Fast Multi-Focus Fusion Based on Deep Learning for Early-Stage Embryo Image Enhancement
Abstract
1. Introduction
2. Related Works
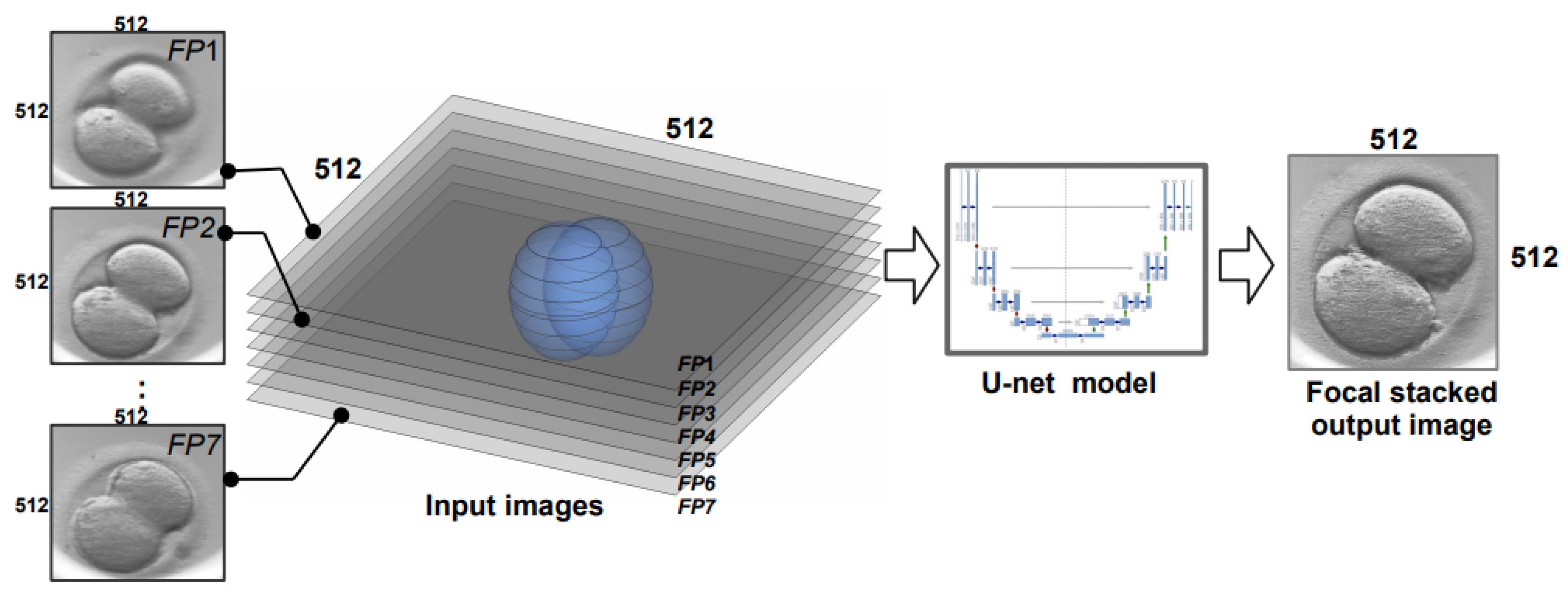
3. Multi-Focus Image Fusion Framework
3.1. Hardware Setup for the Acquisition of Multi-Focus Images
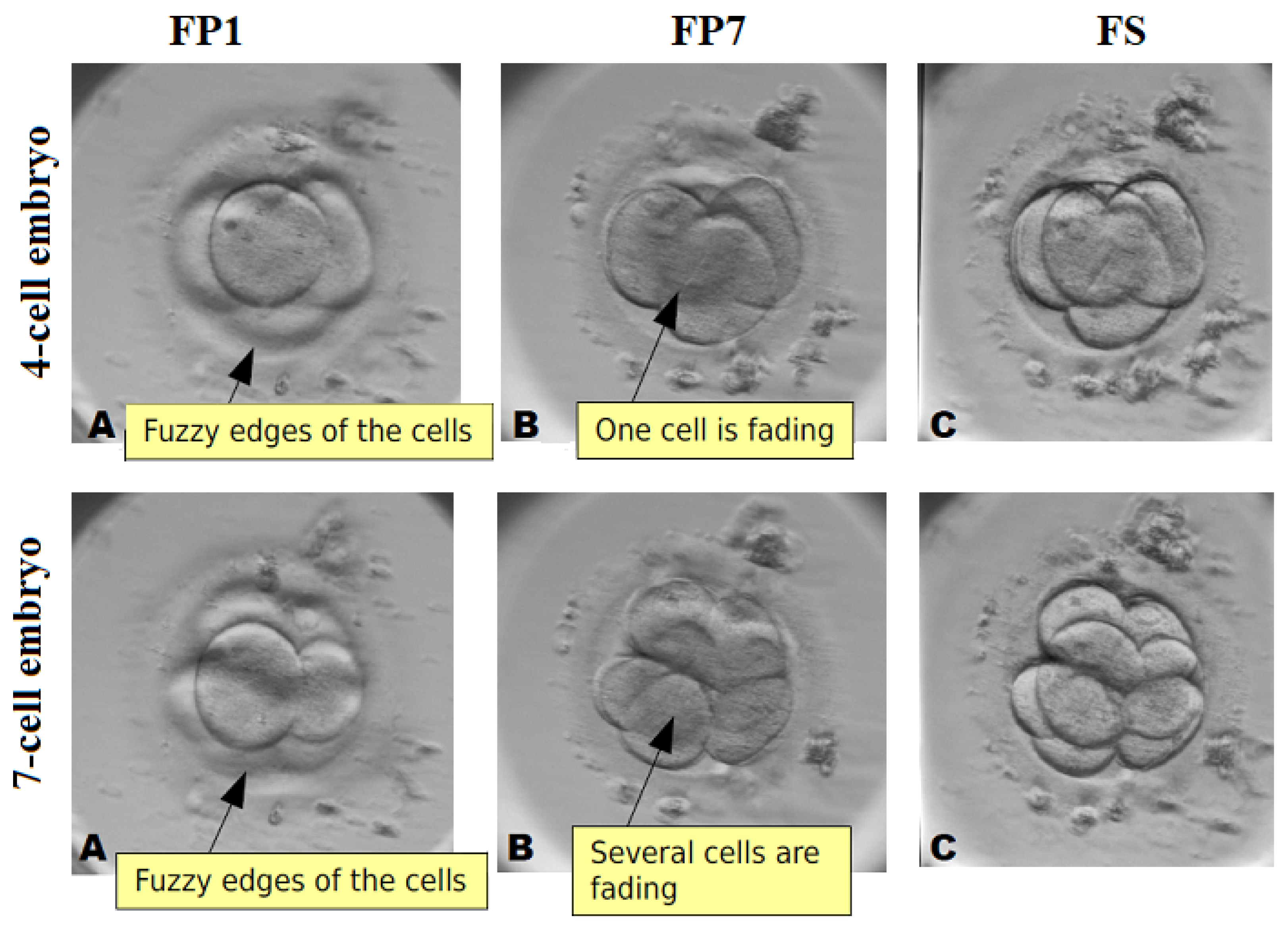
3.2. Data Preparation
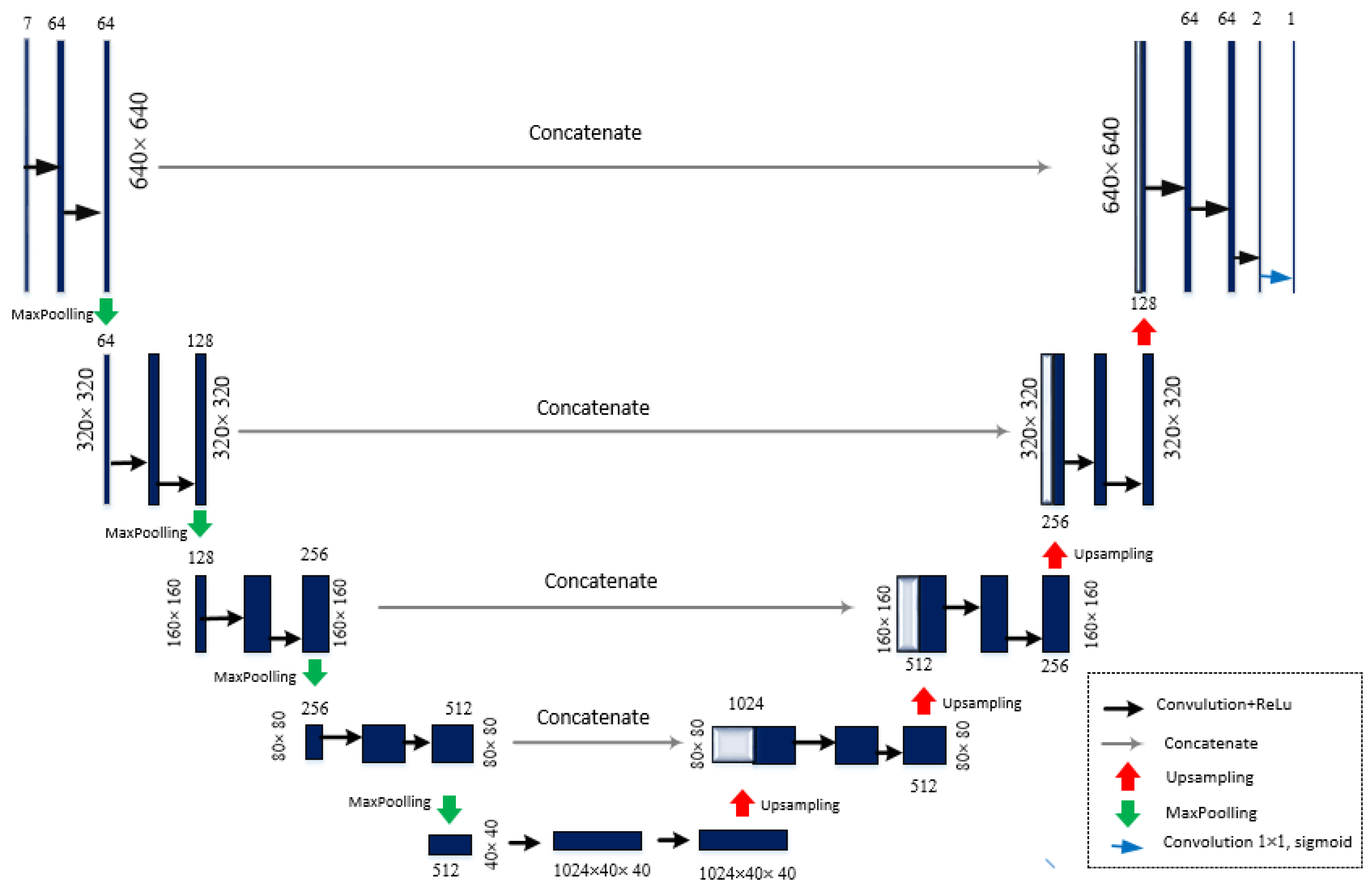
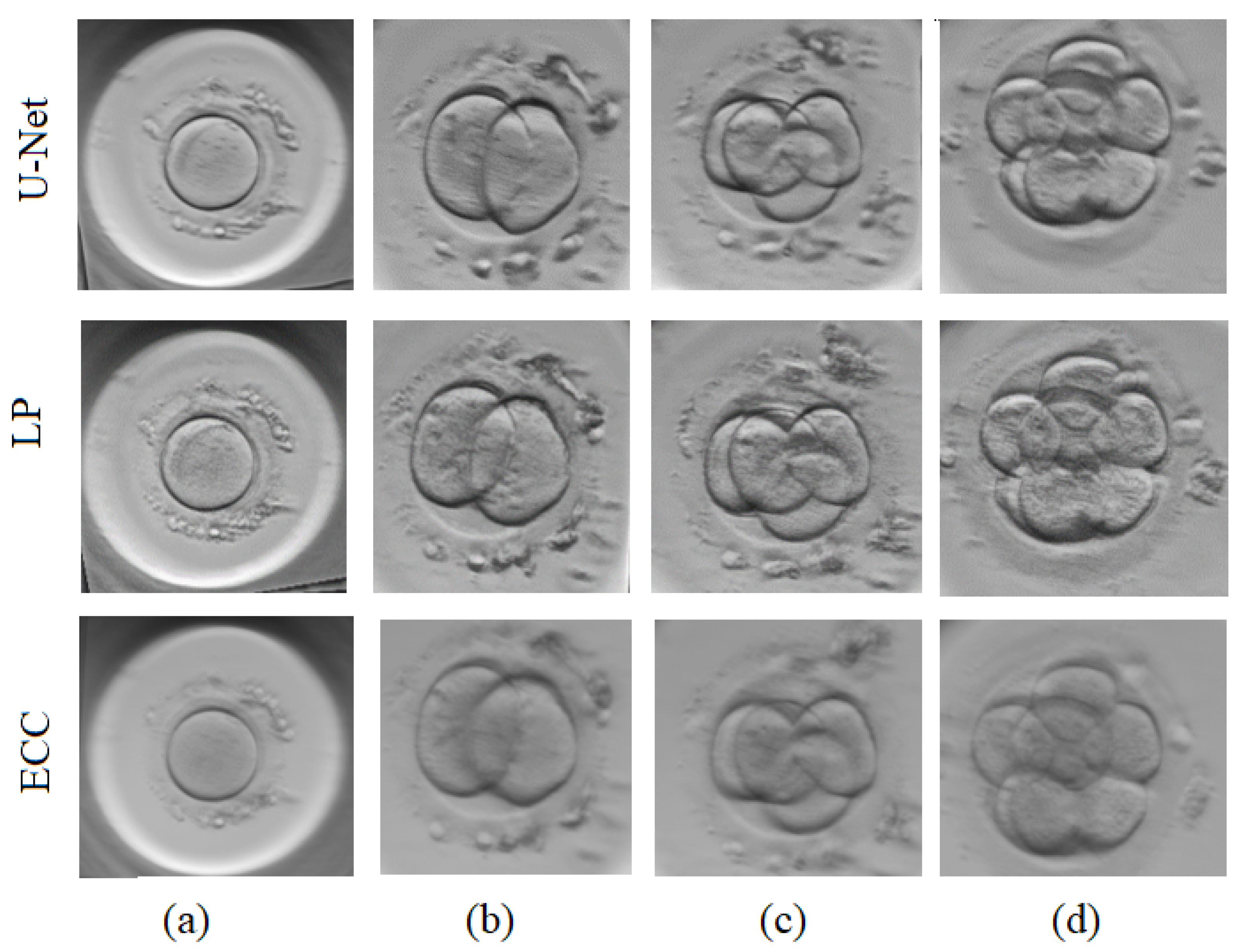
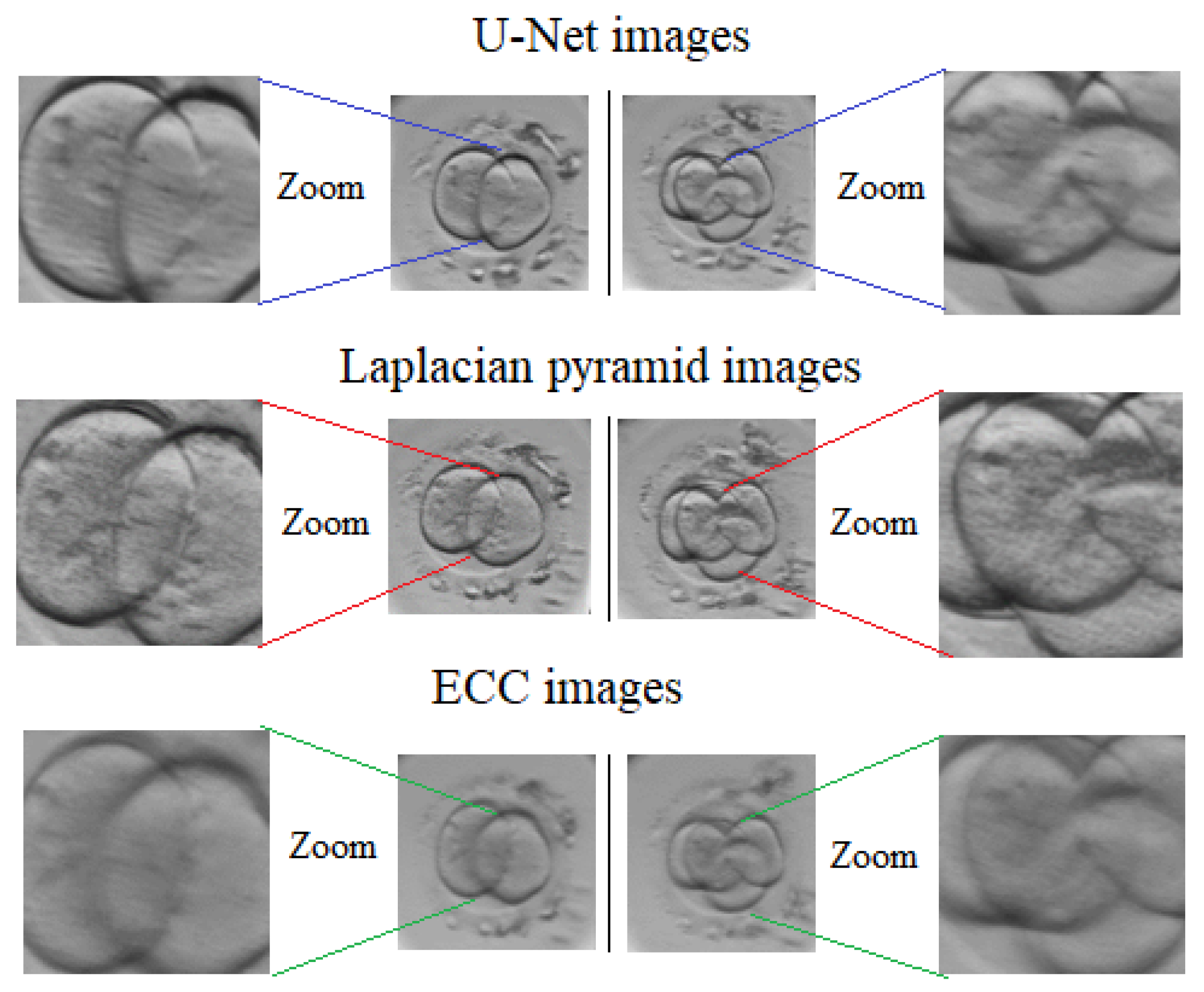
3.3. Multi-Focus Image Fusion Approach Using U-Net Architecture
3.4. Alternative Image Fusion Approaches
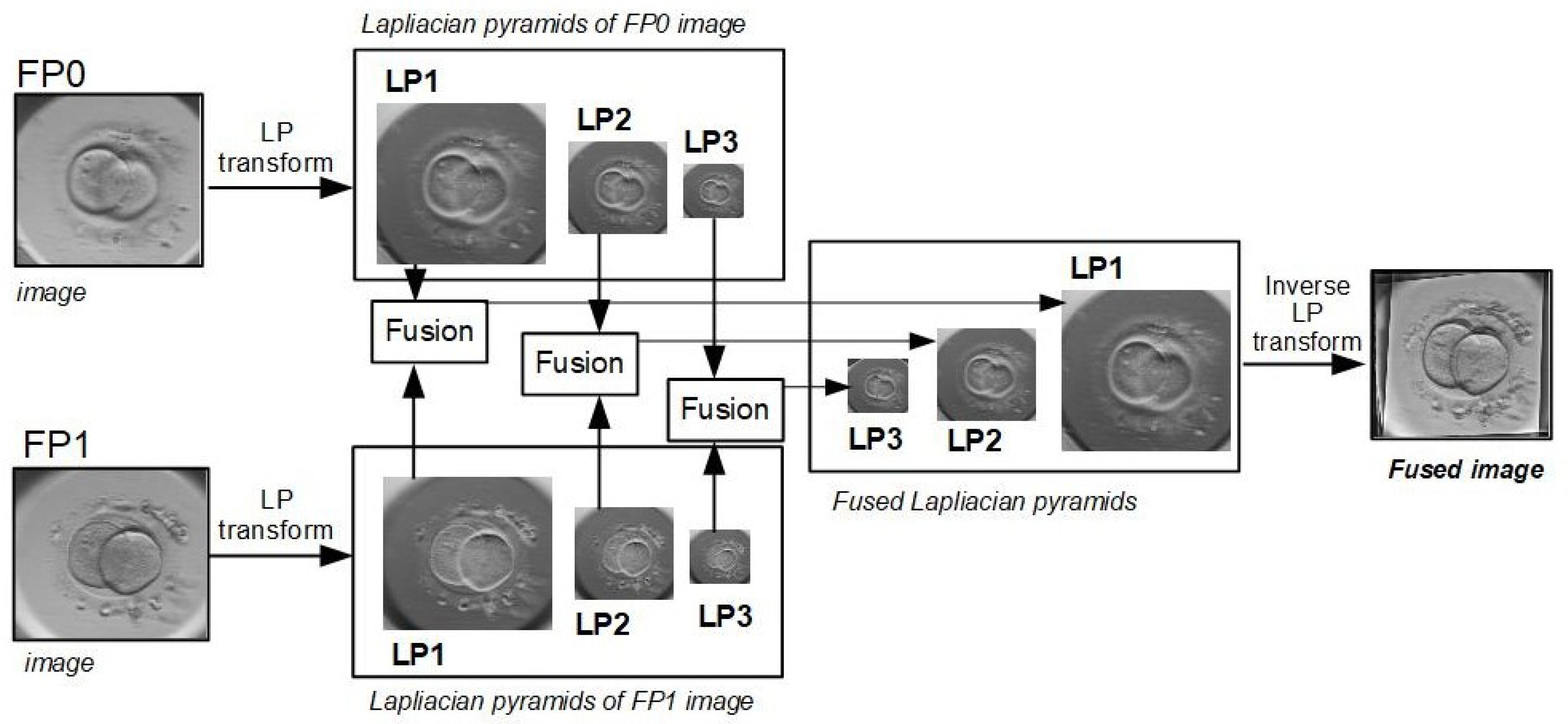
3.4.1. Inverse Laplacian Pyramid Transform
3.4.2. Enhanced Correlation Coefficient Maximization
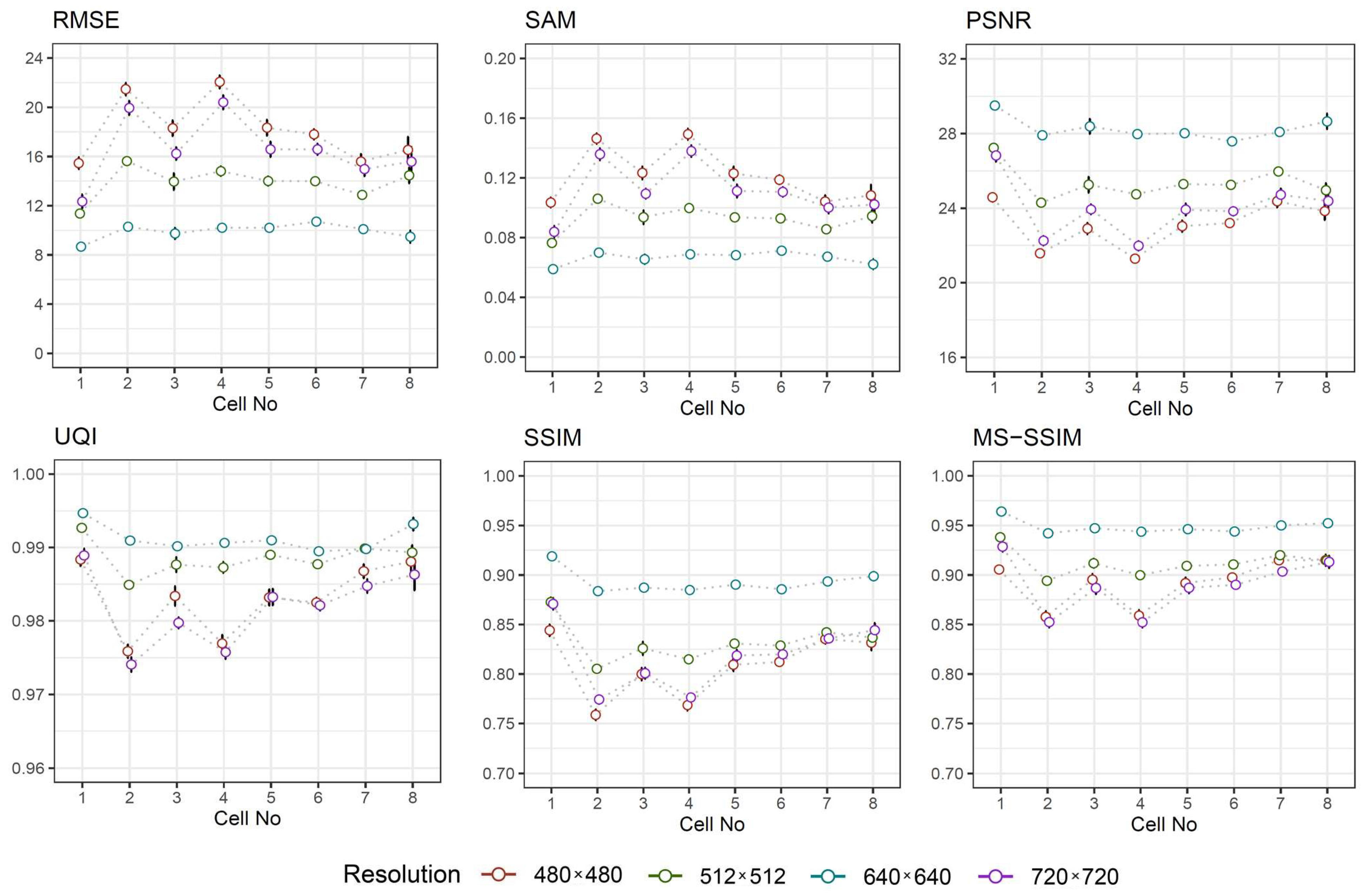
3.5. Image Similarity Metrics
- Root Mean Squared Error (RMSE) is commonly used to estimate the difference between two images by directly computing the variation in pixel values. The smaller value of RMSE represents better similarity [26]. Its value is defined aswhere M and N is the width and the height of image.
- Spectral Angle Mapper (SAM) determines the spectral similarity between two spectra by calculating the angle between the spectra and treating them as vectors in a space with dimensionality equal to the number of bands. Small angles between the two spectrums indicate high similarity, where the ideal value of zero indicates the best spectral quality [27]. It is calculated using the following formulawhere L is the number of bands, u and v are two adjacent spectra.
- Peak Signal-to-Noise Ratio (PSNR) is calculated based on RMSE, taking into account maximum possible pixel value of the image. For 8-bit representation, acceptable values for wireless transmission quality loss are considered to be around 20 dB to 25 dB, while in a lossy image range between 30 and 50 dB, where higher is better [28]. The value of PSNR is obtained usingwhere max is the maximum possible value a pixel can take. The value of is set when the pixels are represented using 8 bits.
- Universal Quality Index (UQI) represents brightness distortion, contrast distortion and correlation difference between two images. The best value is 1 if the images are equal [29]. The mathematical form of UQI iswhere is the mean value of the image, is the standard deviation the image, and is the covariance between two images and .
- Structural Similarity Index Method (SSIM) determines the local patterns of pixel intensities between two images taking into account three estimates of luminance, contrast, and structure [30]. The value ranges between and 1, where the ideal value is 1. The value of SSIM is given bywhere is the mean value of the image, is the standard deviation of image, is the covariance between two images, while and are constants. In this study, the values and have been chosen by default.
- Multi-Scale Structural Similarity Index Method (MS-SSIM), which is more advanced form of SSIM, determines the quality based in the terms of image luminance, contrast and structure at multiple scales. The ideal value is 1. The computations are typically performed in a sliding N × N (by default 11 × 11) Gaussian-weighted window [31].
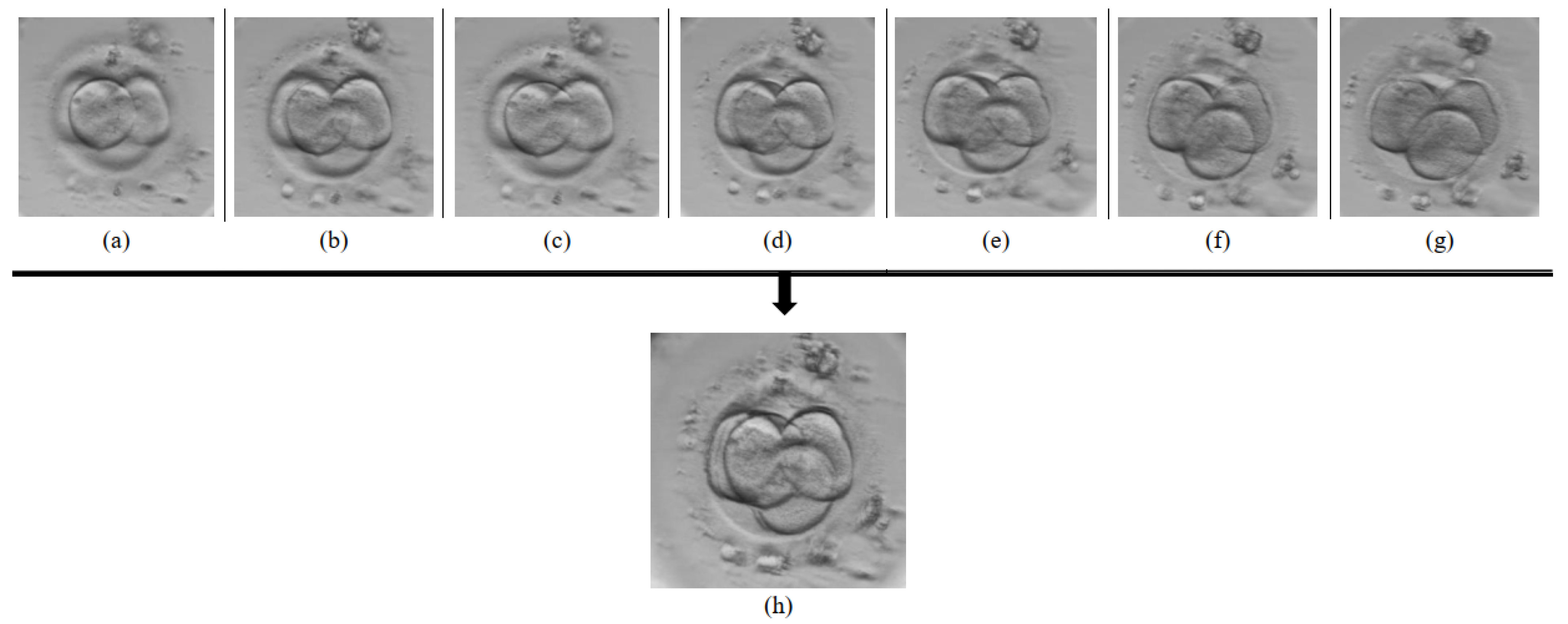
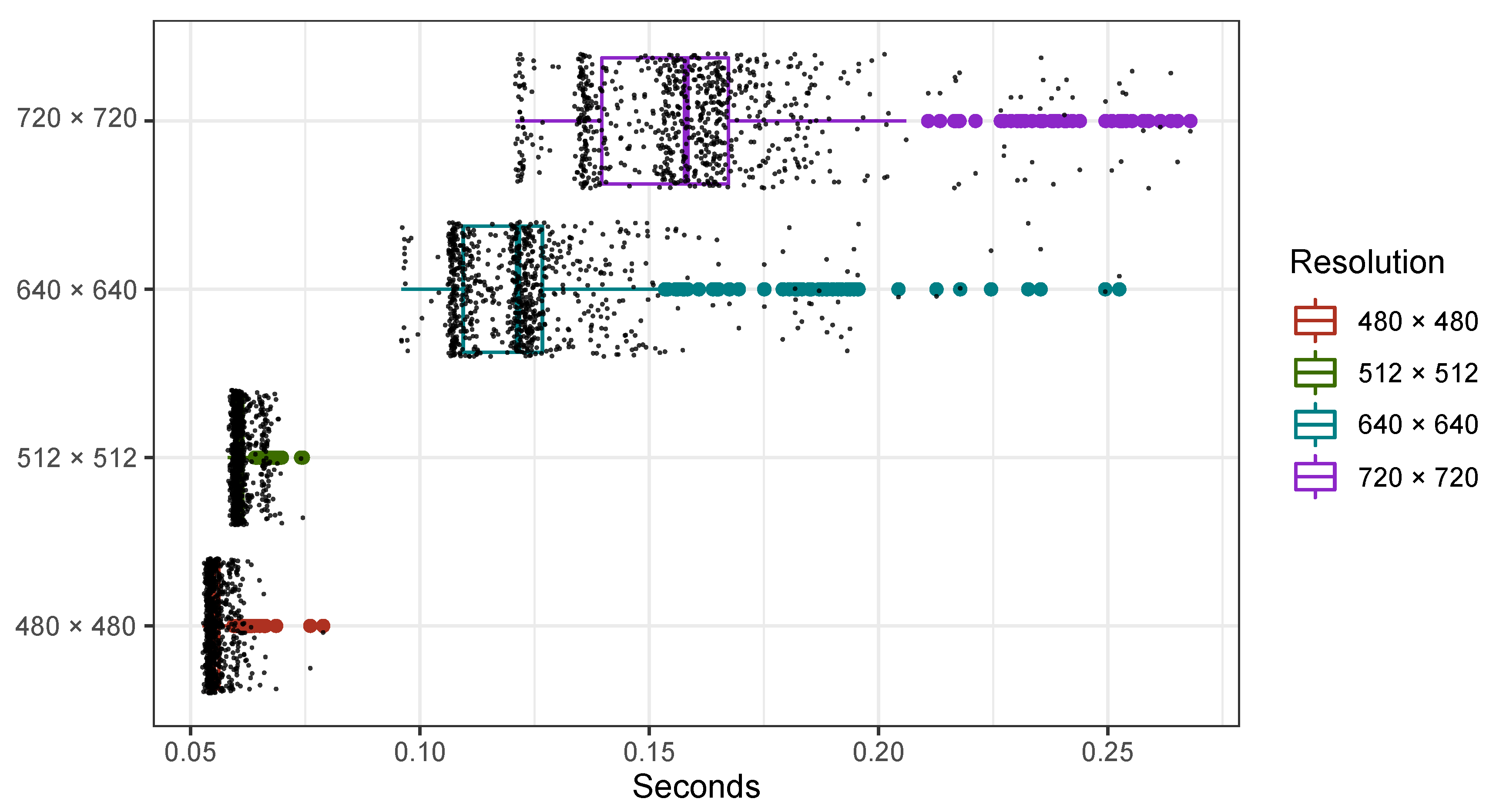
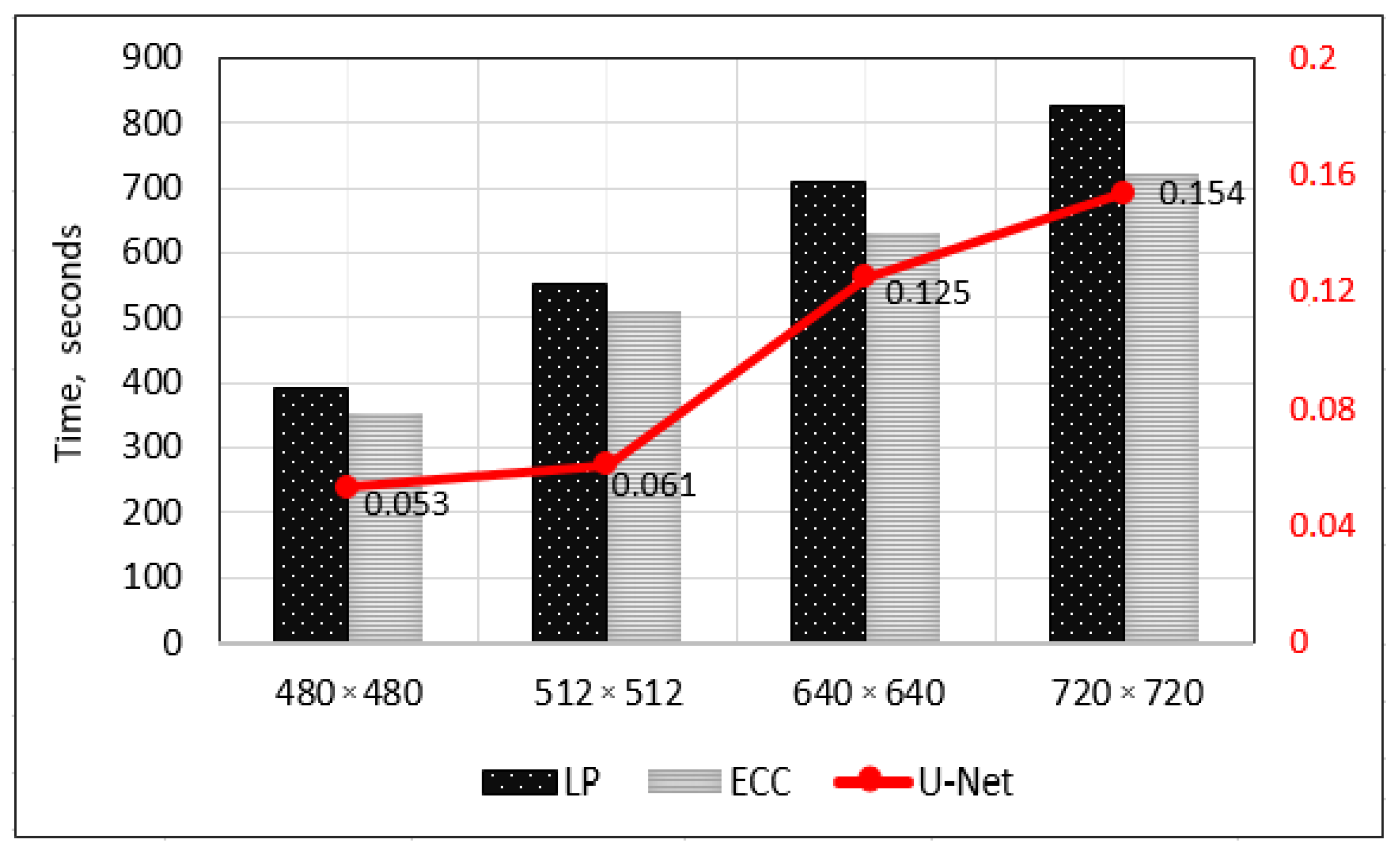
4. Experimental Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CNNs | Convolutional Neural Networks |
| ECC | Enhanced Correlation Coefficient |
| IVF | In-Vitro Fertilization |
| LP | Laplacian pyramid |
| MS-SSIM | Multi-Scale Structural Similarity Index Method |
| PSNR | Peak Signal-to-Noise Ratio |
| SAM | Spectral Angle Mapper |
| SSIM | Structural Similarity Index Method |
| TL | Time-Lapse |
| UQI | Universal Quality Index |
References
- Wang, S.S.; Sun, H.X. Blastocyst transfer ameliorates live birth rate compared with cleavage-stage embryos transfer in fresh in vitro fertilization or intracytoplasmic sperm injection cycles: Reviews and meta-analysis. Yonsei Med. J. 2014, 55, 815–825. [Google Scholar] [CrossRef] [PubMed]
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep Learning Techniques for Medical Image Segmentation: Achievements and Challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Gao, J.; Zhao, D. A review of the application of deep learning in medical image classification and segmentation. Ann. Transl. Med. 2020, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, P.; Kazemi, E.; Zhan, Q.; Malmsten, J.E.; Toschi, M.; Zisimopoulos, P.; Sigaras, A.; Lavery, S.; Cooper, L.A.D.; Hickman, C.; et al. Deep learning enables robust assessment and selection of human blastocysts after in vitro fertilization. NPJ Digit. Med. 2019, 2, 21. [Google Scholar] [CrossRef] [PubMed]
- Iwata, K.; Sato, M.; Matsumoto, I.; Shimura, T.; Yumoto, K.; Negami, A.; Mio, Y. Deep learning based on images of human embryos obtained from high-resolusion time-lapse cinematography for predicting good-quality embryos. Fertil. Steril. 2018, 110, e213. [Google Scholar] [CrossRef]
- Malmsten, J.; Zaninovic, N.; Zhan, Q.; Toschi, M.; Rosenwaks, Z.; Shan, J. Automatic prediction of embryo cell stages using artificial intelligence convolutional neural network. Fertil. Steril. 2018, 110, e360. [Google Scholar] [CrossRef]
- Wang, Y.; Moussavi, F.; Lorenzen, P. Automated Embryo Stage Classification in Time-Lapse Microscopy Video of Early Human Embryo Development. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2013; Springer: Berlin/Heidelberg, Germany, 2013; pp. 460–467. [Google Scholar] [CrossRef]
- Raudonis, V.; Paulauskaite-Taraseviciene, A.; Sutiene, K.; Jonaitis, D. Towards the automation of early-stage human embryo development detection. Biomed. Eng. Online 2019, 18. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, L.; Cheng, J.; Li, C.; Chen, X. Multi-focus image fusion: A Survey of the state of the art. Inf. Fusion 2020, 64, 71–91. [Google Scholar] [CrossRef]
- Aslantas, V.; Toprak, A.N. A pixel based multi-focus image fusion method. Opt. Commun. 2014, 332, 350–358. [Google Scholar] [CrossRef]
- Sigdel, M.S.; Sigdel, M.; Dinc, S.; Dinc, I.; Pusey, M.L.; Aygun, R.S. FocusALL: Focal Stacking of Microscopic Images Using Modified Harris Corner Response Measure. IEEE/ACM Trans. Comput. Biol. Bioinform. 2016, 13, 326–340. [Google Scholar] [CrossRef]
- Lu, H.; Li, Y.; Nakashima, S.; Yang, S.; Zhang, L. Multiframe Medical Images Enhancement on Dual Tree Complex Wavelet Transform Domain. In Proceedings of the 2013 14th ACIS International Conference on Software Engineering, Artificial Intelligence, Networking and Parallel/Distributed Computing, Honolulu, HI, USA, 1–3 July 2013; pp. 611–614. [Google Scholar] [CrossRef]
- Kou, L.; Zhang, L.; Zhang, K.; Sun, J.; Han, Q.; Jin, Z. A multi-focus image fusion method via region mosaicking on Laplacian pyramids. PLoS ONE 2018, 13, e0191085. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chang, F. A Multi-focus Image Fusion Method Based on Laplacian Pyramid. J. Comput. 2011, 6, 2559–2566. [Google Scholar] [CrossRef]
- Tang, H.; Xiao, B.; Li, W.; Wang, G. Pixel convolutional neural network for multi-focus image fusion. Inf. Sci. 2018, 433–434, 125–141. [Google Scholar] [CrossRef]
- Du, C.; Gao, S. Image segmentationbased multi-focus image fusion through multi-scale convolutional neural network. IEEE Access 2017, 5, 15750–15761. [Google Scholar] [CrossRef]
- Prabhakar, K.R.; Srikar, V.S.; Babu, R.V. DeepFuse: A Deep Unsupervised Approach for Exposure Fusion with Extreme Exposure Image Pairs. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 4714–4722. [Google Scholar] [CrossRef]
- Yan, X.; Gilani, S.Z.; Qin, H.; Mian, A. Unsupervised Deep Multi-focus Image Fusion. arXiv 2018, arXiv:1806.07272. [Google Scholar]
- Li, J.; Yuan, G.; Fan, H. Multifocus Image Fusion Using Wavelet-Domain-Based Deep CNN. Comput. Intell. Neurosci. 2019, 2019, 1–23. [Google Scholar] [CrossRef]
- Brown, D.C.C. A Rapid Image Acquisition Method for Focus Stacking in Microscopy. Ultrasound Microsc. Today 2015, 23, 18–25. [Google Scholar] [CrossRef]
- Lahoud, F.; Susstrunk, S. Fast and Efficient Zero-Learning Image Fusion. arXiv 2019, arXiv:1905.03590. [Google Scholar]
- Ho, W.N.L.C. Multi-Focus Image Fusion and Depth Map Estimation Based on Iterative Region Splitting Techniques. J. Imaging 2019, 5, 73. [Google Scholar] [CrossRef]
- Zhang, Q.; Shi, T.; Wang, F.; Blum, R.S.; Han, J. Robust sparse representation based multi-focus image fusion with dictionary construction and local spatial consistency. Pattern Recognit. 2018, 83, 299–313. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Mitchell, H.B. Image Similarity Measures. In Image Fusion: Theories, Techniques and Applications; Springer: Berlin/Heidelberg, Germany, 2010; pp. 167–185. [Google Scholar] [CrossRef]
- Palubinskas, G. Image similarity/distance measures: What is really behind MSE and SSIM? Int. J. Image Data Fusion 2017, 8, 32–53. [Google Scholar] [CrossRef]
- Alparone, L.; Wald, L.; Chanussot, J.; Thomas, C.; Gamba, P.; Bruce, L.M. Comparison of Pansharpening Algorithms: Outcome of the 2006 GRS-S Data-Fusion Contest. IEEE Trans. Geosci. Remote Sens. 2007, 45, 3012–3021. [Google Scholar] [CrossRef]
- Wang, Z.; Hamid, R.; Sheikh, A.C.B. Objective video quality assessment. In Handbook of Video Databases: Design and Applications; CRC Press: Boca Raton, FL, USA, 2003; pp. 1041–1078. [Google Scholar]
- Wang, Z.; Bovik, A.C. A universal image quality index. IEEE Signal Process. Lett. 2002, 9, 81–84. [Google Scholar] [CrossRef]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Simoncelli, E.P.; Bovik, A.C. Multiscale structural similarity for image quality assessment. In Proceedings of the Thrity-Seventh Asilomar Conference on Signals, Systems Computers, Pacific Grove, CA, USA, 9–12 November 2003; Volume 2, pp. 1398–1402. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raudonis, V.; Paulauskaite-Taraseviciene, A.; Sutiene, K. Fast Multi-Focus Fusion Based on Deep Learning for Early-Stage Embryo Image Enhancement. Sensors 2021, 21, 863. https://doi.org/10.3390/s21030863
Raudonis V, Paulauskaite-Taraseviciene A, Sutiene K. Fast Multi-Focus Fusion Based on Deep Learning for Early-Stage Embryo Image Enhancement. Sensors. 2021; 21(3):863. https://doi.org/10.3390/s21030863
Chicago/Turabian StyleRaudonis, Vidas, Agne Paulauskaite-Taraseviciene, and Kristina Sutiene. 2021. "Fast Multi-Focus Fusion Based on Deep Learning for Early-Stage Embryo Image Enhancement" Sensors 21, no. 3: 863. https://doi.org/10.3390/s21030863
APA StyleRaudonis, V., Paulauskaite-Taraseviciene, A., & Sutiene, K. (2021). Fast Multi-Focus Fusion Based on Deep Learning for Early-Stage Embryo Image Enhancement. Sensors, 21(3), 863. https://doi.org/10.3390/s21030863







