Efficient Strike Artifact Reduction Based on 3D-Morphological Structure Operators from Filtered Back-Projection PET Images
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Tomography Equipment and Settings
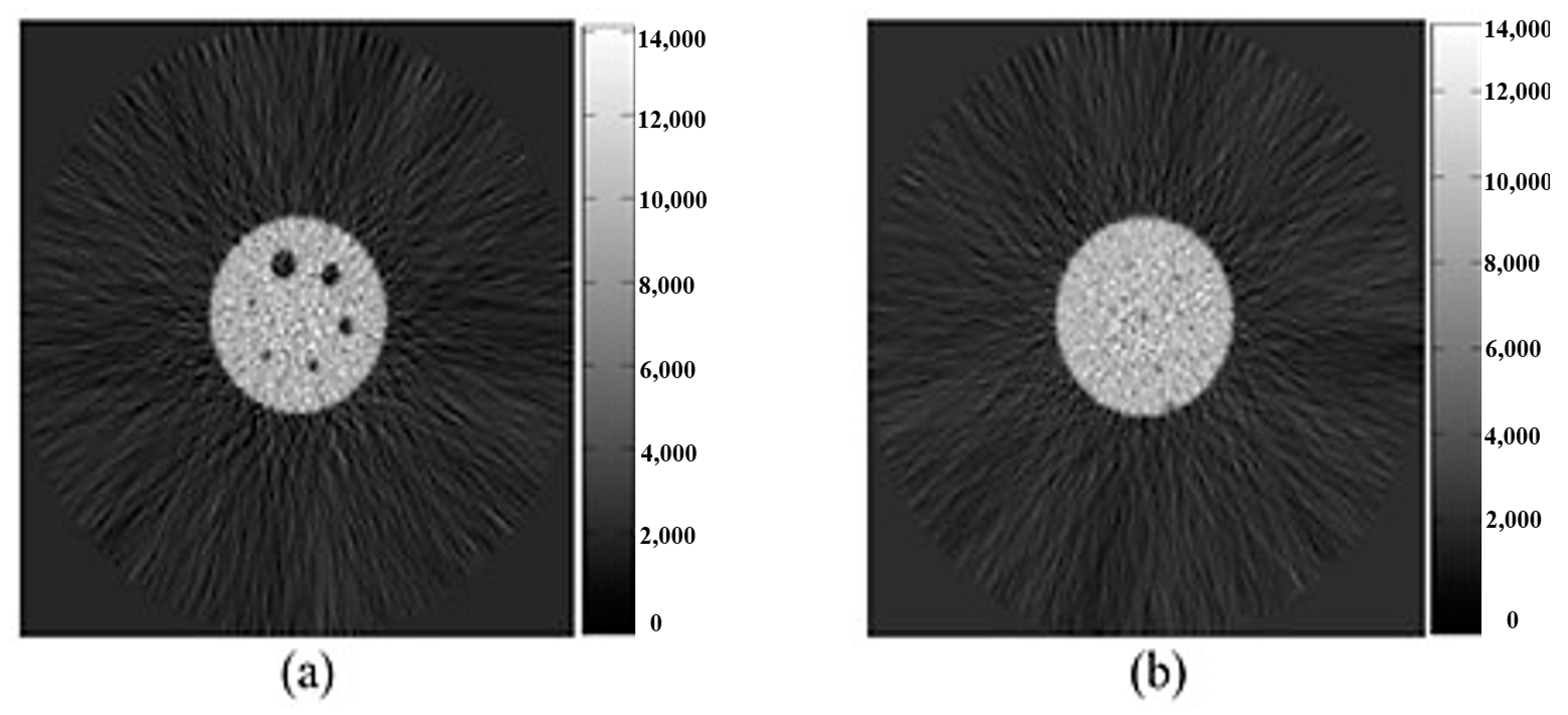
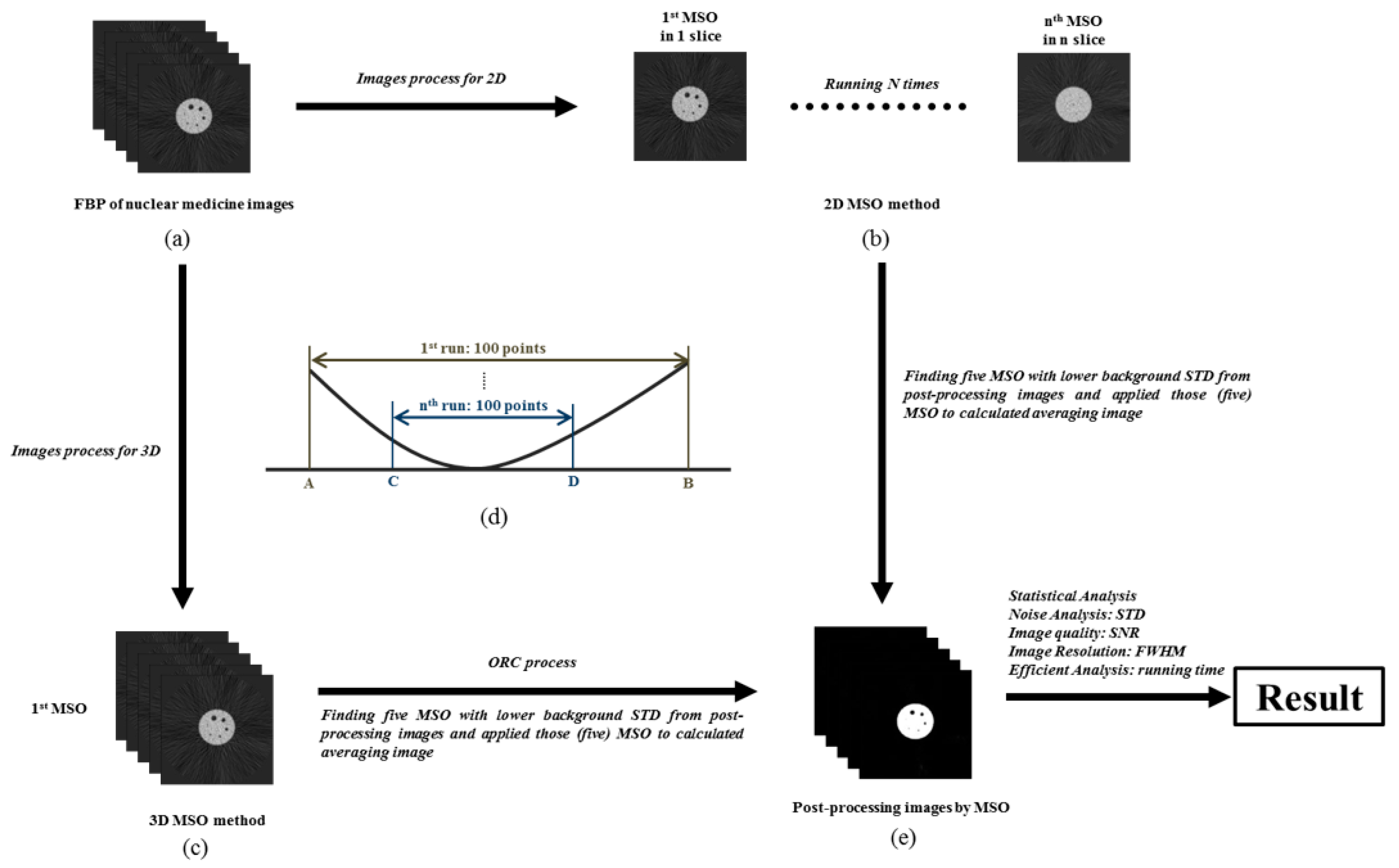
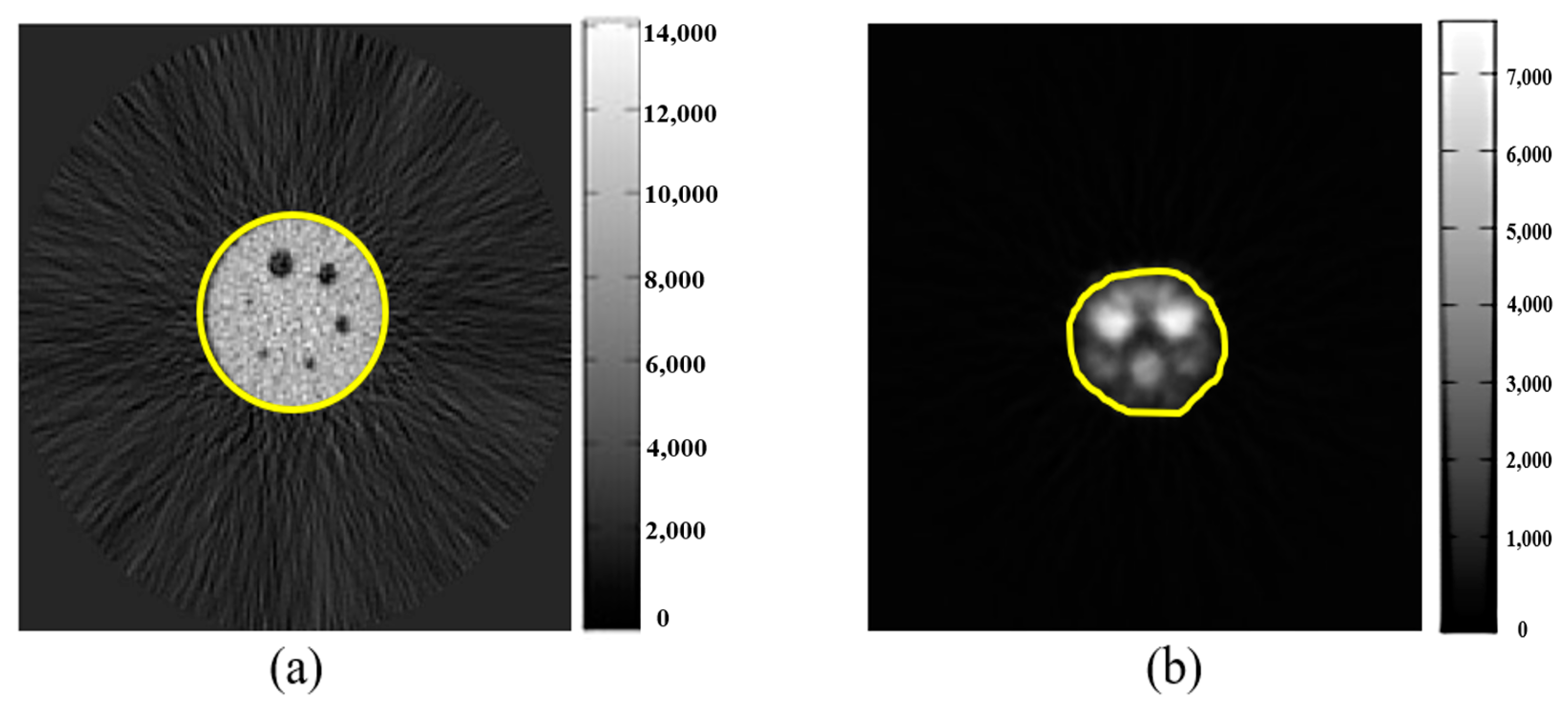
2.3. Research Flowchart and Experimental Design
2.4. Morphological Structure Operation
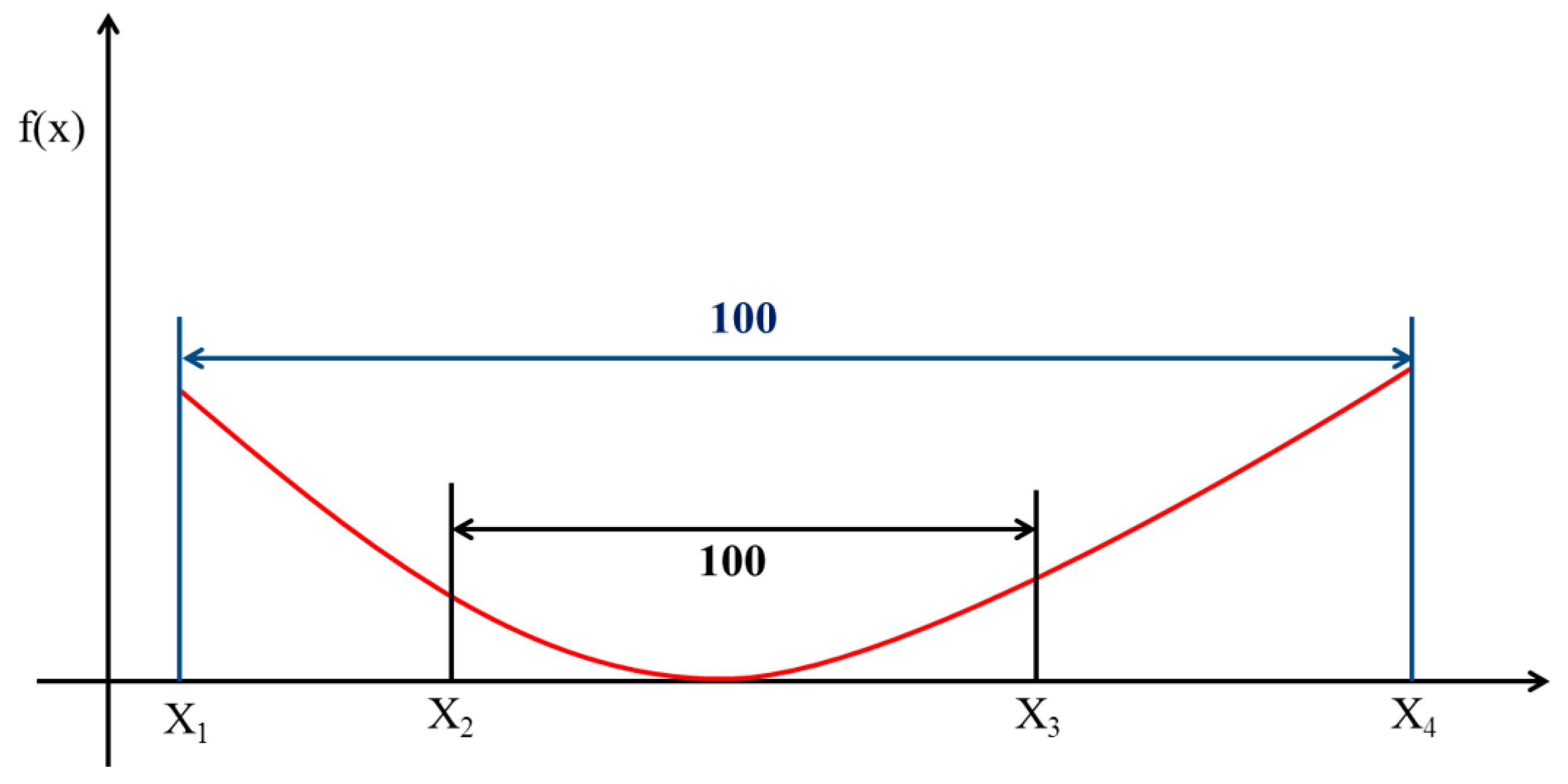
2.5. Optimal Response Curve
2.6. Image Background Value
2.7. Signal-to-Noise Ratio (SNR)
2.8. Image Resolution
2.9. Computation Time Calculation
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morrocchi, M.; Hunter, W.C.; Del Guerra, A.; Lewellen, T.K.; Kinahan, P.E.; MacDonald, L.R.; Bisogni, M.G.; Miyaoka, R.S. Evaluation of event position reconstruction in monolithic crystals that are optically coupled. Phys. Med. Biol. 2016, 61, 8298–8320. [Google Scholar] [CrossRef] [Green Version]
- España, S.; Marcinkowski, R.; Keereman, V.; Vandenberghe, S.; Van Holen, R. DigiPET: Sub-millimeter spatial resolution small-animal PET imaging using thin monolithic scintillators. Phys. Med. Biol. 2014, 59, 3405–3420. [Google Scholar] [CrossRef] [PubMed]
- Vinke, R.; Levin, C.S. A method to achieve spatial linearity and uniform resolution at the edges of monolithic scintillation crystal detectors. Phys. Med. Biol. 2014, 59, 2975–2995. [Google Scholar] [CrossRef] [PubMed]
- Schabel, C.; Gatidis, S.; Bongers, M.; Hüttig, F.; Bier, G.; Kupferschlaeger, J.; Bamberg, F.; La Fougère, C.; Nikolaou, K.; Pfannenberg, C. Improving CT-Based PET Attenuation Correction in the Vicinity of Metal Implants by an Iterative Metal Artifact Reduction Algorithm of CT Data and Its Comparison to Dual-Energy-Based Strategies: A Phantom Study. Investig. Radiol. 2017, 52, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Higashigaito, K.; Angst, F.; Runge, V.M.; Alkadhi, H.; Donati, O.F. Metal Artifact Reduction in Pelvic Computed Tomography With Hip Prostheses: Comparison of Virtual Monoenergetic Extrapolations From Dual-Energy Computed Tomography and an Iterative Metal Artifact Reduction Algorithm in a Phantom Study. Investig. Radiol. 2015, 50, 828–834. [Google Scholar] [CrossRef] [Green Version]
- Sah, B.R.; Stolzmann, P.; Delso, G.; Wollenweber, S.D.; Hüllner, M.; Hakami, Y.A.; Queiroz, M.A.; Barbosa, F.G.; Von Schulthess, G.K.; Pietsch, C.; et al. Clinical evaluation of a block sequential regularized expectation maximization reconstruction algorithm in 18F-FDG PET/CT studies. Nucl. Med. Commun. 2017, 38, 57–66. [Google Scholar] [CrossRef] [Green Version]
- Tian, M.; Jian, H.; Finbarr, O.S. The Gamma Characteristic of Reconstructed PET Images: Implications for ROI Analysis. IEEE Trans. Med. Imaging 2018, 37, 1092–1102. [Google Scholar]
- Lartizien, C.; Kinahan, P.E.; Swensson, R.; Comtat, C.; Lin, M.; Villemagne, V.; Trébossen, R. Evaluating image reconstruction methods for tumor detection in 3-dimensional whole-body PET oncology imaging. J. Nucl. Med. 2003, 44, 276–290. [Google Scholar]
- Abella, M.; Vaquero, J.J.; Soto-Montenegro, M.L.; Lage, E.; Desco, M. Sinogram bow-tie filtering in FBP PET reconstruction. Med. Phys. 2009, 36, 1663–1671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han-Back, S.; Moo-Sub, K.; Martin, L.; Shih-Kien, D.; Min-Geon, C.; Byung Wook, C.; Sungmin, K.; Dong-Wook, K.; Tae Suk, S.; Do-Kun, Y. Application of sigmoidal optimization to reconstruct nuclear medicine image: Comparison with filtered back projection and iterative reconstruction method. Nucl. Eng. Technol. 2020, 53, 258–265. [Google Scholar]
- Bélanger, M.J.; Mann, J.J.; Parsey, R.V. OS-EM and FBP reconstruction satlow count rates: Effecton 3D PET studies of [11C] WAY-100635. Neuroimage 2004, 21, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Boellaard, R.; VanLingen, A.; Lammertsma, A.A. Experimental and clinical evaluatio no fiter ative reconstruction (OSEM) in dynamic PET: Quantitativecharacteristicsand effectson kinetic modeling. J. Nucl. Med. 2001, 42, 808–817. [Google Scholar] [PubMed]
- Krak, N.C.; Boellaard, R.; Hoekstra, O.S.; Twisk, J.W.R.; Hoekstra, C.J.; Lammertsma, A.A. Effects of ROI definition and reconstruction method on quantitative outcome and applicability in a response monitor ingtrial. Eur. J. Nucl. Med. Mol. Imaging 2005, 32, 294–301. [Google Scholar] [CrossRef]
- Lubberink, M.; Boellaard, R.; Anderweerdt, A.P.; Visser, F.C.; Lammertsma, A.A. Quantitative comparison of analytic and iterative reconstruction methods in 2- and 3-dimensional dynamic cardiac 18F-FDG PET. J. Nucl. Med. 2004, 45, 2008–2015. [Google Scholar] [PubMed]
- Mesina, C.T.; Boellaard, R.; Jongbloed, G.; Vandervaart, A.W.; Lammertsma, A.A. Experimental evaluation of iterative reconstruction versus filtered back projection for 3D [15O] water PET activation studies using statistical parametric mapping analysis. Neuroimage 2003, 19, 1170–1179. [Google Scholar] [CrossRef]
- Bruyant, P.P.; Sau, J.; Mallet, J.J. Streak artifact reduction in filtered back-projection using a level line-based interpolation method. J. Nucl. Med. 2000, 41, 1913–1919. [Google Scholar]
- Shih, M.; Amaro, E.J.; Souza, S.E.; Pupo, M.C.; Malta, S.M.; Hoexter, M.Q.; Garrido, G.J.; Bueno, O.A.; Ferraz, H.; Goulart, F.O. Dopamine transporter density by [99mTc]-TRODAT-1 SPECT and neurocognitive performance—A preliminary pilot study. Alasbimn J. 2006, 8, 9. [Google Scholar]
- Angelini, E.; Esser, Y.; Vanheertum, R.; Laine, A. Fusion of brushlet and wavelet denoising methods for nuclear images. In Proceedings of the IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Arlington, VA, USA, 18 April 2004; pp. 1187–1191. [Google Scholar]
- Liow, J.S.; Strother, S.C. Noise and signal decoupling in maximum-likelihood reconstructions and Metz filters for PET brain images. Phys. Med. Biol. 1994, 39, 735–750. [Google Scholar] [CrossRef] [PubMed]
- Le Pogam, A.; Hanzouli, H.; Hatt, M.; Cheze Le Rest, C.; Visvikis, D. Denoising of PET images by combining wavelets and curvelets for improved preservation of resolution and quantitation. Med. Image Anal. 2013, 17, 877–891. [Google Scholar] [CrossRef] [Green Version]
- Gengsheng, L.Z. Maximum-Likelihood Expectation-Maximization Algorithm Versus Windowed Filtered Backprojection Algorithm: A Case Study. J. Nucl. Med. Technol. 2018, 46, 129–132. [Google Scholar]
- Chen, T.B.; Chen, H.Y.; Lin, M.C.; Lin, L.W.; Lu, N.H.; Tsai, F.S.; Huang, Y.H. Delimitated strike artifacts from FBP using a robust morphological structure operation. Radiat. Phys. Chem. 2014, 97, 31–37. [Google Scholar] [CrossRef]
- Zhang, W.; Gao, J.; Yang, Y.; Liang, D.; Liu, X.; Zheng, H.; Hu, Z. Image reconstruction for positron emission tomography based on patch-based regularization and dictionary learning. Med. Phys. 2019, 46, 5014–5026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Liu, Q.; Zhou, C.; Zhang, W.; Wan, Q.; Hu, C.; Gu, Z.; Liang, D.; Liu, X.; Yang, Y.; et al. An improved patch-based regularization method for PET image reconstruction. Quant. Imaging Med. Surg. 2021, 11, 556–570. [Google Scholar] [CrossRef]
- Seo, Y.; Khalighi, M.M.; Wangerin, K.A.; Deller, T.W.; Wang, Y.H.; Jivan, S.; Kohi, M.P.; Aggarwal, R.; Flavell, R.R.; Behr, S.C.; et al. Quantitative and Qualitative Improvement of Low-Count [68Ga] Citrate and [90Y] Microspheres PET Image Reconstructions Using Block Sequential Regularized Expectation Maximization Algorithm. Mol. Imaging Biol. 2020, 22, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Tatsumi, M.; Soeda, F.; Kamiya, T.; Ueda, J.; Katayama, D.; Matsunaga, K.; Watabe, T.; Kato, H.; Tomiyama, N. Effects of New Bayesian Penalized Likelihood Reconstruction Algorithm on Visualization and Quantification of Upper Abdominal Malignant Tumors in Clinical FDG PET/CT Examinations. Front. Oncol. 2021, 11, 707023. [Google Scholar] [CrossRef] [PubMed]
- Leuschner, J.; Schmidt, M.; Ganguly, P.S.; Andriiashen, V.; Coban, S.B.; Denker, A.; Bauer, D.; Hadjifaradji, A.; Batenburg, K.J.; Maass, P.; et al. Quantitative Comparison of Deep Learning-Based Image Reconstruction Methods for Low-Dose and Sparse-Angle CT Applications. J. Imaging 2021, 7, 44. [Google Scholar] [CrossRef]
- Yu, B.; Wang, Y.; Wang, L.; Shen, D.; Zhou, L. Medical Image Synthesis via Deep Learning. Adv. Exp. Med. Biol. 2020, 1213, 23–44. [Google Scholar]
- Sorin, V.; Barash, Y.; Konen, E.; Klang, E. Creating Artificial Images for Radiology Applications Using Generative Adversarial Networks (GANs)—A Systematic Review. Acad. Radiol. 2020, 27, 1175–1185. [Google Scholar] [CrossRef]
- Podgorsak, A.R.; Shiraz Bhurwani, M.M.; Ionita, C.N. CT artifact correction for sparse and truncated projection data using generative adversarial networks. Med. Phys. 2021, 48, 615–626. [Google Scholar] [CrossRef]
- Koshino, K.; Werner, R.A.; Pomper, M.G.; Bundschuh, R.A.; Toriumi, F.; Higuchi, T.; Rowe, S.P. Narrative review of generative adversarial networks in medical and molecular imaging. Ann. Transl. Med. 2021, 9, 821. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, B.; Wang, L.; Zu, C.; Lalush, D.S.; Lin, W.; Wu, X.; Zhou, J.; Shen, D.; Zhou, L. 3D conditional generative adversarial networks for high-quality PET image estimation at low dose. Neuroimage 2018, 174, 550–562. [Google Scholar] [CrossRef] [PubMed]



| Item | Contents |
|---|---|
| Operating system | Windows 64-bit operating system |
| Central processing unit | Intel Core 2 Quad CPU Q6600 |
| Computer memory | 2.00 GB |
| MSO Matrix Size | Background STD in Phantom | Background STD in Rat | Reducing Noise in Phantom | Reducing Noise in Rat |
|---|---|---|---|---|
| Raw 1 | 750.59 | 6.27 | - | - |
| 2 × 2 | 238.94 | 4.91 | 68.2% | 21.7% |
| 2 × 3 | 174.48 | 4.64 | 76.8% | 26.0% |
| 3 × 2 | 171.67 | 4.65 | 77.1% | 25.8% |
| 3 × 3 | 110.64 | 4.19 | 85.3% | 33.2% |
| 3 × 3 ORC 2 | 110.64 | 4.19 | 85.3% | 33.2% |
| 2 × 2 × 2 ORC | 141.15 | 3.65 | 81.2% | 41.8% |
| 2 × 3 × 2 ORC | 112.15 | 3.48 | 85.1% | 44.5% |
| 3 × 2 × 2 ORC | 104.50 | 3.50 | 86.1% | 44.2% |
| 3 × 3 × 2 ORC | 102.98 | 3.25 | 86.3% | 48.2% |
| 2 × 2 × 3 ORC | 112.53 | 3.29 | 85.0% | 47.5% |
| 2 × 3 × 3 ORC | 98.14 | 3.03 | 86.9% | 51.7% |
| 3 × 2 × 3 ORC | 98.20 | 3.24 | 86.9% | 48.3% |
| 3 × 3 × 3 ORC | 96.69 | 2.80 | 87.1% | 55.3% |
| MSO Matrix Size | CR/SNR in Phantom | CR/SNR in Rat | Increasing Contrast/Quality in Phantom | Increasing Contrast/Quality in Rat |
|---|---|---|---|---|
| Raw 1 | 263.44/4.53 | 174.64/14.21 | - | - |
| 2 × 2 | 66.94/5.19 | 157.81/14.53 | −74.59%/14.6% | −9.64%/2.3% |
| 2 × 3 | 110.81/5.41 | 165.53/15.00 | −57.94%/19.4% | −5.22%/5.6% |
| 3 × 2 | 113.30/5.41 | 165.25/14.81 | −56.99%/19.4% | −5.38%/4.2% |
| 3 × 3 | 342.52/5.76 | 182.70/15.38 | 30.02%/27.2% | 4.62%/8.2% |
| 3 × 3 ORC 2 | 342.52/5.76 | 182.70/15.38 | 30.02%/27.2% | 4.62%/8.2% |
| 2 × 2 × 2 ORC | 179.32/5.51 | 209.00/15.39 | −31.93%/21.6% | 19.67%/8.3% |
| 2 × 3 × 2 ORC | 320.34/5.68 | 221.07/15.84 | 21.60%/25.4% | 26.59%/11.5% |
| 3 × 2 × 2 ORC | 378.69/5.73 | 218.28/15.89 | 43.75%/26.5% | 24.99%/11.8% |
| 3 × 3 × 2 ORC | 429.41/5.78 | 240.26/16.51 | 63.00%/27.6% | 37.57%/16.2% |
| 2 × 2 × 3 ORC | 284.67/5.55 | 254.07/15.85 | 8.06%/22.5% | 45.48%/11.5% |
| 2 × 3 × 3 ORC | 448.64/5.69 | 286.22/16.08 | 70.30%/25.6% | 63.89%/13.2% |
| 3 × 2 × 3 ORC | 455.88/5.66 | 251.22/15.67 | 73.05%/24.9% | 43.85%/10.3% |
| 3 × 3 × 3 ORC | 584.64/5.80 | 328.22/15.91 | 121.93%/28.0% | 87.94%/12.0% |
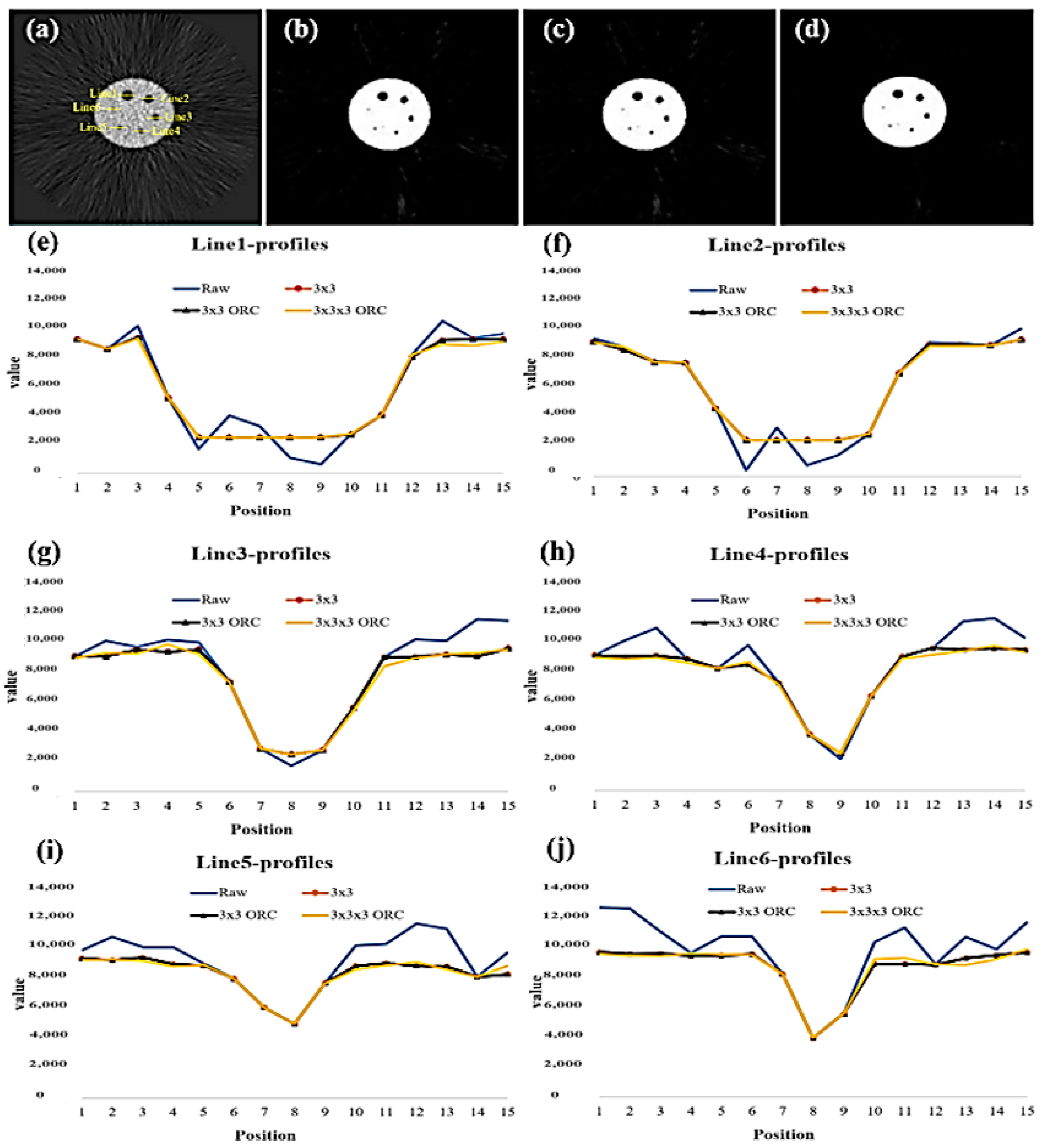
| FWHM of Line# | Aperture Size with/without MSO (mm) | ||||
|---|---|---|---|---|---|
| Reality | Raw | 3 × 3 | 3 × 3 ORC | 3 × 3 × 3 ORC | |
| Line1 | 31.80 | 29.17 | 33.33 | 33.33 | 29.17 |
| Line2 | 25.40 | 20.83 | 25.00 | 25.00 | 25.00 |
| Line3 | 19.10 | 14.58 | 16.67 | 16.67 | 18.75 |
| Line4 | 15.90 | 12.50 | 12.50 | 12.50 | 12.50 |
| Line5 | 12.70 | 10.42 | 12.50 | 12.50 | 12.50 |
| Line6 | 9.50 | 10.42 | 8.33 | 8.33 | 10.42 |
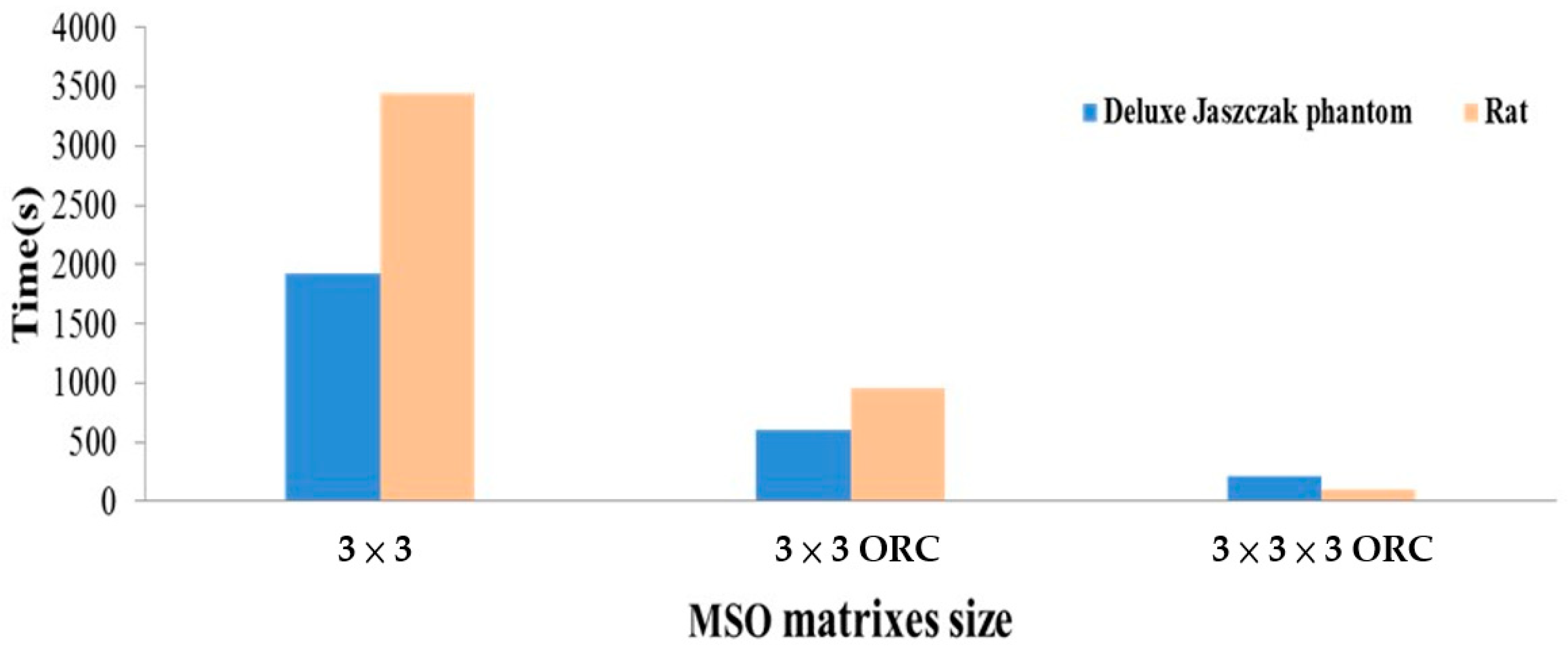
| MSO Matrix Size | Processing Time(s) | |
|---|---|---|
| Deluxe Jaszczak Phantom | Rat | |
| 2 × 2 | 37.86 | 53.73 |
| 2 × 3 | 184.08 | 288.88 |
| 3 × 2 | 184.29 | 287.39 |
| 3 × 3 | 1925.38 | 3445.9 |
| 3 × 3 ORC 1 | 603.81 | 953.08 |
| 2 × 2 × 2 ORC | 26.38 | 25.83 |
| 2 × 3 × 2 ORC | 33.57 | 36.95 |
| 3 × 2 × 2 ORC | 64.36 | 36.89 |
| 3 × 3 × 2 ORC | 40.47 | 172.97 |
| 2 × 2 × 3 ORC | 34.26 | 80.10 |
| 2 × 3 × 3 ORC | 70.87 | 91.28 |
| 3 × 2 × 3 ORC | 36.66 | 91.24 |
| 3 × 3 × 3 ORC | 219.09 | 100.08 |
| Authors | Year | Modality | Task | Method | Finding |
|---|---|---|---|---|---|
| Gao et al. [23,24] | 2020 | PET | Reduce noise | PBRA | It is difficult to reduce noise and keep or improve quality of images due to encounter the low SNR of images or low count rates |
| Seo et al. [25] | 2020 | PET | Improvement quality | BSREMA | To enhances the quality of images and accuracy of qualification as per occurred in low-count rates of PET scanning |
| Tatsumi et al. [26] | 2021 | PET/CT | Improvement quality of image | BPLRA | To promote the gray levels of image as per low count rates condition |
| Leuschner et al. [27] | 2021 | CT | Improvement quality of image | Deep learning methods | The experimental results were shown and demonstrated to be able to improve the quality of noise images |
| Yu et al. [28] | 2020 | Medical image synthesis | Reduce noise, enhance quality | Deep learning methods, 3D GAN | To promote the quality of image under low count rates or low dose |
| Soren et al. [29] | 2020 | CT, MRI, PET/MRI, PET/CT, | Reduce noise, enhance quality | GAN (GANs) | These novel models made a great impact on the computer vision field |
| Podgorsak et al. [30] | 2021 | CT | CT artifact correction | GANs | Improvement of reconstructed image quality under sparse angles |
| Koshino et al. [31] | 2021 | Medical and molecular imaging | Reduce noise, enhance quality | GANs | GANs are promising tools for medical and molecular imaging for promoted quality of images |
| Wang et al. [32] | 2018 | PET | Reduce noise, enhance quality | 3D GANs | GANs are promising tools for improvement quality of image under low count rates |
| Presented Methods | 2021 | PET | Reduce noise, enhance quality | 2D and 3D MSO with ORC | Demonstrated efficiently perform denoising processing and obtained acceptable quality of images |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiu, C.-Y.; Huang, Y.-H.; Du, W.-C.; Wang, C.-Y.; Chen, H.-Y.; Shiu, Y.-S.; Lu, N.-H.; Chen, T.-B. Efficient Strike Artifact Reduction Based on 3D-Morphological Structure Operators from Filtered Back-Projection PET Images. Sensors 2021, 21, 7228. https://doi.org/10.3390/s21217228
Chiu C-Y, Huang Y-H, Du W-C, Wang C-Y, Chen H-Y, Shiu Y-S, Lu N-H, Chen T-B. Efficient Strike Artifact Reduction Based on 3D-Morphological Structure Operators from Filtered Back-Projection PET Images. Sensors. 2021; 21(21):7228. https://doi.org/10.3390/s21217228
Chicago/Turabian StyleChiu, Chun-Yi, Yung-Hui Huang, Wei-Chang Du, Chi-Yuan Wang, Huei-Yong Chen, Yun-Shiuan Shiu, Nan-Han Lu, and Tai-Been Chen. 2021. "Efficient Strike Artifact Reduction Based on 3D-Morphological Structure Operators from Filtered Back-Projection PET Images" Sensors 21, no. 21: 7228. https://doi.org/10.3390/s21217228
APA StyleChiu, C.-Y., Huang, Y.-H., Du, W.-C., Wang, C.-Y., Chen, H.-Y., Shiu, Y.-S., Lu, N.-H., & Chen, T.-B. (2021). Efficient Strike Artifact Reduction Based on 3D-Morphological Structure Operators from Filtered Back-Projection PET Images. Sensors, 21(21), 7228. https://doi.org/10.3390/s21217228






