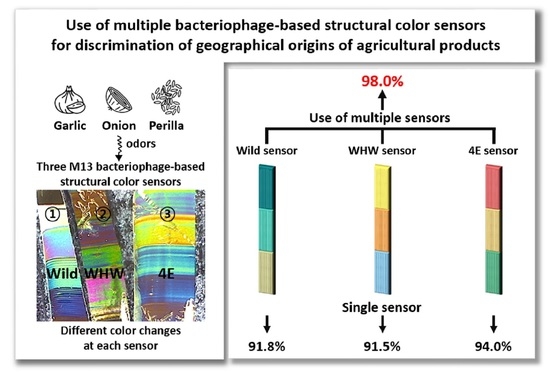
Use of Multiple Bacteriophage-Based Structural Color Sensors to Improve Accuracy for Discrimination of Geographical Origins of Agricultural Products
Abstract
1. Introduction
2. Materials and Methods
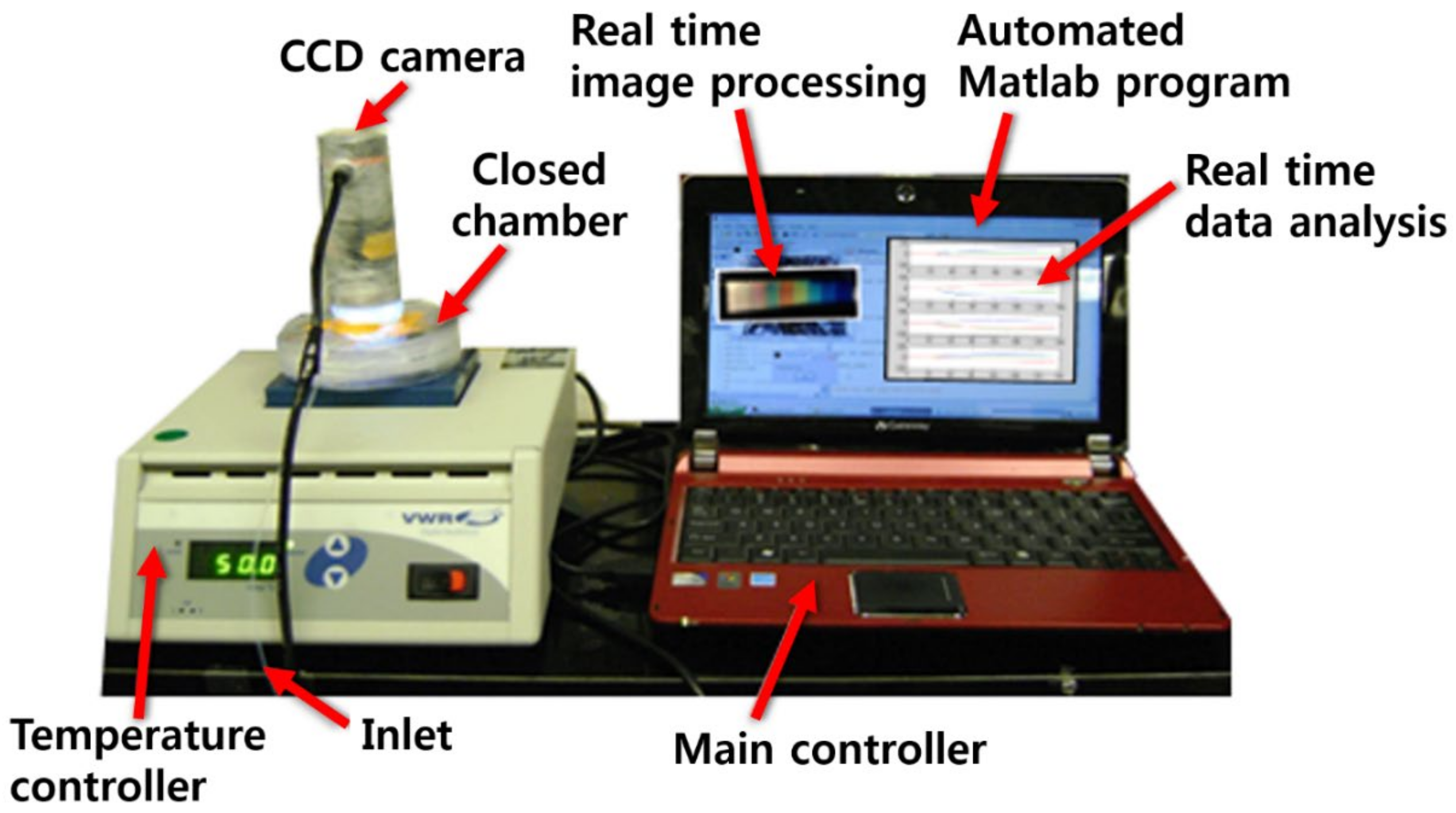
2.1. Sample Preparation and Measurement of Samples Using Color Sensors
2.2. Discriminant Analysis Using SVM
3. Results
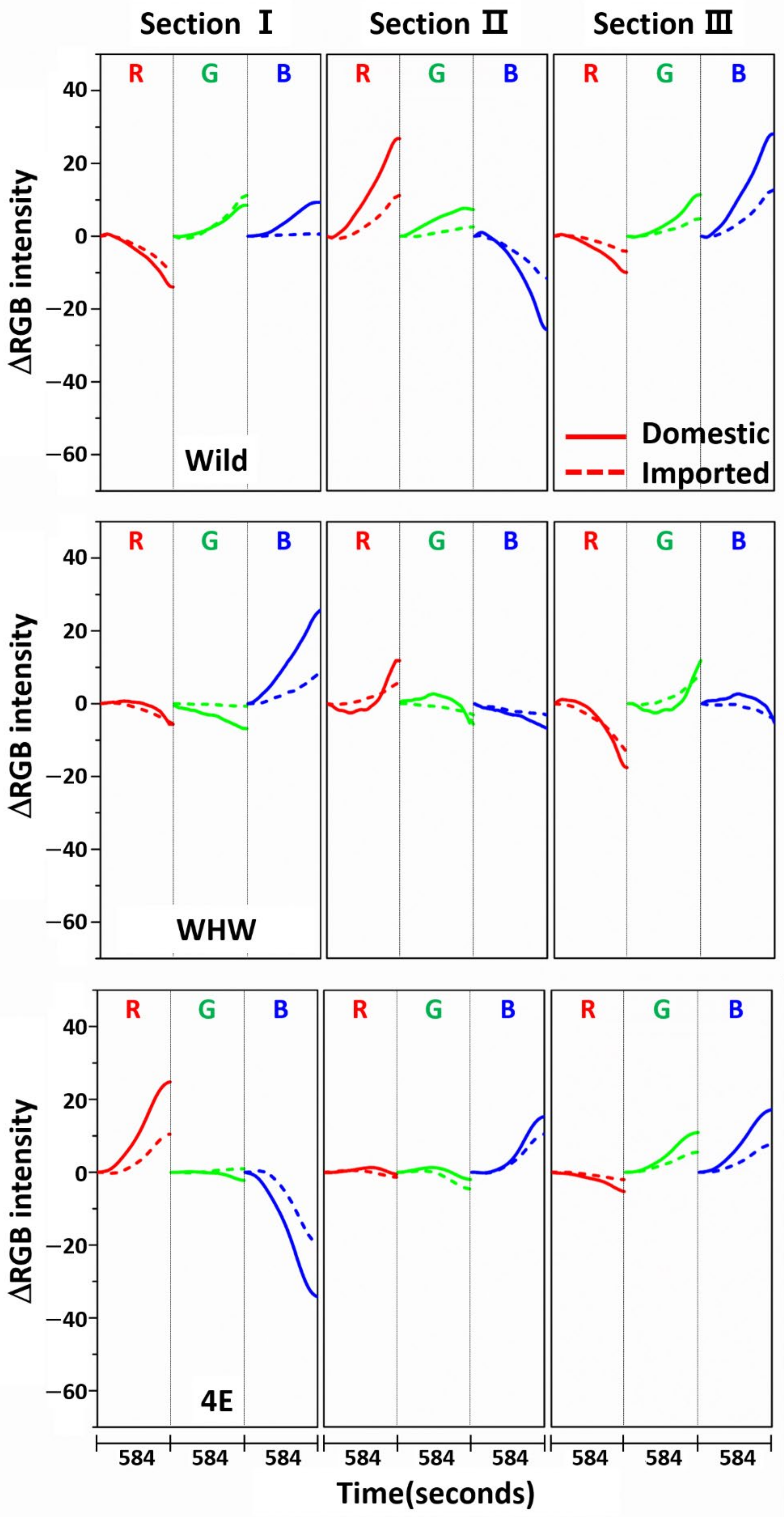
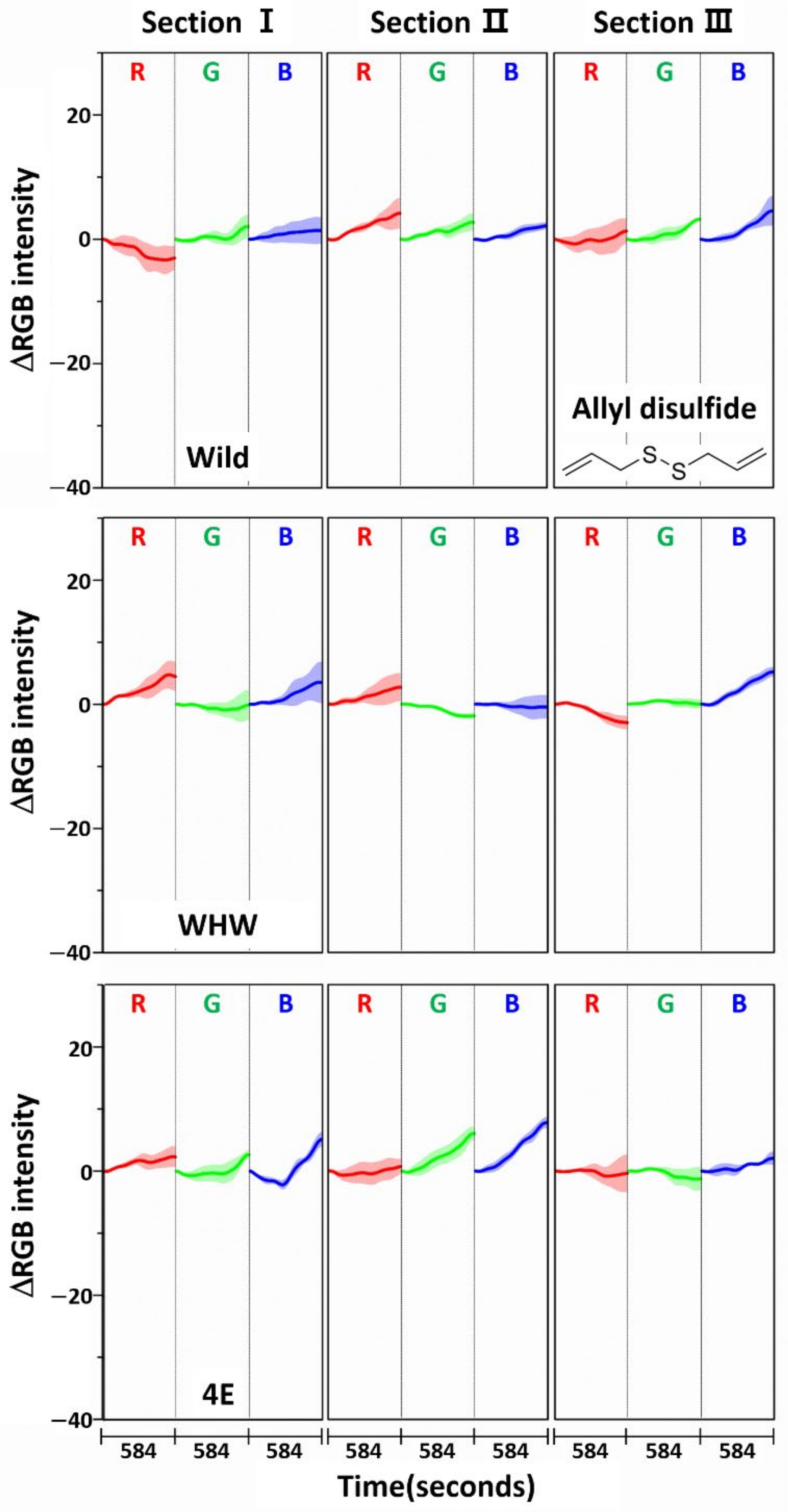
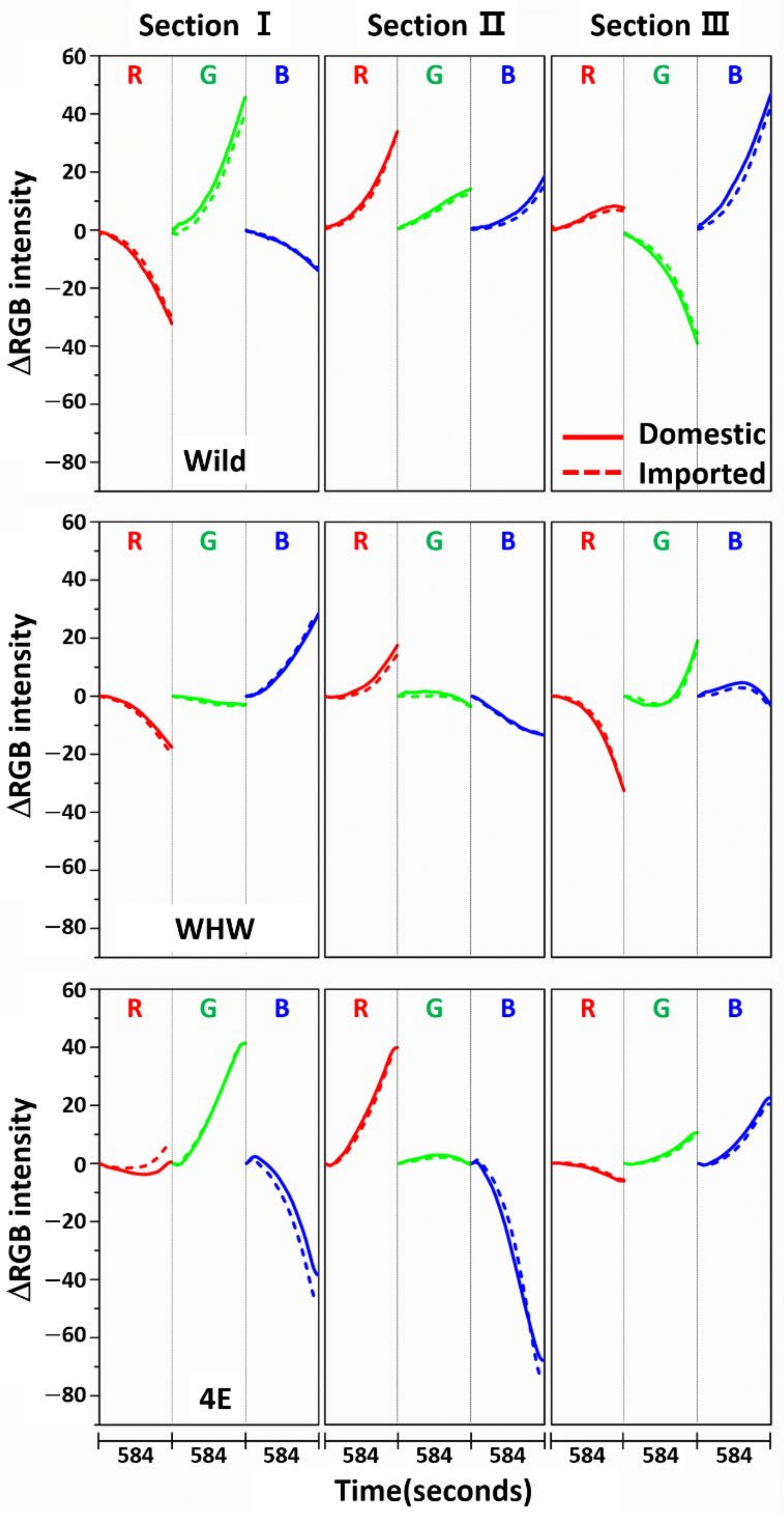
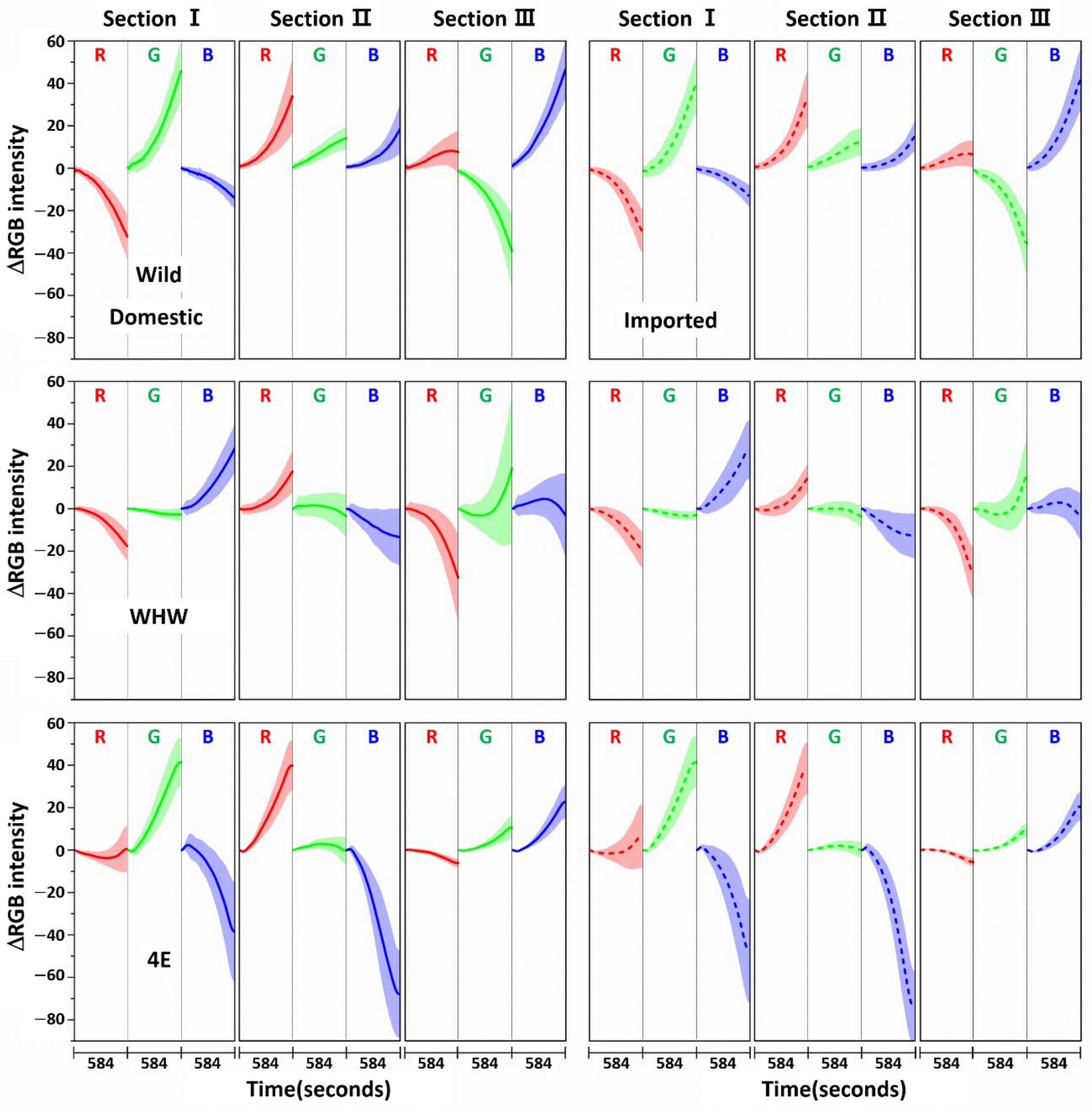
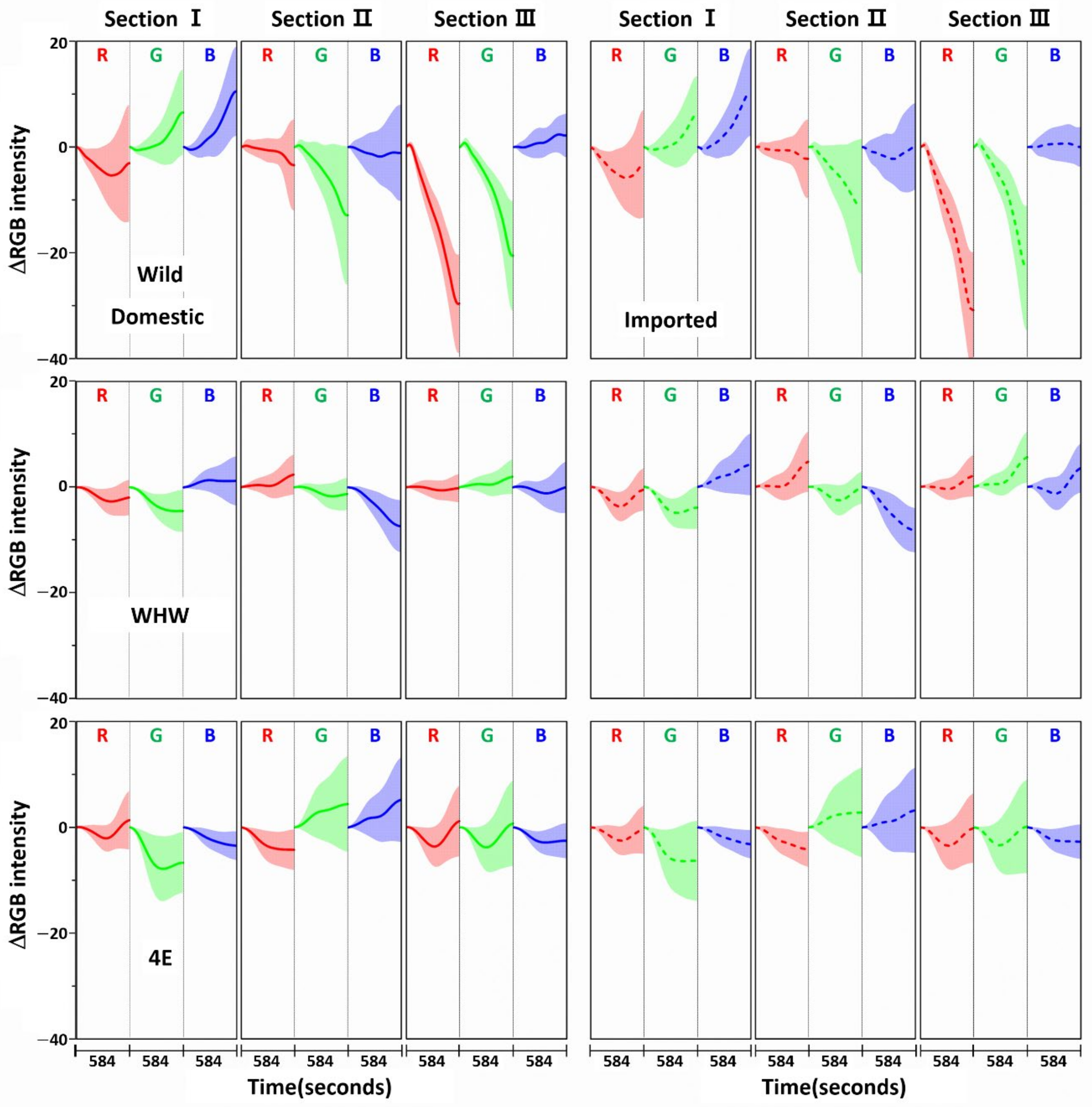
3.1. Examination of RGB Signals of Garlic Samples Measured with Each Color Sensor
3.2. Examination of RGB Signals of Onion Samples Measured with Each Color Sensor
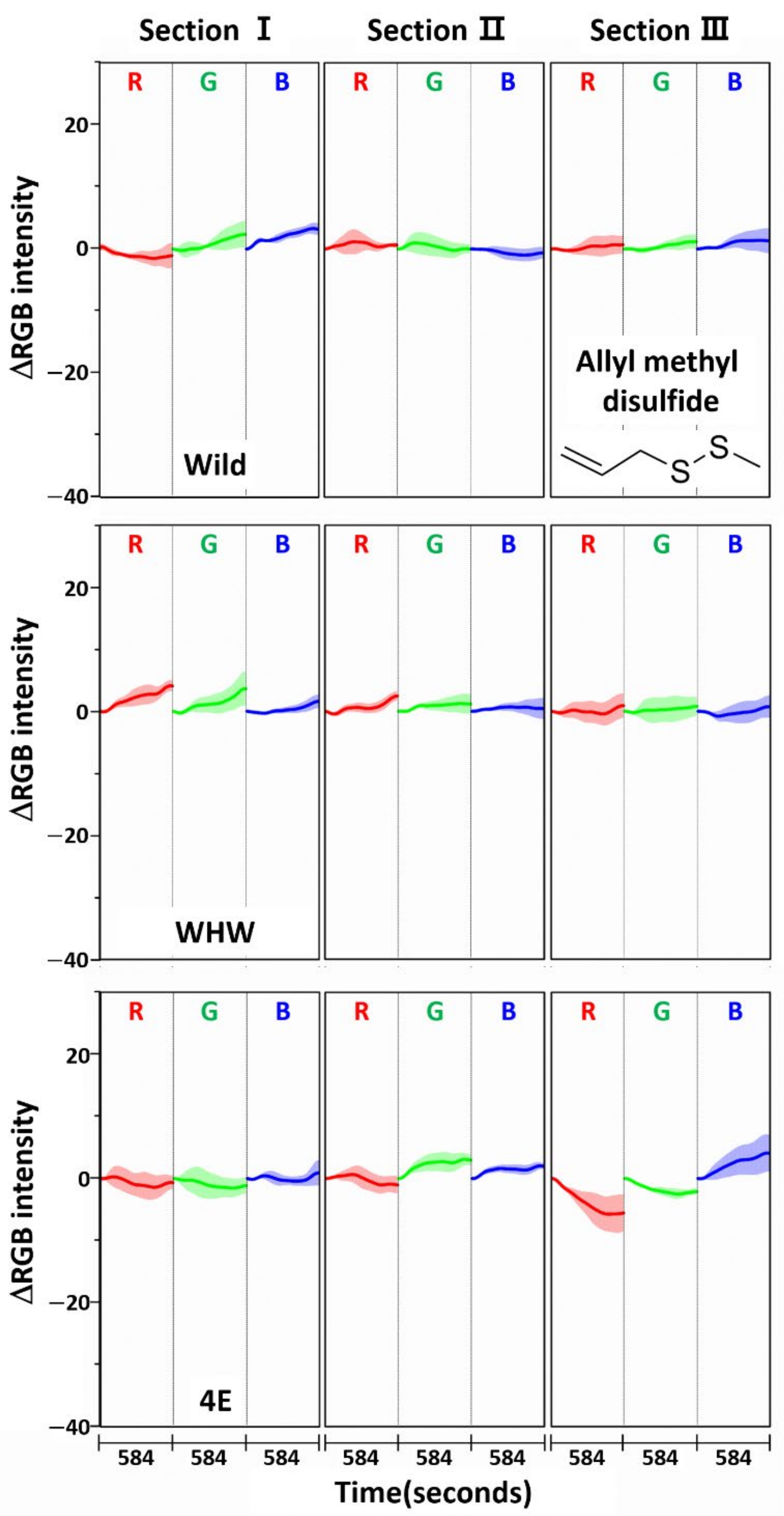
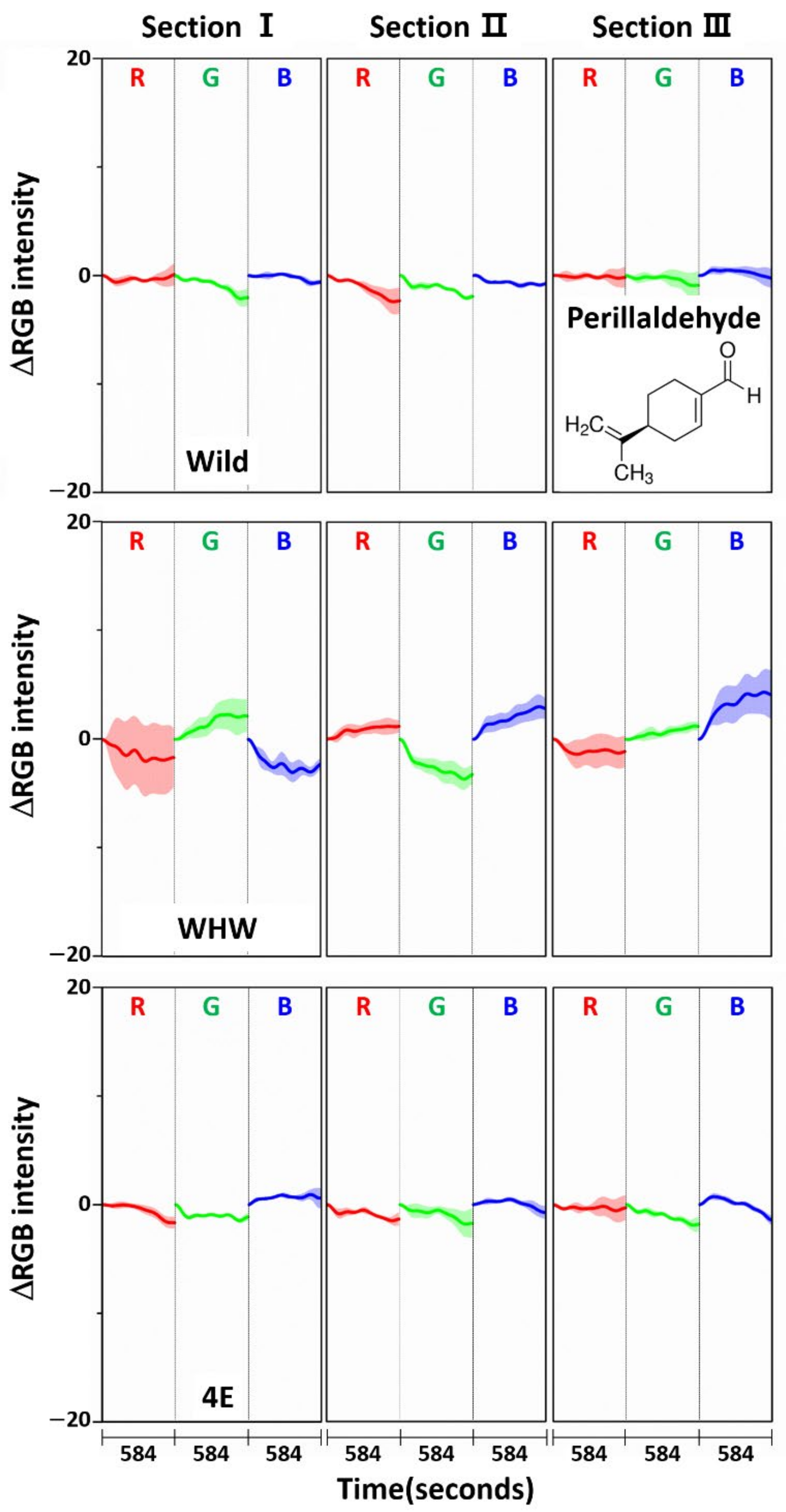
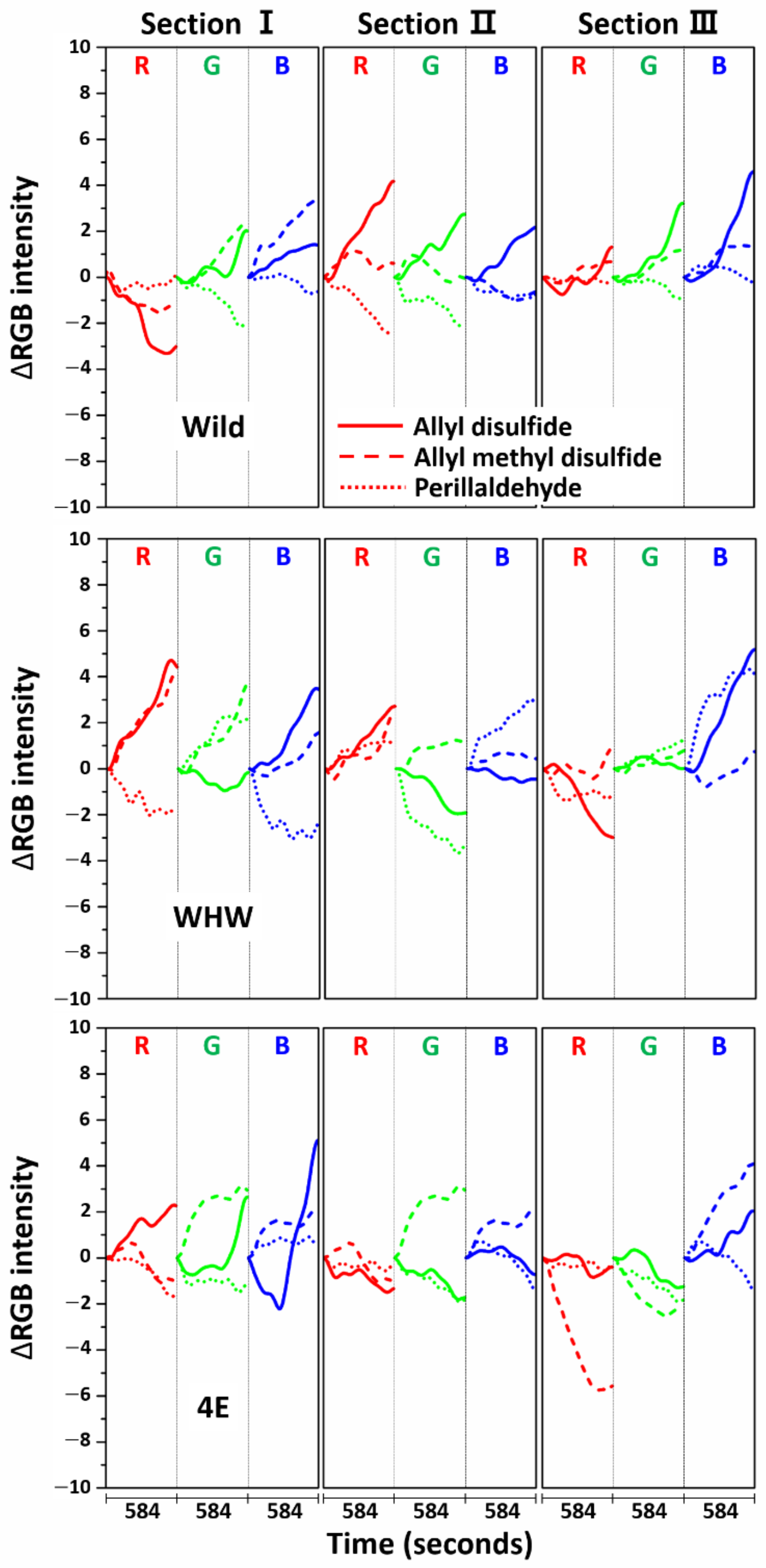
3.3. Examination of RGB Signals of Perilla Samples Measured with Each Color Sensor
3.4. Examination of Discrimination Accuracy Using SVM
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bajoub, A.; Carrasco-Pancorbo, A.; Ajal, E.A.; Ouazzani, N.; Fernández-Gutiérrez, A. Potential of LC–MS phenolic profiling combined with multivariate analysis as an approach for the determination of the geographical origin of north Moroccan virgin olive oils. Food Chem. 2015, 166, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Jaitz, L.; Siegl, K.; Eder, R.; Rak, G.; Abranko, L.; Koellensperger, G.; Hann, S. LC–MS/MS analysis of phenols for classification of red wine according to geographic origin, grape variety and vintage. Food Chem. 2010, 122, 366–372. [Google Scholar] [CrossRef]
- Lopez, M.I.; Pettis, J.S.; Smith, I.B.; Chu, P.-S. Multiclass Determination and Confirmation of Antibiotic Residues in Honey Using LC-MS/MS. J. Agric. Food Chem. 2008, 56, 1553–1559. [Google Scholar] [CrossRef]
- Seol, D.; Moon, J.S.; Lee, Y.; Han, J.; Jang, D.; Kang, D.J.; Moon, J.; Jang, E.; Oh, J.W.; Chung, H. Feasibility of using a bacteriophage-based structural color sensor for screening the geographical origins of agricultural products. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 197, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.W.; Chung, W.J.; Heo, K.; Jin, H.-E.; Lee, B.Y.; Wang, E.; Zueger, C.; Wong, W.; Meyer, J.; Kim, C.; et al. Biomimetic virus-based colourimetric sensors. Nat. Commun. 2014, 5, 3043. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.S.; Kim, W.G.; Shin, D.M.; Lee, S.Y.; Kim, C.; Lee, Y.; Han, J.; Kim, K.; Yoo, S.Y.; Oh, J.W. Bioinspired M-13 bacteriophage-based photonic nose for differential cell recognition. Chem. Sci. 2017, 8, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.S.; Lee, Y.; Shin, D.M.; Kim, C.; Kim, W.G.; Park, M.; Han, J.; Song, H.; Kim, K.; Oh, J.W. Identification of Endocrine Disrupting Chemicals using a Virus-Based Colorimetric Sensor. Chem. Asian J. 2016, 11, 3097–3101. [Google Scholar] [CrossRef]
- Seol, D.; Jang, D.; Oh, J.W.; Cha, K.; Chung, H. Discrimination of phthalate species using a simple phage-based colorimetric sensor in conjunction with hierarchical support vector machine. Environ. Res. 2019, 170, 238–242. [Google Scholar] [CrossRef]
- Lee, S.-W.; Mao, C.; Flynn, C.E.; Belcher, A.M. Ordering of quantum dots using genetically engineered viruses. Science 2002, 296, 892–895. [Google Scholar] [CrossRef]
- Wang, J.; Yang, M.; Zhu, Y.; Wang, L.; Tomsia, A.P.; Mao, C. Phage nanofibers induce vascularized osteogenesis in 3D printed bone scaffolds. Adv. Mater. 2014, 26, 4961–4966. [Google Scholar] [CrossRef]
- Jaworski, J.; Yokoyama, K.; Zueger, C.; Chung, W.J.; Lee, S.W.; Majumdar, A. Polydiacetylene incorporated with peptide receptors for the detection of trinitrotoluene explosives. Langmuir 2011, 27, 3180–3187. [Google Scholar] [CrossRef] [PubMed]
- Merzlyak, A.; Lee, S.-W. Engineering Phage Materials with Desired Peptide Display: Rational Design Sustained through Natural Selection. Bioconjug. Chem. 2009, 20, 2300–2310. [Google Scholar] [CrossRef]
- Lee, J.H.; Fan, B.; Samdin, T.D.; Monteiro, D.A.; Desai, M.S.; Scheideler, O.; Jin, H.E.; Kim, S.; Lee, S.-W. Phage-Based Structural Color Sensors and Their Pattern Recognition Sensing System. ACS Nano 2017, 11, 3632–3641. [Google Scholar] [CrossRef] [PubMed]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- John, C. Platt Sequential Minimal Optimization: A Fast Algorithm for Training Support Vector Machines. 1998. Available online: https://www.microsoft.com/en-us/research/publication/sequential-minimal-optimization-a-fast-algorithm-for-training-support-vector-machines/ (accessed on 16 June 2020).
- Christopher, J.C.; Burges, A. Tutorial on Support Vector Machines for Pattern Recognition. Data Min. Knowl. Discov. 1998, 2, 121–167. [Google Scholar]
- Chung, W.J.; Oh, J.W.; Kwak, K.; Lee, B.Y.; Meyer, J.; Wang, E.; Hexemer, A.; Lee, S.-W. Biomimetic self-templating supramolecular structures. Nature 2011, 478, 364–368. [Google Scholar] [CrossRef]
- Moon, J.-S.; Park, M.; Kim, W.-G.; Kim, C.; Hwang, J.; Seol, D.; Kim, C.-S.; Sohn, J.-R.; Chung, H.; Oh, J.-W. M-13 bacteriophage based structural color sensor for detecting antibiotics. Sens. Actuators B Chem. 2017, 240, 757–762. [Google Scholar] [CrossRef]
- Sch¨olkopf, B.; Sung, K.K.; Burges, C.J.C.; Girosi, F.; Niyogi, P.; Poggio, T.; Vapnik, V. Comparing Support Vector Machines with Gaussian Kernels to Radial Basis Function Classifiers. IEEE Trans. Signal Process. 1997, 45, 2758–2765. [Google Scholar] [CrossRef]
- Kavzoglu, T.; Colkesen, I. A kernel functions analysis for support vector machines for land cover classification. Int. J. Appl. Earth Obs. 2009, 11, 352–359. [Google Scholar] [CrossRef]
- Park, J.; Lee, J.-M.; Chun, H.; Lee, Y.; Hong, S.-J.; Jung, H.; Kim, Y.-J.; Kim, W.-G.; Devaraj, V.; Choi, E.J.; et al. Optical bioelectronic nose of outstanding sensitivity and selectivity toward volatile organic compounds implemented with genetically engineered bacteriophage: Integrated study of multi-scale computational prediction and experimental validation. Biosens. Bioelectron. 2021, 177, 112979. [Google Scholar] [CrossRef]
- Jirovetz, L.; Buchbauer, G.; Ngassoum, M.; Geissler, M. Analysis of the headspace aroma compounds of the seeds of the Cameroonian “garlic plant” Hua gabonii using SPME/GC/FID, SPME/GC/MS and olfactometry. Eur. Food Res. Technol. 2002, 214, 212–215. [Google Scholar] [CrossRef]
- Colina-Coca, C.; Gonzalez-Pena, D.; Vega, E.; de Ancos, B.; Sanchez-Moreno, C. Novel approach for the determination of volatile compounds in processed onion by headspace gas chromatography-mass spectrometry (HS GC-MS). Talanta 2013, 103, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Gwari, G.; Lohani, H.; Bhandari, U.; Haider, S.Z.; Singh, S.; Andola, H.; Chauhan, N. Chemical diversity in the volatiles of Perilla frutescens (L.) Britt. populations from Uttarakhand Himalaya (India). J. Essent. Oil Res. 2015, 28, 49–54. [Google Scholar] [CrossRef]
- Bumblauskiené, L.J.V.; Janulis, V.; Mazdzieriené, R.; Ragazinskiené, O. Preliminary analysis on essential oil composition of Perilla L. cultivated in Lithuania. Acta Pol. Pharm 2009, 66, 409–413. [Google Scholar] [PubMed]
- Huang, B.; Lei, Y.; Tang, Y.; Zhang, J.; Qin, L.; Liu, J. Comparison of HS-SPME with hydrodistillation and SFE for the analysis of the volatile compounds of Zisu and Baisu, two varietal species of Perilla frutescens of Chinese origin. Food Chem. 2011, 125, 268–275. [Google Scholar] [CrossRef]
- Nishimura, T.; Ohyama, K.; Goto, E.; Inagaki, N. Concentrations of perillaldehyde, limonene, and anthocyanin of Perilla plants as affected by light quality under controlled environments. Sci. Hortic. 2009, 122, 134–137. [Google Scholar] [CrossRef]
- Feng, L.; Musto, C.J.; Suslick, K.S. A Simple and Highly Sensitive Colorimetric Detection Method for Gaseous Formaldehyde. J. Am. Chem. 2010, 132, 4046–4047. [Google Scholar] [CrossRef]
- Wang, Y.Z.; Huo, X.; Zhao, D.; Geng, Y.; Fa, X.; Luo, H.; Yang, X.; Hou, M.C. Fast recognition of trace volatile compounds with a nanoporous dyes-based colorimetric sensor array. Talanta 2019, 192, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhuang, J.; Wei, G. Recent advances in the design of colorimetric sensors for environmental monitoring. Environ. Sci. Nano 2020, 7, 2195–2213. [Google Scholar] [CrossRef]
- Alberti, G.; Zanoni, C.; Magnaghi, L.R.; Biesuz, R. Disposable and Low-Cost Colorimetric Sensors for Environmental Analysis. Int. J. Environ. Res. Public Health 2020, 17, 8331. [Google Scholar] [CrossRef]



| Sample | Sensor | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|
| Garlic | Wild | 91.8% (0.4%) | 97.5% (0.5%) | 86.1% (0.9%) |
| WHW | 91.5% (0.5%) | 95.3% (0.7%) | 87.9% (0.9%) | |
| 4E | 94.0% (0.3%) | 97.6% (0.4%) | 90.4% (0.9%) | |
| Wild + WHW + 4E | 98.0% (0.3%) | 98.6% (0.4%) | 97.4% (0.4%) | |
| Onion | Wild | 89.8% (0.5%) | 89.8% (0.9%) | 90.0% (1.1%) |
| WHW | 90.3% (0.5%) | 90.3% (1.0%) | 90.3% (1.0%) | |
| 4E | 88.6% (0.5%) | 86.4% (1.1%) | 91.0% (1.0%) | |
| Wild + WHW + 4E | 97.5% (0.3%) | 97.7% (0.4%) | 97.5% (0.5%) | |
| Perilla | Wild | 89.8% (0.4%) | 86.7% (1.9%) | 92.6% (1.6%) |
| WHW | 92.1% (0.5%) | 87.1% (1.0%) | 96.9% (0.9%) | |
| 4E | 91.6% (0.4%) | 94.2% (1.1%) | 89.1% (1.2%) | |
| Wild + WHW + 4E | 97.1% (0.4%) | 97.4% (1.4%) | 96.8% (1.4%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seol, D.; Jang, D.; Cha, K.; Oh, J.-W.; Chung, H. Use of Multiple Bacteriophage-Based Structural Color Sensors to Improve Accuracy for Discrimination of Geographical Origins of Agricultural Products. Sensors 2021, 21, 986. https://doi.org/10.3390/s21030986
Seol D, Jang D, Cha K, Oh J-W, Chung H. Use of Multiple Bacteriophage-Based Structural Color Sensors to Improve Accuracy for Discrimination of Geographical Origins of Agricultural Products. Sensors. 2021; 21(3):986. https://doi.org/10.3390/s21030986
Chicago/Turabian StyleSeol, Daun, Daeil Jang, Kyungjoon Cha, Jin-Woo Oh, and Hoeil Chung. 2021. "Use of Multiple Bacteriophage-Based Structural Color Sensors to Improve Accuracy for Discrimination of Geographical Origins of Agricultural Products" Sensors 21, no. 3: 986. https://doi.org/10.3390/s21030986
APA StyleSeol, D., Jang, D., Cha, K., Oh, J.-W., & Chung, H. (2021). Use of Multiple Bacteriophage-Based Structural Color Sensors to Improve Accuracy for Discrimination of Geographical Origins of Agricultural Products. Sensors, 21(3), 986. https://doi.org/10.3390/s21030986