1. Introduction
Sanitation inspection is an ongoing concern for food distributors, restaurant owners, and others within the food industry. These individuals must prevent potential contamination and infection from spreading among workers and to consumers. The failure to meet legal requirements can result in the damage to institution or restaurant reputations, the loss of trust between the establishment and its workers and customers, and financial repercussions. All U.S. meat, poultry, and egg processing establishments are regulated by the Food Safety and Inspection Service (FSIS) and are required to have Sanitation Standard Operating Procedures (SSOPs). SSOPs are written procedures that the establishment develops and implements to prevent the direct contamination of food products. It is an establishment’s responsibility to execute the procedures as written in the SSOPs. Each establishment must maintain a daily documentation of the application and monitoring of the SSOPs, as well as any necessary corrective action. It is critically important to maintain the required documents as evidence of SSOP compliance and keep these up to date.
According to the Asia Pacific Society of Infection Control (APSIC) guidelines, “there are several methods for assessing environmental cleanliness: (1) a conventional program of direct and indirect observation (e.g., visual assessment, observation of performance, customer/staff satisfaction surveys); (2) an enhanced program of monitoring residual bioburden (e.g., environmental culture, adenosine triphosphate (ATP) bioluminescence); (3) and environmental marking tools (e.g., fluorescent dust marking of surfaces)” [
1,
2,
3].
The visualization of fluorescence emission has great utility for food safety inspection. Food-related biological residues have been shown to have characteristic fluorescence emission spectra in visible (VIS) and near-infrared (NIR) wavelengths. Dairy cow feces show red fluorescence emissions peaking at 680 nm when excited by ultraviolet (UV) radiation (360 nm) [
4,
5]. Chlorophyll (Chl) in green plants has unique fluorescence emissions in the red and far-red regions, peaking at 685 nm and 730 nm [
6,
7]. Additionally, a number of plant constituents have been reported to have a UV emission at 340 nm and blue and green emissions peaking near 450 nm and 530 nm [
6,
7,
8]. Meat products have been shown to have fluorescence emission in UV, blue, and green wavelengths. Proteins are known to emit UV fluorescence, and a variety of aromatic compounds emit fluorescence in blue and green wavelengths [
8,
9,
10,
11,
12].
Multiple automatic imaging inspection techniques and systems have been used for food safety inspection. The online inspection of poultry carcasses for fecal contamination has been developed using a multispectral imaging system to visualize reflectance spectra features of visible wavelength regions [
13,
14]. A hyperspectral imaging system to detect fecal contamination on apples was developed, and this system can measure both reflectance and fluorescence in the visible to near-infrared [
15,
16]. A portable hyperspectral imaging system has also been developed to monitor sanitation procedures in food processing facilities. It showed the ability to detect minute quantities of juice from produce on food processing equipment [
17,
18]. An imaging device that is portable and capable of fluorescence-based contaminant detection on food products and food processing equipment can easily be integrated with workflows and sanitation audits in food handling facilities.
Some efforts have been made to commercialize line scan spectral imaging systems without disinfection capability, and with some documentation capabilities. Headwall Photonics, Inc. [
19] has commercialized a line scan system after licensing a patent based on the research of one of our coauthors’ references cited in [
15,
16,
17,
18]. P&P Optica [
20], Inc. has commercialized a similar line scan spectral imaging system that they claim is “able to detect, identify, and remove many types of foreign objects on production lines” as well as using “artificial intelligence to provide insights about shelf life, product composition, flavor, fat content, quality variation and much more”. VERITIDE Ltd. [
21] is commercializing a fluorescence-based point measurement system to detect fecal contamination on meat carcasses, as well as a fluorescence-based production line imaging system for meat carcasses.
During the COVID-19 pandemic, many restaurants, dining facilities, kitchens, as well as food processing facilities, were heavily affected by virus contamination resulting in multiple facility shutdowns [
22]. A need has emerged for new cleaning procedures and protocols to ensure all staff and customer high-touch areas are cleaned and monitored. Saliva and respiratory droplets can be one of the major contamination carriers for many diseases, including influenza, coronavirus, and Ebola [
23,
24]. Previous scientific studies directed to the detection of saliva and respiratory droplets have shown that short-wavelength UV excitation at 282 nm can be used to indicate the presence of saliva stains in which the α amylase enzyme gives off a characteristic emission spectrum at 345–355 nm, which can be identified using fluorescence imaging. The presence of the fluorescence emission at 345–355 nm with excitation at 282 nm proves to be a strong indicator of saliva, even when deposited on human skin [
25].
In this paper, for the first time, we introduce a fast, convenient, and easy-to-use handheld system for “contamination, sanitization inspection, and disinfection (CSI-D)” that enables the rapid detection of saliva and respiratory droplets, and other organic residues that are present in kitchens, dining areas, and food processing facilities. The system provides the immediate disinfection and documentation of contaminants on surfaces that may cause disease spread. CSI-D can wirelessly communicate the inspection process, which allows remotely located personnel to immediately provide oversight and respond to inspection issues. The CSI-D system is not intended to be a primary disinfection or cleaning tool; instead, it acts as a post-cleaning audit solution complementary to other post-cleaning auditing tools (ATP, FT-IR, etc.), as well as providing documentation of cleanliness. The system’s disinfection capability is intended to provide spot disinfection only during audits or incident response and is not employed as a large area disinfection method (e.g., fogging).
The key innovations of this device encompass the visualization of contamination using fluorescence imaging, the disinfection of the contamination using UVC illumination, and the documentation of cleanliness. The combination of detection, disinfection, documentation, and verification is the core innovation from an operator’s point of view. Specific technical innovations include the ability to capture fluorescence images under bright ambient light situations in food processing facilities, institutional kitchens, and dining facilities. Previous systems (described above) had difficulties with bright ambient light and, often, could only function in darker rooms or under shrouding. Other innovations related to the UVC germicidal LEDs include the integration of safety systems based on sensors and software (LIDAR, gyroscope, motion detector, etc.) that help protect the operator and other personnel from accidental UVC light exposure. These sensors are also used to monitor motion and distance during the image capture process to ensure images are free from motion artifacts, such as image blur, and provide software-based guidance for operators when they are moving the camera too quickly or are too far away or too close. Finally, the image database and records of contamination for each location at each facility, combined with the local hazards and disease prevalence, will enable the future delivery of intelligent dynamic risk assessment associated with each surface to guide cleaning and inspection processes.
2. Materials and Methods
2.1. System Description
As shown in
Figure 1, the CSI-D system consists of a handheld device that incorporates illumination, imaging, battery power, display, and processing units in a single system. The illumination module includes the 405 nm and 275 nm LED arrays, heat management, and driver circuits. The 275 nm LEDs were chosen because they were very close to the 282 nm excitation maximum wavelength of salivary amylase, were commercially available, and had high optical power. This wavelength is also a very effective germicidal wavelength. The 405 nm LEDs were selected because we previously used them for the detection of other organic residues, such as food residues containing fluorophores like collage; flavins; bacterial porphyrins; and chlorophyll. They are not used for the detection of saliva and respiratory droplets.
During the fluorescence imaging mode, the 405 nm or 275 nm LED arrays are turned on and off sequentially via electronic signals. During the disinfection mode, the 275 nm LEDs are turned on continuously for 2–5 s. The imaging system includes an RGB camera and a UV camera that communicate with the processing unit, which triggers the image acquisition and storage of fluorescence images of organic residues (RGB camera), or saliva and respiratory droplets and certain organic residues (UV camera) during fluorescence imaging. The RGB camera is also used in “ViewFinder” mode, whereby an operator can locate the area of interest to be scanned. The camera systems include lenses and spectral bandpass filters that select wavelengths specific to the contamination emission wavelength ranges.
The processing unit includes a system on module (SOM) board that controls the illumination and imaging modules to capture the fluorescence and background images under the appropriate illumination. The SOM processes images to provide meaningful information to the operator and for inspection records. The CSI-D system also includes a LIDAR module that communicates with the SOM module, which initializes and controls the LIDAR module and receives distance information from the rangefinder and temperature information from its temperature sensor. We are using an Android device as a smart display to provide an operator interface. The CSI-D system is designed to communicate with a dedicated cloud server in which all task lists are assembled, and inspection reports and video data are stored and managed.
The operator can select a disinfection mode using the hand controls and user interface. The system calculates how long the UVC illumination should be activated by calculating the surface distance using the LIDAR module.
2.2. Contamination Detection Algorithm
During the contamination detection procedure, we captured images that demonstrate the presence or absence of contamination on surfaces. Two fluorescence images were captured under blue/violet excitation using a color camera with a dual bandpass filter that passed selected green and red wavebands. We also captured an ultraviolet B (UVB) fluorescence image under UVC excitation using a UV-sensitive monochrome camera with a single bandpass filter. The image capture sequence comprised continuous image capture and background subtraction. For each fluorescence acquisition, we captured two frames: one frame with the LED illumination “ON” and one frame with the LED illumination “OFF.” The LED “ON” frame contained fluorescence emissions as well as any background light that could leak through the bandpass filter. The LED “OFF” frame contained only the background light. We implemented real-time background subtraction software to reduce the contribution of the background light to the final fluorescence image. The contamination detection algorithm was then applied to the background-subtracted fluorescence image frames. The UV camera image sensor was 2048 × 2048 pixels, and to increase the sensitivity and speed of the image capture, the pixels were binned to an image of 256 × 256 pixels. The RGB camera image sensor was 1280 × 960 pixels, but we selected a region of interest to read out that was 1024 × 768 pixels. At a distance of 20 cm, the field of view (FOV) of the RGB camera was 30 cm, and the FOV of the UV camera was 10.5 cm. To obtain reliable segmentation results for each image frame that are independent of fluorescence and background image intensity variations, due to the device movement during the inspection, the detection algorithm has to continuously adapt itself to changing fluorescence and background intensity levels. While imaging with a fixed-mount camera can allow simple conventional thresholding (e.g., Otsu’s method), we constantly moved the camera across different scenes, and therefore we adopted adaptive thresholding to change the threshold dynamically over each frame. This more sophisticated version of thresholding can accommodate the varying background and fluorescence conditions in each frame. Whenever intensities between an object’s fluorescence and the image background are significant, but their exact magnitude or location in the image is unknown, segmentation is possible by threshold optimization through the image intensity histogram. The image histogram represents the distribution and frequency of occurrence of the intensity for every pixel in the image. Because each image/frame varies in the frequency and distribution of these pixel intensities, the histogram can be normalized by the number of pixels for convenience of analysis.
During the first step of designing the algorithm, we generated a dataset from various contamination biofilms, such as olive oil, spinach juice, and egg white, on various surfaces, including steel, wood, and plastic. The ambient light is an important variable that needs to be considered in real-world applications of the device. We tried a wide range of ambient light from 0 to 90 foot-candle (FC) at intervals of 10 FCs, when capturing the images from each contamination on the different surfaces. We chose this range because the facilities we are targeting for CSI-D inspections can have ambient light up to 80 FCs.
After capturing the images, we manually segmented the areas of the contamination. We then used a similarity measurement metric called “dice similarity” [
26] and attempted to adjust the threshold level by minimizing the error between the manually segmented regions and the regions that resulted from the algorithm for the range of ambient light and surfaces. The method of thresholding we used was to create a histogram of a fluorescence image. We then normalized the histogram so that each bin was expressed as a percentage of the total number of pixels in the image. We then set a threshold: if the bin contained more than 0.2% of the pixels in the image, its pixel values were set to zero, and if the bin contained less than 0.2% of the pixels in the image but more than zero, its values were set to one. We selected 0.2% because it gave a good compromise between the detection of contaminant fluorescence and the detection of artifacts without applying additional spatial filters for size, etc. It also provided a good dice similarity (74.22 ± 0.32) to the manually segmented images. This is an interim segmentation method that is “good enough” for our present purposes, but we are working on applying more advanced machine learning-based segmentation.
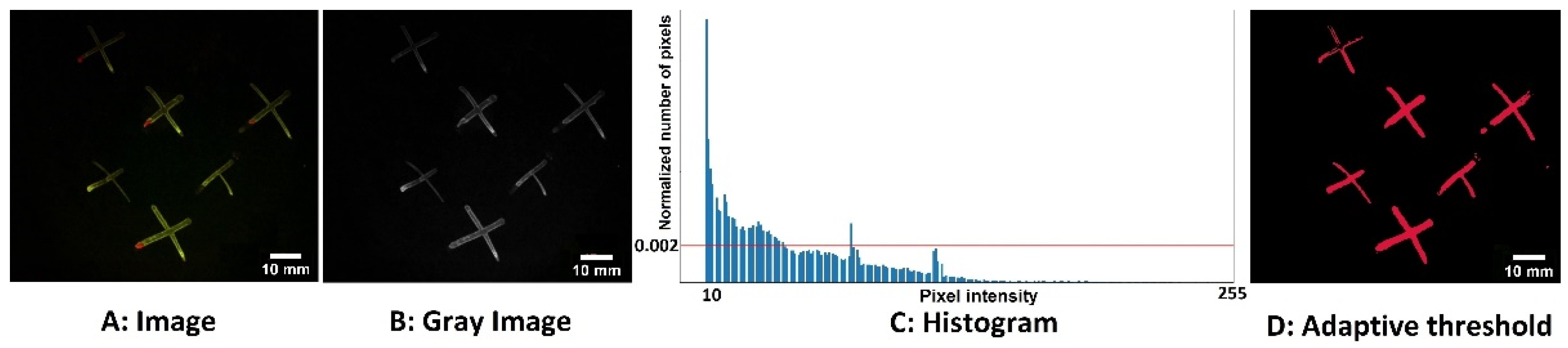
Figure 2 shows how we applied our adaptive threshold segmentation method to the green channel fluorescence with excitation 405 nm and a green emission band filtered to 510–560 nm. Specifically,
Figure 2A demonstrates the color fluorescence image captured by the camera with both green and red bands, while
Figure 2B is the extracted green band monochrome image. The histogram analysis described above is shown in
Figure 2C, and
Figure 2D displays the resulting binary image mask.
The segmentation algorithm has been implemented in the processing unit of the CSI-D system. After the color camera, fluorescence images were captured, and background intensity was subtracted; the red and green channels were separated to provide individual monochrome fluorescence images. Since the UV camera is a monochrome camera, color separation is not necessary. Currently, each channel is segmented separately to identify areas of potential contamination. In future work, we will investigate fluorescence color ratio analysis and other more sophisticated and unsupervised analyses that will use all three fluorescence signals to distinguish between different types of contamination and provide a better segmentation and classification of contamination.
2.3. Microbial/Viral Strain Preparation
We are using strains of bacteria, virus, and mold to test the effectiveness of disinfection by the CSI-D system. The experiments have been conducted in a biosafety level 2 laboratory, at the University of North Dakota School of Medicine & Health Sciences, with the following pathogens.
Aspergillus fumigatus: A. fumigatus [NIH 5233] was grown for 7 days at 37 °C on Sabouraud dextrose agar (SDA). The mature fungus was harvested using 0.1% Tween 80 in sterile phosphate-buffered saline (PBS), filtered through a sterile gauze, and resuspended in sterile PBS containing 0.1% Tween 80. The conidia were quantitated using a hemocytometer and resuspended at a final concentration of 5 × 106/mL. The surface was decontaminated using 100% pure concentrated Cavicide and sterile Kimwipes, followed by 70% EtOH and sterile Kimwipes. After vortexing to mix, a 20 µL droplet of conidia was placed at the center of the surface for UVC light treatment (or no treatment for controls) using a P20 micropipette.
Streptococcus pneumoniae: S. pneumoniae serotype 6A strain BG7322 was provided by Rochester General Hospital Research Institute and was originally obtained from Sanofi Pasteur [
27,
28].
S. pneumoniae was resuspended to a final volume of 5.0 × 10
6/mL or 2.5 × 10
6/mL. The surface was decontaminated using 70% EtOH and sterile Kimwipes. After vortexing to mix, a 20 uL droplet was placed at the center of the surface using a P20 micropipette.
Influenza A virus: A mouse-adapted strain of influenza A virus strain A/PR/8/34 H1N1 (Charles River) was resuspended in an infection medium containing 1μg/mL TPCK-trypsin (Sigma) to obtain a virus concentration at 7 × 106 PFU/μL (107 TCID50/μL), where PFU is plaque forming units and TCID is the median tissue culture infectious dose. TCID50 was calculated using Equation (2), while PFU was calculated using Equation (3). The platform was disinfected using 70% EtOH and sterile Kimwipes. After vortexing to mix, a 20 μL droplet (containing 1.4 × 105 PFUs, TCID50 2 × 105) was placed at the center of the platform using a P20 micropipette.
2.4. Disinfection Procedure
For the UVC irradiation by the CSI-D system illumination module, the specimen was placed anywhere within a 22.5 cm2 area that was marked on the surface of the platform. This area in the center of the illumination field has relatively uniform illumination power (>85%) from the UVC LEDs. Disinfection efficacy testing was performed by positioning the system at the designated distance from the contamination sample height, and manually pressing the two disinfection buttons to trigger the UVC illumination for the specified time duration. The illumination power density was measured with a UV radiometer (Opsytec Dr. Grobel GmBH) sensor, as the distances between the device and the sample were changed.
2.5. Post-Disinfection Treatment
After the disinfection treatment, the remaining pathogens were noted and used to calculate logarithmic reductions, as shown in Equations (4) and (5). The samples were observed and counted following the specified methods below.
A. fumigatus: For all samples exposed to UVC light or unexposed controls, a new sterile pipette tip was used to pipette the droplet and then resuspend it in 480 µL of sterile PBS. Samples were immediately placed on ice. For quantitation, the conidia were serially diluted 3 times at a ratio of 1:10 in sterile PBS and plated on fresh SDA. After incubation at 37 °C for 24 h, the colonies on the plate were counted, and the colony-forming units (CFU) were calculated using Equation (1).
S. pneumoniae: For all samples exposed to UVC light or unexposed controls, a new sterile pipette tip was used to pipette the droplet and then resuspend it in 480 µL of sterile PBS. Samples were immediately placed on ice. After UVC light exposure, the bacteria were serially diluted 3 times at a ratio of 1:10 in sterile PBS and plated in a volume of 10 µL on Trypticase soy agar II supplemented with 5% sheep’s blood (BD 221239). After incubation at 37 °C for 24 h, the colonies on the plate were counted, and the CFU was calculated using Equation (1).
Influenza A virus: For all samples exposed to UVC light or unexposed controls, the droplet was pipetted using a new sterile pipette tip and collected in 80 μL of infection medium in a fresh tube. After a quick vortex, the samples were kept on ice until further steps were taken. Test (UVC exposed) and control (not exposed) samples were subjected to TCID50 viral titration assay to determine the infectivity of the influenza A virus using Madin–Darby canine kidney (MDCK) cells. Each sample was 10-fold serially diluted and overlaid on a confluent monolayer of MDCK cells and then incubated at 37 °C in a CO2 incubator. On Day 5, the MDCK monolayers were fixed with 4% formaldehyde and stained with 0.5% crystal violet stain. The cells were observed for the presence or absence of cytopathic effect and scored as positive or negative events, respectively.
Colony-forming units (CFUs) were calculated using the following equation:
where CFU is the number of colony-forming units in a 0.5 mL sample, n is equal to the number of countable colonies per 20 μL volume plated, and d is the dilution level yielding countable colonies.
TCID
50/mL was calculated by:
where x
0 is the decimal logarithm of the initial dilution factor; d is the decimal logarithm of serial dilution factor; x
i is the score of positive events; n is the number of replicates; and v is the decimal logarithm of the reciprocal inoculation volume in mL.
Plaque forming units (PFUs)/mL were calculated using the following equation:
Logarithmic reductions for
A. fumigatus and
S. pneumoniae were calculated by:
Logarithmic reductions for the influenza A virus were calculated by:
Percent killing for
A. fumigatus and
S. pneumoniae were calculated by:
Percent killing for the influenza A virus was calculated by:
4. Conclusions
We have demonstrated a new handheld imaging system, CSI-D, which can rapidly detect, disinfect, and document possible organic residues which may host pathogens. The system uses light at two fluorescence excitation wavelengths (ultraviolet C at 275 nm and violet at 405 nm) for the detection of organic residues, including saliva and respiratory droplets. The 275 nm light is also used to disinfect pathogens commonly found in contamination residues.
We have demonstrated that the system can capture and segment fluorescence images of saliva and respiratory droplets, as well as organic residues, such as vegetable residue (spinach); fats (olive oil); and proteins (egg albumen), all under ambient room lighting conditions.
We have demonstrated that the UVC light in the system can deactivate bacterial, viral, and mold pathogens using A. fumigatus, S. pneumoniae, and the influenza A virus (a fungus, a bacterium, and a virus, respectively, which are each commonly found in saliva and respiratory droplets). After the exposure to UVC light from the CSI-D, all three pathogens experienced 100% sterilization or deactivation in under 10 seconds. Up to 5-log reductions (99.999%) have also been shown within 10 s of UVC irradiation from the CSI-D system.
When used as an auditing tool to verify and document cleanliness following sanitization, the CSI-D system can improve safety for food processing, preparation, and serving facilities and protect their staff and customers.