-4970 @ 3.60 GHz 4 cores, 8 threads | NVIDIA® GeForce™ RTX 2060 6 GB GDDR6 |
| Patch | WSI | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Read | Score | Stain Normalization | Prediction | Read | Score | Stain Normalization | Prediction | |||||||||
| Device | Avg | Std | Avg | Std | Avg | Std | Avg | Std | Avg | Std | Avg | Std | Avg | Std | Avg | Std |
| A | 0.00120757 | 0.00311363 | 0.00585298 | 0.00502059 | 0.00512905 | 0.00418173 | 0.01730045 | 0.00850617 | 25.8035576 | 4.33829335 | 7.68691321 | 2.50054828 | 6.74114185 | 2.1823579 | 22.1192591 | 6.90573801 |
| B | 0.00068973 | 0.00150109 | 0.00306787 | 0.0037015 | 0.00288733 | 0.00062438 | 0.01441587 | 0.00175146 | 13.3892811 | 2.29629633 | 3.59321811 | 1.04956324 | 3.38756321 | 0.98944353 | 16.8213335 | 4.91918301 |
| C | 0.00182337 | 0.00503892 | 0.00807697 | 0.00628436 | 0.00729532 | 0.00634226 | 0.02249318 | 0.0100416 | 45.2693313 | 8.99897452 | 12.1239614 | 3.77223369 | 10.8517522 | 3.36864663 | 31.9982855 | 9.74059672 |
| D | 0.00103901 | 0.00228084 | 0.00437608 | 0.00080403 | 0.00404258 | 0.00098854 | 0.01435914 | 0.00211298 | 19.5256697 | 3.39723828 | 4.94245166 | 1.44889791 | 4.59097219 | 1.34596044 | 16.6959336 | 4.87815493 |
| E | 0.00082695 | 0.00167953 | 0.00421429 | 0.00068222 | 0.00400594 | 0.00090984 | 0.01223286 | 0.00216942 | 16.0484505 | 2.76278471 | 4.94652829 | 1.4439207 | 4.70010431 | 1.37218657 | 14.3399619 | 4.1861481 |
| F | 0.00083281 | 0.00172759 | 0.00413688 | 0.00075105 | 0.0038354 | 0.00094666 | 0.01479934 | 0.00293383 | 15.8376234 | 2.74964356 | 4.7714866 | 1.3976735 | 4.42655776 | 1.29581397 | 16.9997911 | 4.98414625 |
| G | 0.00062777 | 0.00133961 | 0.00292148 | 0.00038463 | 0.00270451 | 0.00067159 | 0.0102172 | 0.00150291 | 12.1361434 | 2.0832663 | 3.42516114 | 1.00071198 | 3.17680098 | 0.92726679 | 11.9159923 | 3.48494303 |
| H | 0.00084291 | 0.00175864 | 0.00398322 | 0.00065638 | 0.0037566 | 0.00093803 | 0.03768491 | 0.00818776 | 15.8986914 | 2.81638821 | 4.5458395 | 1.34199731 | 4.29495389 | 1.26743003 | 42.2879135 | 12.595224 |
| I | 0.00069517 | 0.0015282 | 0.00299673 | 0.0003936 | 0.00285997 | 0.00066557 | 0.00345098 | 0.01023957 | 13.4663605 | 2.32710305 | 3.51854302 | 1.02719857 | 3.35297541 | 0.97965636 | 4.29145549 | 1.29811287 |
| J | 0.00062976 | 0.00137508 | 0.00428461 | 0.00027188 | 0.00246943 | 0.00060632 | 0.00275394 | 0.00906872 | 12.3392324 | 2.12166155 | 5.039479 | 1.47022453 | 2.91945068 | 0.85082711 | 3.35472003 | 1.00963885 |
| K | 0.00084153 | 0.00173514 | 0.00417708 | 0.00059019 | 0.00397734 | 0.00091936 | 0.01973523 | 0.01713999 | 16.462836 | 2.81694585 | 4.89897847 | 1.430121 | 4.67706547 | 1.36512005 | 23.4278429 | 6.84671918 |
| L | 0.00091354 | 0.00194007 | 0.00449805 | 0.00163909 | 0.00421455 | 0.00178876 | 0.0420623 | 0.01316229 | 17.743488 | 3.08314193 | 5.29275112 | 1.55927849 | 4.97026951 | 1.46232287 | 5.16441015 | 1.56004368 |
| M | 0.0006116 | 0.00129119 | 0.00286777 | 0.00039014 | 0.00265452 | 0.00068521 | 0.0030545 | 0.03559015 | 11.8833887 | 2.0393166 | 3.36354507 | 0.98195641 | 3.11514971 | 0.90982144 | 4.20128196 | 1.7395392 |
| N | 0.0008502 | 0.00498445 | 0.00405446 | 0.0007176 | 0.00375849 | 0.00097299 | 0.04150535 | 0.01602984 | 16.6332854 | 2.85486699 | 4.75020676 | 1.39576506 | 4.41236681 | 1.2946882 | 49.1934053 | 14.4530511 |
Appendix B. Comparison between Different CNN Architectures
| Patch | WSI | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Read | Score | Stain Normalization | Prediction | Read | Score | Stain Normalization | Prediction | |||||||||
| Architecture | Avg | Std | Avg | Std | Avg | Std | Avg | Std | Avg | Std | Avg | Std | Avg | Std | Avg | Std |
| PROMETEO | 0.000612 | 0.001291 | 0.002868 | 0.00039 | 0.002655 | 0.000685 | 0.003054 | 0.004845 | 11.88339 | 2.039317 | 3.363545 | 0.981956 | 3.11515 | 0.909821 | 4.201282 | 1.739539 |
| ResNet34 | 0.000612 | 0.001279 | 0.002844 | 0.000405 | 0.002676 | 0.000686 | 0.008982 | 0.010086 | 11.92095 | 2.045551 | 3.333316 | 0.973793 | 3.148634 | 0.918751 | 10.71205 | 3.134596 |
| InceptionV3 | 0.000621 | 0.001317 | 0.002915 | 0.00041 | 0.002691 | 0.001136 | 0.039772 | 0.013828 | 12.10135 | 2.076446 | 3.415152 | 0.997178 | 3.168544 | 0.925316 | 46.72138 | 13.64997 |
| VGG16 | 0.000635 | 0.001341 | 0.002931 | 0.000427 | 0.002785 | 0.000691 | 0.028664 | 0.009241 | 12.37371 | 2.140448 | 3.45475 | 1.007557 | 3.280768 | 0.957531 | 34.92197 | 10.16074 |
| VGG19 | 0.000628 | 0.001313 | 0.002931 | 0.000425 | 0.00278 | 0.000682 | 0.029931 | 0.009305 | 12.34846 | 2.116793 | 3.44631 | 1.005834 | 3.266729 | 0.953449 | 36.25006 | 10.5361 |
| MobileNet | 0.000612 | 0.001278 | 0.00285 | 0.00042 | 0.002688 | 0.001111 | 0.025689 | 0.010986 | 11.96025 | 2.044115 | 3.383497 | 0.986745 | 3.208441 | 0.936362 | 31.11017 | 9.030854 |
| DenseNet121 | 0.000611 | 0.001284 | 0.002879 | 0.000389 | 0.002687 | 0.000683 | 0.042489 | 0.01686 | 11.82392 | 2.035413 | 3.373127 | 0.985149 | 3.148681 | 0.919557 | 51.48291 | 14.94588 |
| Xception | 0.0006 | 0.001261 | 0.00288 | 0.000374 | 0.002758 | 0.000655 | 0.03405 | 0.011789 | 11.68313 | 1.987459 | 3.375536 | 0.986863 | 3.235289 | 0.945187 | 41.76486 | 12.17527 |
| ResNet101 | 0.000607 | 0.001265 | 0.002839 | 0.000398 | 0.002701 | 0.00067 | 0.043287 | 0.014679 | 11.84637 | 2.035472 | 3.327598 | 0.971357 | 3.171104 | 0.925785 | 52.51713 | 15.26661 |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P. Epidemiology of prostate cancer. World J. Oncol. 2019, 10, 63. [Google Scholar] [CrossRef] [PubMed]
- Borley, N.; Feneley, M.R. Prostate cancer: Diagnosis and staging. Asian J. Androl. 2009, 11, 74. [Google Scholar] [CrossRef] [PubMed]
- Hamet, P.; Tremblay, J. Artificial intelligence in medicine. Metabolism 2017, 69, S36–S40. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, A.S. The impact of artificial intelligence in medicine on the future role of the physician. PeerJ 2019, 7, e7702. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.; Wu, G.; Suk, H.I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zheng, X.; Li, Y.; Zhang, Q.; Ying, S. Multimodal neuroimaging feature learning with multimodal stacked deep polynomial networks for diagnosis of Alzheimer’s disease. IEEE J. Biomed. Health Inform. 2017, 22, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Dou, Q.; Chen, H.; Yu, L.; Zhao, L.; Qin, J.; Wang, D.; Mok, V.C.; Shi, L.; Heng, P.A. Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans. Med. Imaging 2016, 35, 1182–1195. [Google Scholar] [CrossRef] [PubMed]
- Duran-Lopez, L.; Dominguez-Morales, J.P.; Corral-Jaime, J.; Vicente-Diaz, S.; Linares-Barranco, A. COVID-XNet: A custom deep learning system to diagnose and locate COVID-19 in chest X-ray images. Appl. Sci. 2020, 10, 5683. [Google Scholar] [CrossRef]
- Bahrami, K.; Shi, F.; Rekik, I.; Shen, D. Convolutional neural network for reconstruction of 7T-like images from 3T MRI using appearance and anatomical features. In Deep Learning and Data Labeling for Medical Applications; Springer: Berlin/Heidelberg, Germany, 2016; pp. 39–47. [Google Scholar]
- Ström, P.; Kartasalo, K.; Olsson, H.; Solorzano, L.; Delahunt, B.; Berney, D.M.; Bostwick, D.G.; Evans, A.J.; Grignon, D.J.; Humphrey, P.A.; et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: A population-based, diagnostic study. Lancet Oncol. 2020, 21, 222–232. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Silva, V.W.K.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Duran-Lopez, L.; Dominguez-Morales, J.P.; Conde-Martin, A.F.; Vicente-Diaz, S.; Linares-Barranco, A. PROMETEO: A CNN-Based Computer-Aided Diagnosis System for WSI Prostate Cancer Detection. IEEE Access 2020, 8, 128613–128628. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Reinhard, E.; Adhikhmin, M.; Gooch, B.; Shirley, P. Color transfer between images. IEEE Comput. Graph. Appl. 2001, 21, 34–41. [Google Scholar] [CrossRef]
- Magee, D.; Treanor, D.; Crellin, D.; Shires, M.; Smith, K.; Mohee, K.; Quirke, P. Colour normalisation in digital histopathology images. In Proceedings of the Optical Tissue Image analysis in Microscopy, Histopathology and Endoscopy (MICCAI Workshop), London, UK, 24 September 2009; pp. 100–111. [Google Scholar]
- Zhuang, F.; Qi, Z.; Duan, K.; Xi, D.; Zhu, Y.; Zhu, H.; Xiong, H.; He, Q. A comprehensive survey on transfer learning. arXiv 2019, arXiv:1911.02685. [Google Scholar] [CrossRef]
- Zhang, Q.; Bai, C.; Chen, Z.; Li, P.; Yu, H.; Wang, S.; Gao, H. Deep learning models for diagnosing spleen and stomach diseases in smart Chinese medicine with cloud computing. Concurr. Comput. Pract. Exp. 2019, e5252. [Google Scholar] [CrossRef]
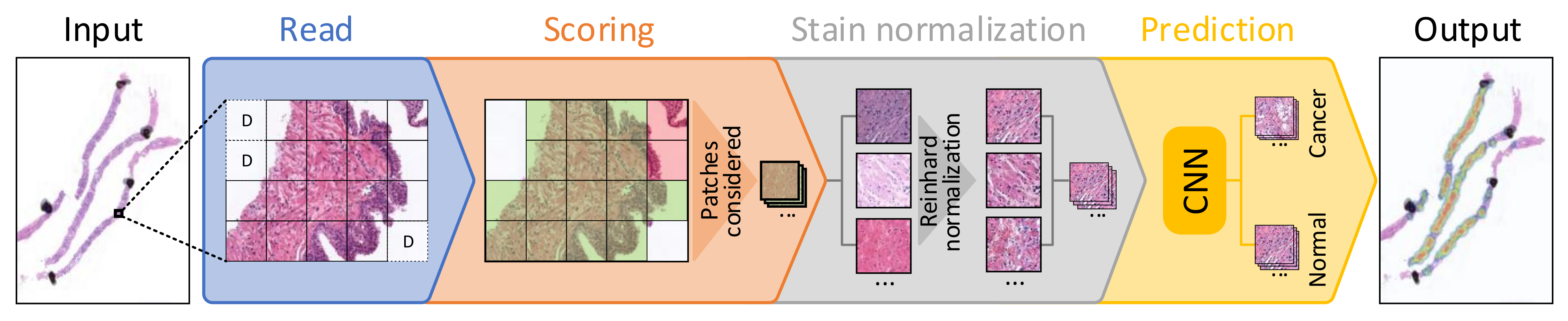


| Hospital | No. of WSIs | ||
|---|---|---|---|
| Normal | Malignant | Total | |
| Virgen de Valme Hospital | 27 | 70 | 97 |
| Clínic Hospital | 100 | 129 | 229 |
| Puerta del Mar Hospital | 79 | 65 | 144 |
| Model | Avg. Prediction Time (patch) | Avg. Prediction Time (WSI) | Slowdown * | Trainable Parameters |
|---|---|---|---|---|
| PROMETEO | 3.054 ± 4.845 ms | 4.201 ± 1.739 s | 1× | 1,107,010 |
| ResNet34 | 8.982 ± 10.086 ms | 10.712 ± 3.134 s | 2.55× | 21,800,107 |
| InceptionV3 | 41.301 ± 44.282 ms | 49.076 ± 14.353 s | 11.68× | 23,851,784 |
| VGG16 | 28.664 ± 9.241 ms | 34.921 ± 10.160 s | 8.31× | 138,357,544 |
| VGG19 | 29.931 ± 9.305 ms | 36.250 ± 10.536 s | 8.63× | 143,667,240 |
| MobileNet | 25.689 ± 10.986 ms | 31.110 ± 9.030 s | 7.41× | 4,253,864 |
| DenseNet121 | 42.489 ± 16.859 ms | 51.483 ± 14.945 s | 12.25× | 8,062,504 |
| Xception | 34.050 ± 11.789 ms | 41.764 ± 12.175 s | 9.94× | 22,910,480 |
| ResNet101 | 43.287 ± 14.679 ms | 52.517 ± 15.266 s | 12.50× | 44,707,176 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duran-Lopez, L.; Dominguez-Morales, J.P.; Rios-Navarro, A.; Gutierrez-Galan, D.; Jimenez-Fernandez, A.; Vicente-Diaz, S.; Linares-Barranco, A. Performance Evaluation of Deep Learning-Based Prostate Cancer Screening Methods in Histopathological Images: Measuring the Impact of the Model’s Complexity on Its Processing Speed. Sensors 2021, 21, 1122. https://doi.org/10.3390/s21041122
Duran-Lopez L, Dominguez-Morales JP, Rios-Navarro A, Gutierrez-Galan D, Jimenez-Fernandez A, Vicente-Diaz S, Linares-Barranco A. Performance Evaluation of Deep Learning-Based Prostate Cancer Screening Methods in Histopathological Images: Measuring the Impact of the Model’s Complexity on Its Processing Speed. Sensors. 2021; 21(4):1122. https://doi.org/10.3390/s21041122
Chicago/Turabian StyleDuran-Lopez, Lourdes, Juan P. Dominguez-Morales, Antonio Rios-Navarro, Daniel Gutierrez-Galan, Angel Jimenez-Fernandez, Saturnino Vicente-Diaz, and Alejandro Linares-Barranco. 2021. "Performance Evaluation of Deep Learning-Based Prostate Cancer Screening Methods in Histopathological Images: Measuring the Impact of the Model’s Complexity on Its Processing Speed" Sensors 21, no. 4: 1122. https://doi.org/10.3390/s21041122
APA StyleDuran-Lopez, L., Dominguez-Morales, J. P., Rios-Navarro, A., Gutierrez-Galan, D., Jimenez-Fernandez, A., Vicente-Diaz, S., & Linares-Barranco, A. (2021). Performance Evaluation of Deep Learning-Based Prostate Cancer Screening Methods in Histopathological Images: Measuring the Impact of the Model’s Complexity on Its Processing Speed. Sensors, 21(4), 1122. https://doi.org/10.3390/s21041122











