Neural Network Analysis for Microplastic Segmentation
Abstract
:1. Introduction
- Kernel weights histogram shows the distribution of weight values of each layer. By using this histogram, the neural network designer can determine the degree of layer utilization, and by classifying unnecessary layers according to the layer utilization degree, the neural network structure can be optimized. This paper demonstrates this optimization process;
- A neural network suitable for tiny object segmentation was proposed by deriving from the existing NNs (Neural Networks), and it shows better performance with only 10–20% of the amount of computation over the existing NNs.
2. Related Works
3. Neural Networks for Microplastic Segmentation
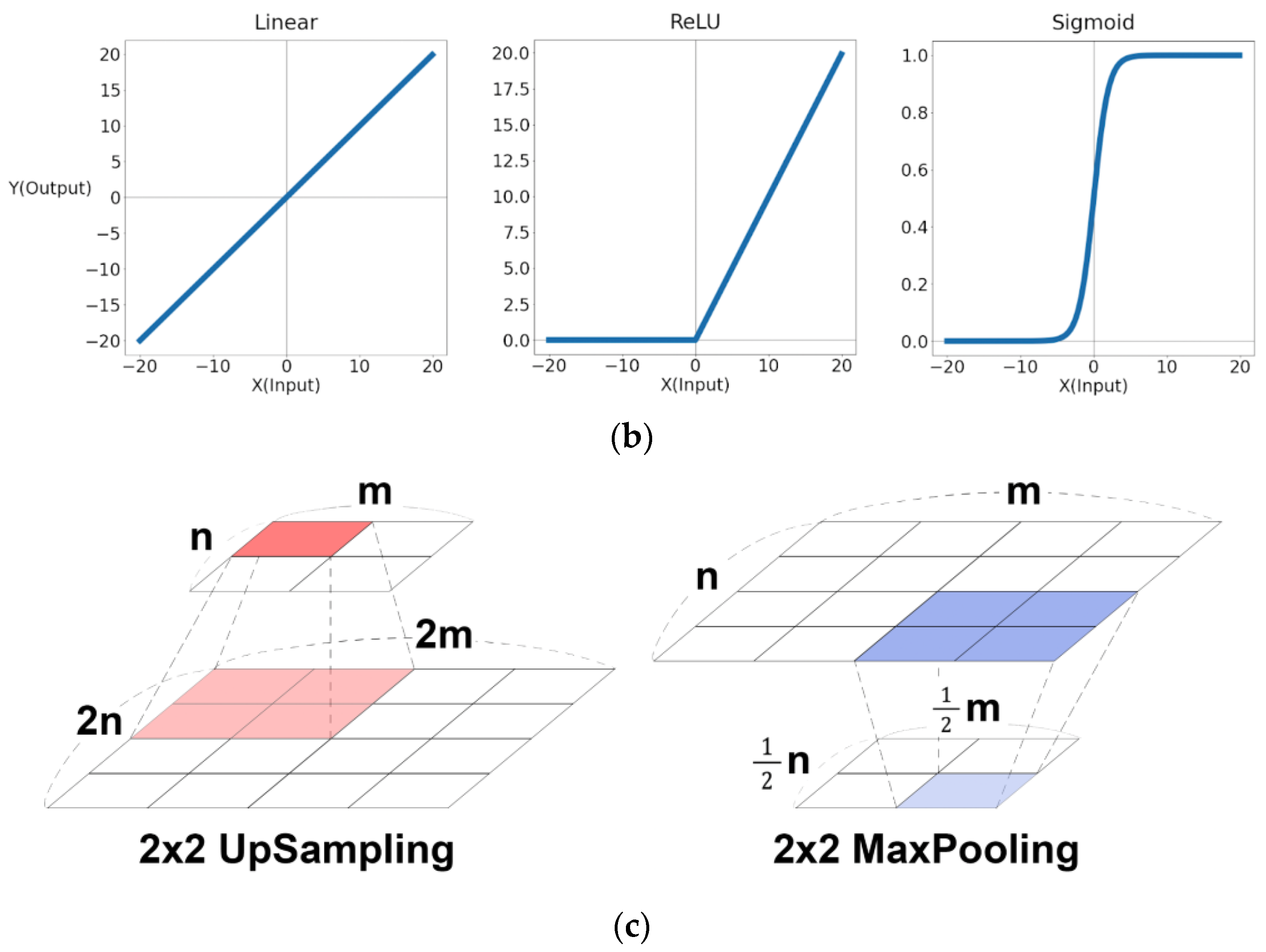
- Equation (1) is the convolutional layer expression, where and are the kernel size and the stride of the kernel. In addition, the term means the function applied to each layer, and the terms and the horizontal and vertical position of an element of a layer output. The term means (,)-th kernel matrix of layer, and the position of matrix element is expressed as . The term means (,)-th bias of layer. The term means a row position of a kernel, and the term a column position of the kernel;
- Equation (2) is an expression of 2x2 MaxPooling. The result of is the maximum value among the 2x2 submatrix with its upper left position of the matrix ;
- Equation (3) is an expression of 2x2 UpSampling. The result of positions to in the matrix is set to the value;
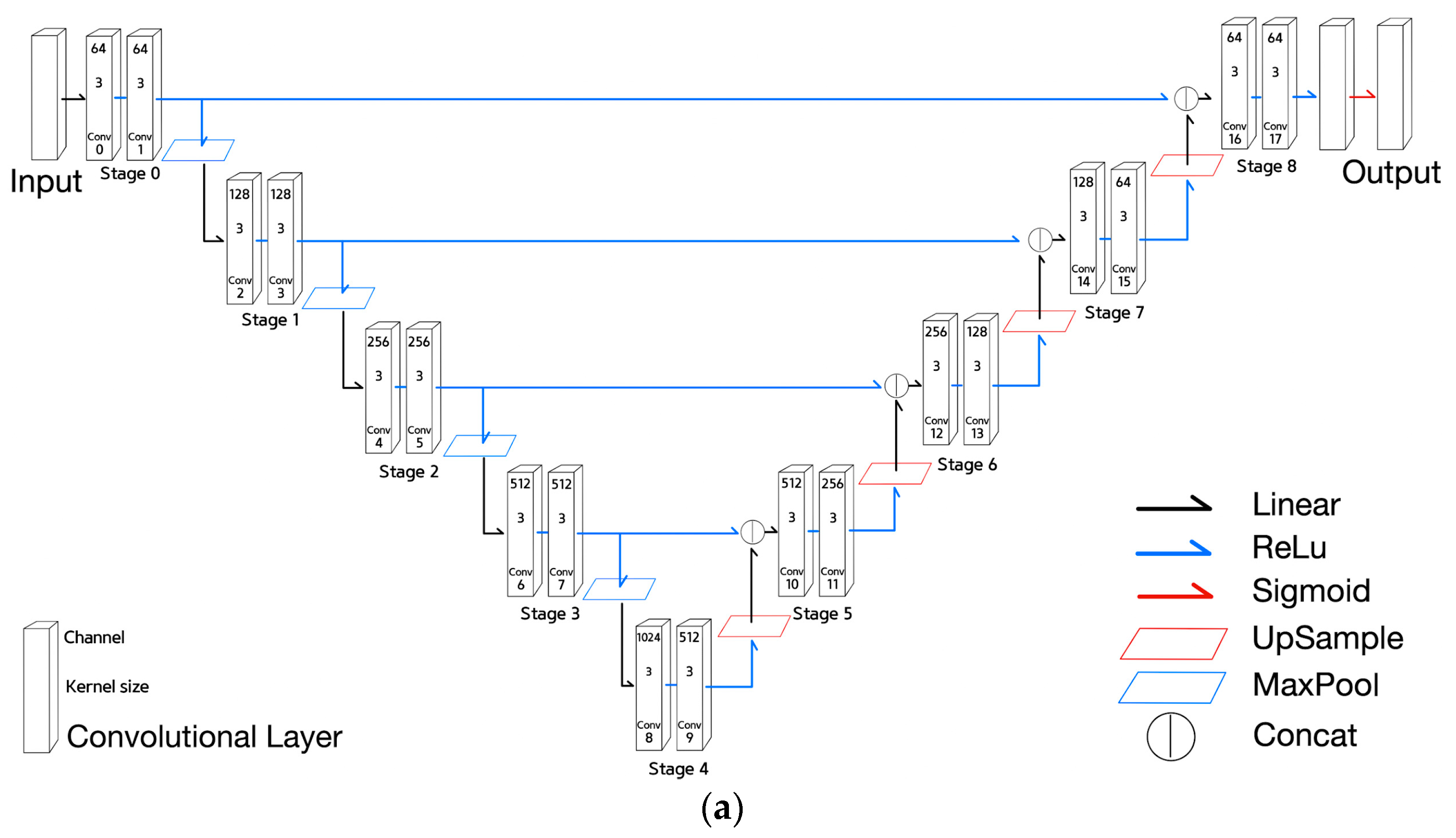
- The stages of U-net can be expressed by Equation (4) to (6):
- Equation (4) is the equation of the 0th stage that indicates the application of two convolutional layers with kernel size of three on the input and then ReLU() activation function expressed by is applied. The term means an input tensor of a model;
- Equation (5) is the encoder stage equation that indicates the application of maxpooling to the previous stage, followed by two convolutional layers and then final ReLU activation function. The term used in this equation means 2x2 maxpooling;
- Equation (6) is the decoder stage equation. The equation means the concatenation of the corresponding encoder stage output with the upsampled previous stage output, followed by two convolutional layers and then the final ReLU activation. The term used in Equation (6) means 2x2 upsampling.
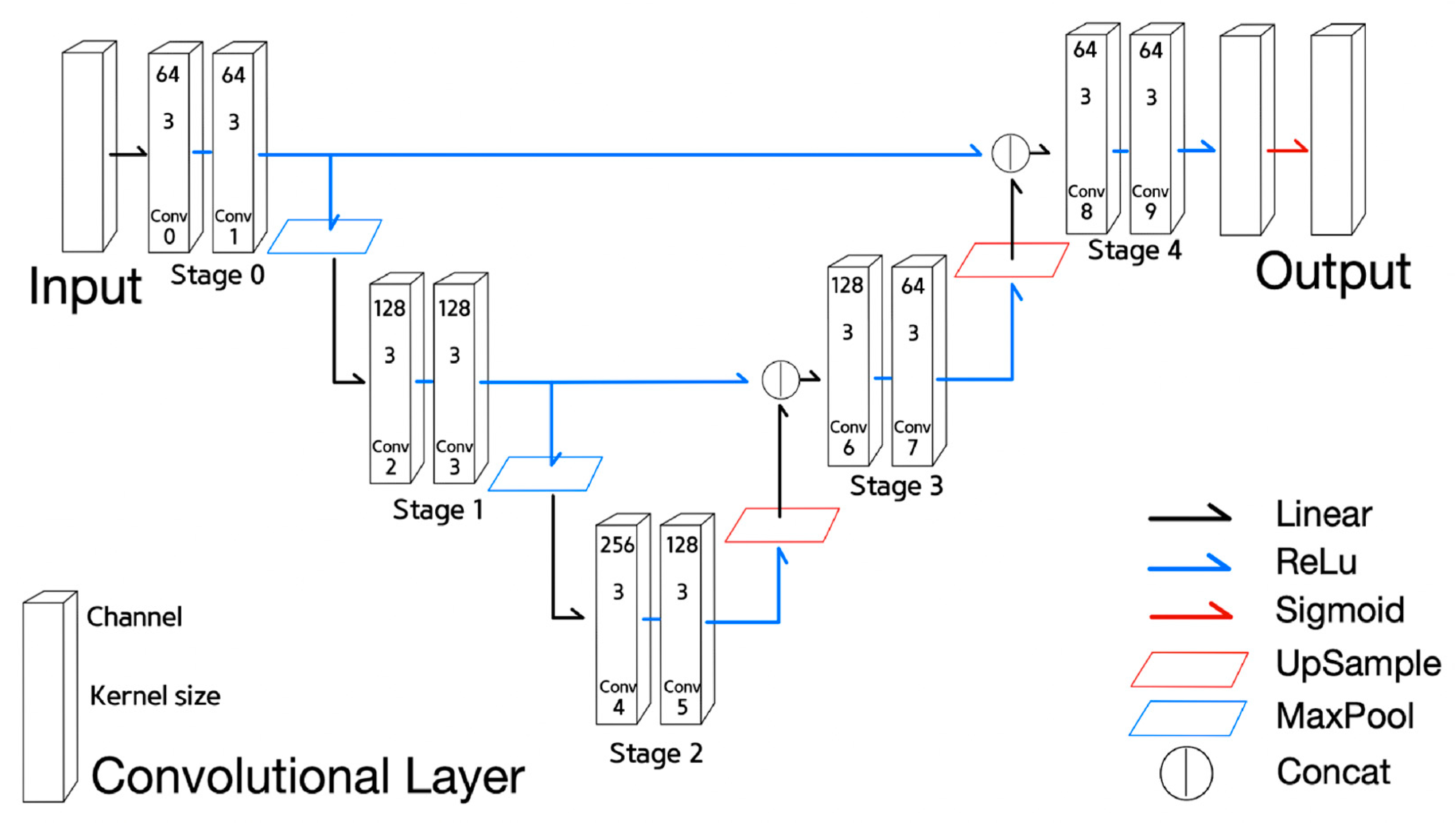
- States of Half U-net can be described in Equations (7)–(9);
- Equations (7)–(9) are defined similarly to Equations (4)–(6) of U-net, respectively. However, Equation (8) is for the encoder stage corresponding to the 1st or the 2nd stage and Equation (9) for the decoder stage corresponding to the 3rd and the 4th stage.
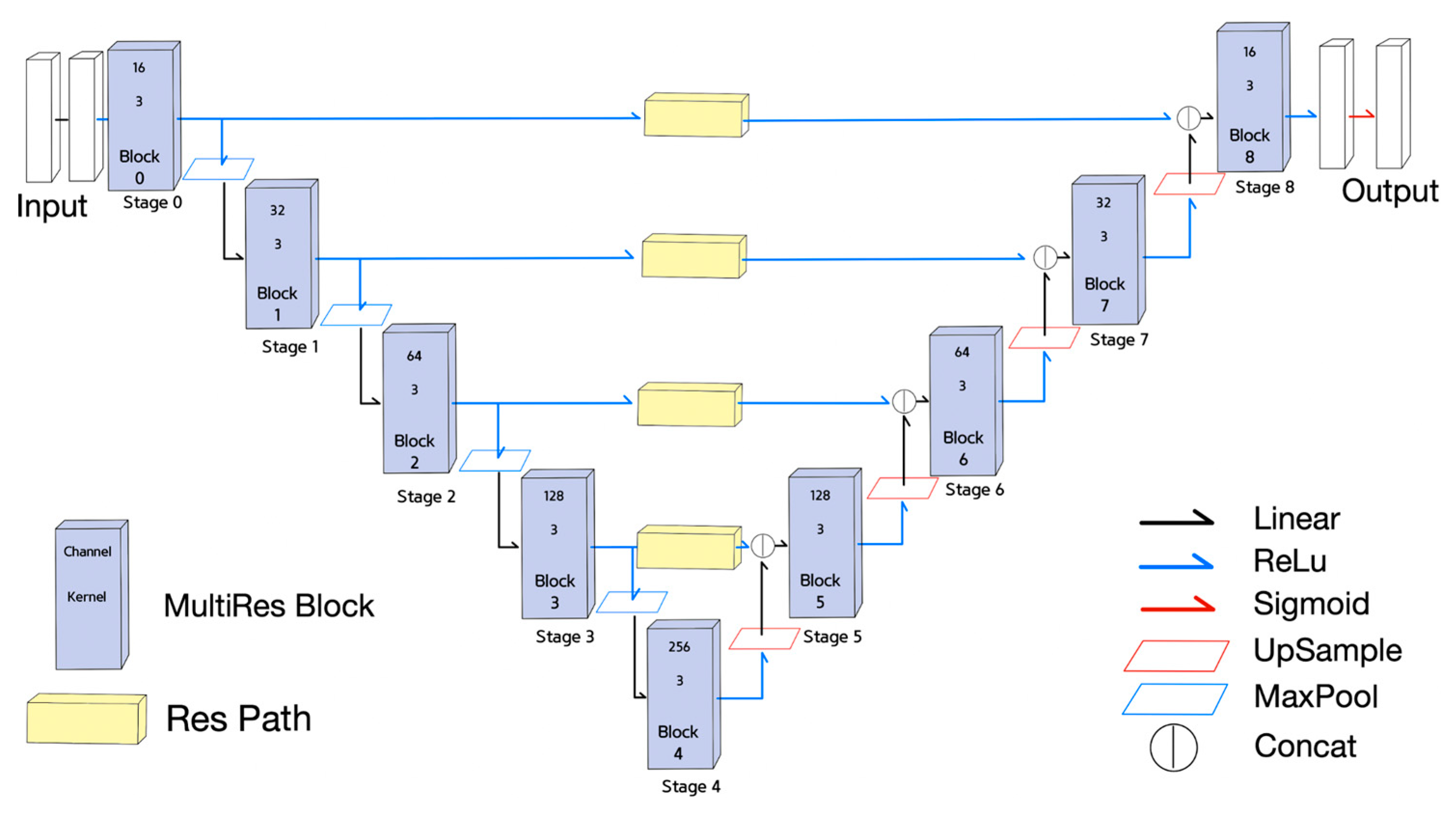
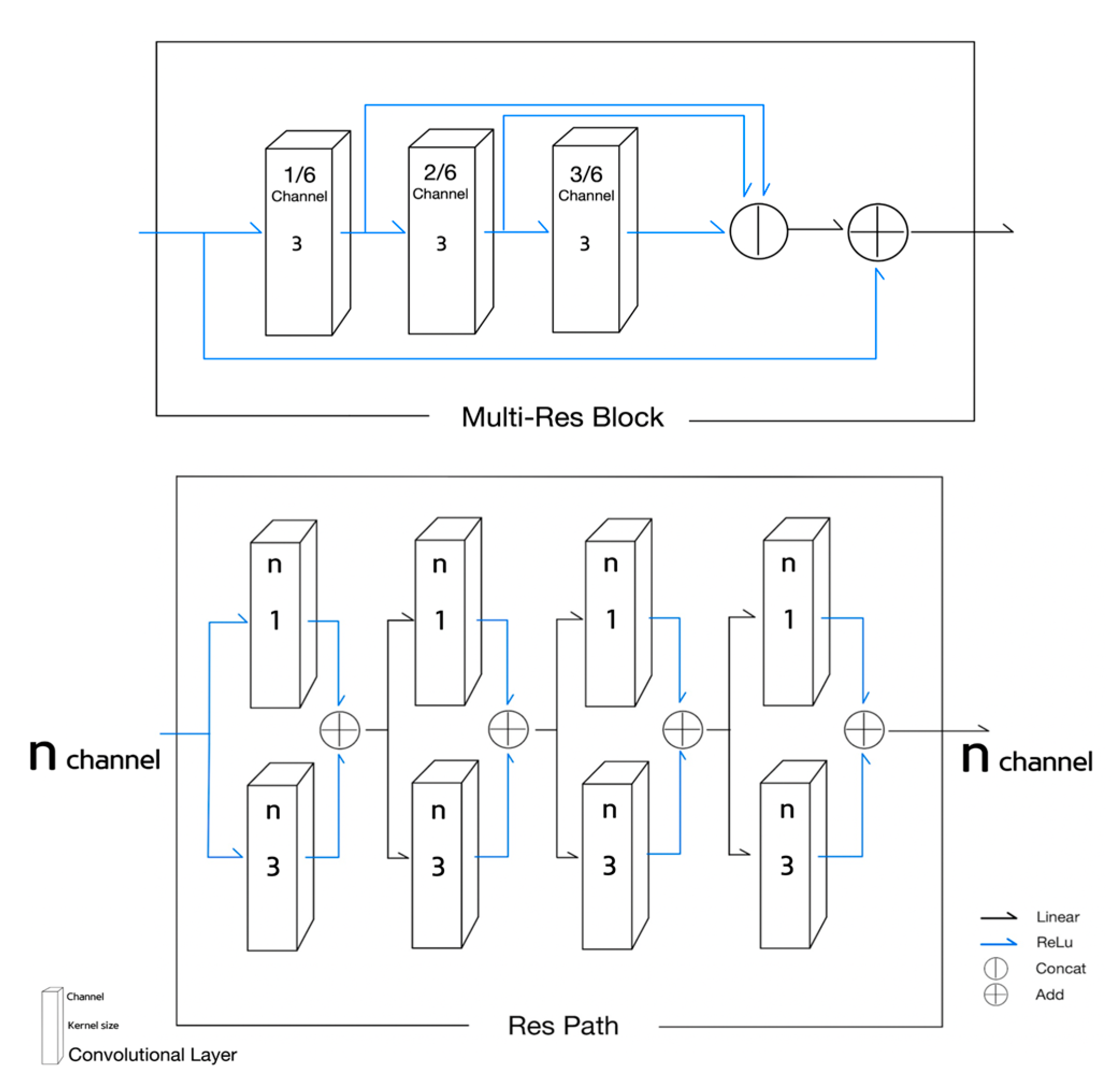
- Equations (10)–(13) is for the MultiRes Block ();
- The Equations (10)–(12) correspond to the 1st, the 2nd, and the 3rd convolutional block of the MultiRes Block illustrated in Figure 4. The term in Equations (10)–(12) means the number of channels in an input ;
- MRB Equation (13) indicates is equal to the concatenation of the outputs of Equations (10)–(12) plus the input . Concatenating (10) to (12) is taken as the result of MultiRes Block;
- The Equations (14) and (15) describe the residual path in Figure 4;
- The basic block of the residual path in Figure 4 described in Equation (14) is composed of the addition of the results of two convolutional layers of kernel sizes of 1 and 3;
- The four times repetition of the basic block of Equation (14) becomes the result of ResPath() as described in Equation (15);
- Stages of MultiResUNet can be expressed in Equations (16)–(18);
- Equation (16) is for the 0th stage whose result is activated with ReLU after passing through the ;
- Equation (17) is for the encoder stage ( = 1, 2, 3, 4), and the max pooled previous stage is activated with ReLU after passing through ;
- Equation (18) is for the decoder stage ( = 5, 6, 7, 8) that concatenates the upsampled previous stage output with the corresponding encoder stage output at the same pooling level passed through the . The concatenation output is activated with ReLU after passing through the .
- The stages of Half MultiResUNet can be expressed by Equations (19)–(21);
- Equations (19)–(21) are described similarly to Equations (16)–(18) of MultiResUNet. However, the number of stages is half. Equation (20) is for the encoder stage (= 1, 2) and Equation (21) for the decoder stage (= 3, 4).
- Stages of Quarter MultiResUNet can be represented in Equations (22)–(24);
- Equations (22)–(24) are similarly described as Equations (16)–(18) of MultiResUNet, but the number of stages is quarter. Equation (23) is for the encoder stage (= 1) and Equation (24) for the decoder stage (= 2).
4. Experiment
4.1. Dataset
4.2. Loss Function
- In the Equations (25)–(29), is the batch of the ground truth set and is the batch of the data predicted by the neural network. One specific ground truth (prediction) image in the batch is expressed in ;
- The weights for both classes, true (microplastic) and false (background), assigned to reflect the class imbalance for the training images with 512 × 512 resolution are shown in Equations (25) and (26), respectively;
- The original cross-entropy loss formula is given in Equation (27);
- To accommodate the class imbalance, we multiplied the formula ‘WeightMat’ shown in Equation (28), with the CrossEntropy Equation (27);
- The final loss function with batch size ‘n’ is given by Equation (29).
4.3. Training and Validation
4.4. Segmentation Performance Comparison and Analysis
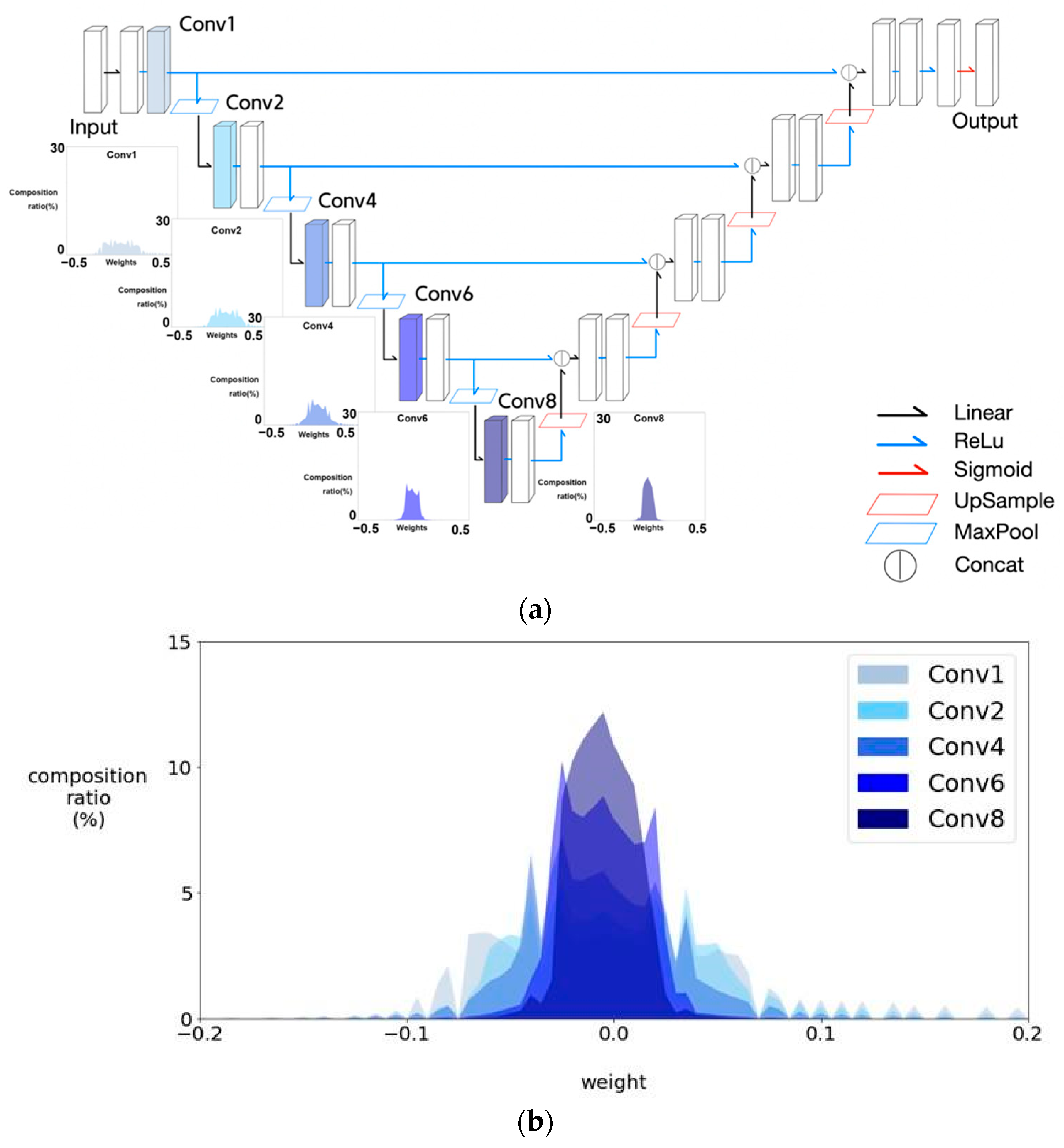
4.5. Observations on Weight Histograms of U-Net and MultiResUNet
4.6. Observations on Weight Histograms of Half and Quarter MultiResUNet
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rogers, K. Microplastics. Encyclopedia Britannica. 8 September 2020. Available online: https://www.britannica.com/technology/microplastic (accessed on 20 September 2021).
- Barboza, L.G.A.; Cózar, A.; Gimenez, B.C.; Barros, T.L.; Kershaw, P.J.; Guilhermino, L. Microplastics Pollution in the Marine Environment. World Seas Environ. Eval. 2019, 329–351. [Google Scholar] [CrossRef]
- Chatterjee, S.; Sharma, S. Microplastics in Our Oceans And Marine Health. Field Actions Sci. Rep. 2019, 19, 54–61. Available online: https://journals.openedition.org/factsreports/5257 (accessed on 20 September 2021).
- Cowger, W.; Gray, A.; Christiansen, S.; DeFrond, H.; Deshpande, A.; Hemabessiere, L.; Lee, E.; Mill, L.; Munno, K.; Oßmann, B.; et al. Critical Review of Processing and Classification Techniques for Images and Spectra in Microplastic Research. Appl. Spectrosc. 2020, 74, 989–1010. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Navarro, J.; Castrillón-Santana, M.; Gómez, M.; Herrera, A.; Marín-Reyes, P. Automatic Counting and Classification of Microplastic Particles. In Proceedings of the 7th International Conference on Pattern Recognition Applications and Methods—ICPRAM, Funchal, Portugal, 16–18 January 2018; pp. 646–652, ISBN 978-989-758-276-9. ISSN 2184-4313. [Google Scholar] [CrossRef]
- Lorenzo-Navarro, J.; Castrillón-Santana, M.; Sánchez-Nielsen, E.; Zarco, B.; Herrera, A.; Martínez, I.; Gómez, M. Deep learning approach for automatic microplastics counting and classification. Sci. Total Environ. 2021, 765, 142728. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. May 2015, Volume 9351, pp. 234–241. Available online: https://arxiv.org/abs/1505.04597v1 (accessed on 30 July 2021).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning For Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- TensorBoard. Available online: https://www.tensorflow.org/tensorboard (accessed on 30 July 2021).
- Li, X.; Zhang, L.; You, A.; Yang, M.; Yang, K.; Tong, Y. Global Aggregation Then Local Distribution In Fully Convolutional Networks. arXiv 2019, arXiv:1909.07229. Available online: https://arxiv.org/abs/1909.07229v1 (accessed on 20 August 2021).
- Choi, S.; Kim, J.T.; Choo, J. Cars Can’t Fly Up In The Sky: Improving Urban-Scene Segmentation Via Height-Driven Attention Networks. arXiv 2020, arXiv:2003.05128. Available online: https://arxiv.org/abs/2003.05128v3 (accessed on 20 August 2021).
- Mohan, R.; Valada, A. Efficientps: Efficient Panoptic Segmentation. arXiv 2020, arXiv:2004.02307. Available online: https://arxiv.org/abs/2004.02307 (accessed on 20 September 2021).
- Cheng, B.; Collins, M.D.; Zhu, Y.; Liu, T.; Huang, T.S.; Adam, H.; Chen, L. Panoptic-Deeplab: A Simple, Strong, And Fast Baseline For Bottom-Up Panoptic Segmentation. arXiv 2019, arXiv:1911.10194. Available online: https://arxiv.org/abs/1911.10194v3 (accessed on 20 September 2021).
- Chen, L.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic Image Segmentation With Deep Convolutional Nets, Atrous Convolution, And Fully Connected Crfs. arXiv 2016, arXiv:1606.00915. Available online: https://arxiv.org/abs/1606.00915v2 (accessed on 20 August 2021).
- Chen, L.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-Decoder With Atrous Separable Convolution For Semantic Image Segmentation. arXiv 2018, arXiv:1802.02611. Available online: https://arxiv.org/abs/1802.02611v3 (accessed on 20 August 2021).
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid Scene Parsing Network. arXiv 2016, arXiv:1612.01105. Available online: https://arxiv.org/abs/1612.01105v2 (accessed on 20 August 2021).
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Ibtehaz, N.; Rahman, M.S. MultiResU-net: Rethinking U-Net Architecture for Multimodal Biomedical Image Segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Semantic Image Segmentation With Deep Convolutional Nets And Fully Connected Crfs. arXiv 2014, arXiv:1412.7062. Available online: https://arxiv.org/abs/1412.7062v4 (accessed on 20 August 2021).
- Chen, L.-C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking Atrous Convolution for Semantic Image Segmentation. arXiv 2017, arXiv:1706.05587. Available online: https://arxiv.org/abs/1706.05587v3 (accessed on 30 July 2021).
- Suárez-Paniagua, V.; Segura-Bedmar, I. Evaluation of Pooling Operations in Convolutional Architectures for Drug-Drug Interaction Extraction. BMC Bioinform. 2018, 19, 39–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks For Semantic Segmentation. arXiv 2016, arXiv:1605.06211. Available online: https://arxiv.org/abs/1605.06211v1 (accessed on 20 August 2021).
- Liu, J.; Chao, F.; Lin, C. Task Augmentation By Rotating For Meta-Learning. arXiv 2020, arXiv:2003.00804. Available online: https://arxiv.org/abs/2003.00804v1 (accessed on 20 August 2021).
- Zhong, Z.; Zheng, L.; Kang, G.; Li, S.; Yang, Y. Random Erasing Data Augmentation. arXiv 2017, arXiv:1708.04896. Available online: https://arxiv.org/abs/1708.04896 (accessed on 20 August 2021). [CrossRef]
- Ho, Y.; Wookey, S. The Real-World-Weight Cross-Entropy Loss Function: Modeling the Costs of Mislabeling. IEEE Access 2020, 8, 4806–4813. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method For Stochastic Optimization. arXiv 2014, arXiv:1412.6980. Available online: https://arxiv.org/abs/1412.6980v9 (accessed on 20 August 2021).
- Amendola, G.; Fabrizio, M.; Golden, J.M. Exponential Decay. In Thermodynamics of Materials with Memory; Springer: Boston, MA, USA, 2012. [Google Scholar] [CrossRef]
- Rodriguez, J.D.; Perez, A.; Lozano, J.A. Sensitivity Analysis of k-Fold Cross Validation in Prediction Error Estimation. Pattern Analysis and Machine Intelligence. IEEE Trans. 2010, 32, 569–575. [Google Scholar]
- Lipton, Z.C.; Elkan, C.; Narayanaswamy, B. Thresholding Classifiers To Maximize F1 Score. arXiv 2014, arXiv:1402.1892. Available online: https://arxiv.org/abs/1402.1892v2 (accessed on 20 August 2021).
- Rahman, M.A.; Wang, Y. Optimizing Intersection-Over-Union In Deep Neural Networks For Image Segmentation. Adv. Vis. Comput. 2016, 234–244. [Google Scholar] [CrossRef]
- Minaee, S.; Boykov, Y.; Porikli, F.; Plaza, A.; Kehtarnavaz, N.; Terzopoulos, D. Image Segmentation Using Deep Learning: A Survey. arXiv 2020, arXiv:2001.05566. Available online: https://arxiv.org/abs/2001.05566 (accessed on 20 August 2021).









| Model (150 Eps) | 1st Case | 2nd Case | 3rd Case | |||
| Precision | Recall | Precision | Recall | Precision | Recall | |
| U-net | 0 | 0 | 0.0692 | 0.9644 | 0 | 0 |
| Half U-net | 0.0565 | 0.9370 | 0.0437 | 0.9470 | 0 | 0 |
| MultiResUNet | 0.1583 | 0.9386 | 0.0190 | 0.9448 | 0.0329 | 0.9557 |
| Half MultiResUNet | 0.2899 | 0.9180 | 0.3175 | 0.9322 | 0.2771 | 0.9404 |
| Quarter MultiResUNet | 0.1065 | 0.9517 | 0.1451 | 0.9772 | 0.1828 | 0.9624 |
| Model (150 Eps) | 4th Case | 5th Case | Average | |||
| Precision | Recall | Precision | Recall | Precision | Recall | |
| U-net | 0.0178 | 0.8138 | 0 | 0 | 0.0174 | 0.3556 |
| Half U-net | 0.0357 | 0.8547 | 0 | 0 | 0.0272 | 0.5478 |
| MultiResUNet | 0.0092 | 0.8813 | 0.0138 | 0.8972 | 0.0466 | 0.9235 |
| Half MultiResUNet | 0.2828 | 0.9447 | 0.2219 | 0.9832 | 0.2778 | 0.9437 |
| Quarter MultiResUNet | 0.2214 | 0.9456 | 0.1316 | 0.9817 | 0.1575 | 0.9637 |
| Model (150 Eps) | 1st Case | 2nd Case | 3rd Case | |||
| F1 Score | mIoU | F1 Score | mIoU | F1 Score | mIoU | |
| U-net | 0 | 0 | 0.1291 | 0.0690 | 0 | 0 |
| Half U-net | 0.1067 | 0.0563 | 0.0836 | 0.0436 | 0 | 0 |
| MultiResUNet | 0.2710 | 0.1567 | 0.0372 | 0.0189 | 0.0636 | 0.0328 |
| Half MultiResUNet | 0.4407 | 0.2826 | 0.4737 | 0.3103 | 0.4281 | 0.2723 |
| Quarter MultiResUNet | 0.1916 | 0.1060 | 0.2526 | 0.1446 | 0.3072 | 0.1815 |
| Model (150 Eps) | 4th Case | 5th Case | AVERAGE | |||
| F1 Score | mIoU | F1 Score | mIoU | F1 Score | mIoU | |
| U-net | 0.0349 | 0.0178 | 0 | 0 | 0.0328 | 0.0173 |
| Half U-net | 0.0685 | 0.0355 | 0 | 0 | 0.0517 | 0.0270 |
| MultiResUNet | 0.0181 | 0.0091 | 0.0272 | 0.0138 | 0.0834 | 0.0462 |
| Half MultiResUNet | 0.4352 | 0.2782 | 0.3621 | 0.2210 | 0.4279 | 0.2728 |
| Quarter MultiResUNet | 0.3588 | 0.2186 | 0.2321 | 0.1313 | 0.2685 | 0.1563 |
| Model (150 Eps) | 1st Case | 2nd Case | 3rd Case | |||
| r.w. F1 Score | r.w. mIoU | r.w. F1 Score | r.w.mIoU | r.w. F1 Score | r.w. mIoU | |
| U-net | 0 | 0 | 0.1245 | 0.0665 | 0 | 0 |
| Half U-net | 0.0999 | 0.0528 | 0.0792 | 0.0413 | 0 | 0 |
| MultiResUNet | 0.2544 | 0.1471 | 0.0351 | 0.0179 | 0.0608 | 0.0313 |
| Half MultiResUNet | 0.4046 | 0.2594 | 0.4416 | 0.2893 | 0.4026 | 0.2561 |
| Quarter MultiResUNet | 0.1823 | 0.1009 | 0.2468 | 0.1413 | 0.2956 | 0.1747 |
| Model (150 Eps) | 4th Case | 5th Case | Average | |||
| r.w. F1 Score | r.w. mIoU | r.w. F1 Score | r.w. mIoU | r.w. F1 Score | r.w. mIoU | |
| U-net | 0.0284 | 0.0145 | 0 | 0 | 0.0306 | 0.0162 |
| Half U-net | 0.0585 | 0.0303 | 0 | 0 | 0.0475 | 0.0249 |
| MultiResUNet | 0.0159 | 0.0080 | 0.0244 | 0.0124 | 0.0781 | 0.0433 |
| Half MultiResUNet | 0.4111 | 0.2628 | 0.3560 | 0.2173 | 0.4032 | 0.2570 |
| Quarter MultiResUNet | 0.3393 | 0.2067 | 0.2279 | 0.1289 | 0.2584 | 0.1505 |
| Model (Input Size) | FLOPs | Parameters |
|---|---|---|
| U-net (512 × 512) | 329.7 B | 21.9776 M |
| Half U-net (512 × 512) | 204.0 B | 1.7724 M |
| MultiResUNet (512 × 512) | 204.8 B | 13.5469 M |
| Half MultiResUNet (512 × 512) | 42.9 B | 0.2149 M |
| Quarter MultiResUNet (512 × 512) | 21.9 B | 0.0445 M |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, G.; Jhang, K. Neural Network Analysis for Microplastic Segmentation. Sensors 2021, 21, 7030. https://doi.org/10.3390/s21217030
Lee G, Jhang K. Neural Network Analysis for Microplastic Segmentation. Sensors. 2021; 21(21):7030. https://doi.org/10.3390/s21217030
Chicago/Turabian StyleLee, Gwanghee, and Kyoungson Jhang. 2021. "Neural Network Analysis for Microplastic Segmentation" Sensors 21, no. 21: 7030. https://doi.org/10.3390/s21217030
APA StyleLee, G., & Jhang, K. (2021). Neural Network Analysis for Microplastic Segmentation. Sensors, 21(21), 7030. https://doi.org/10.3390/s21217030






