W-TSS: A Wavelet-Based Algorithm for Discovering Time Series Shapelets
Abstract
:1. Introduction
2. Materials and Methods
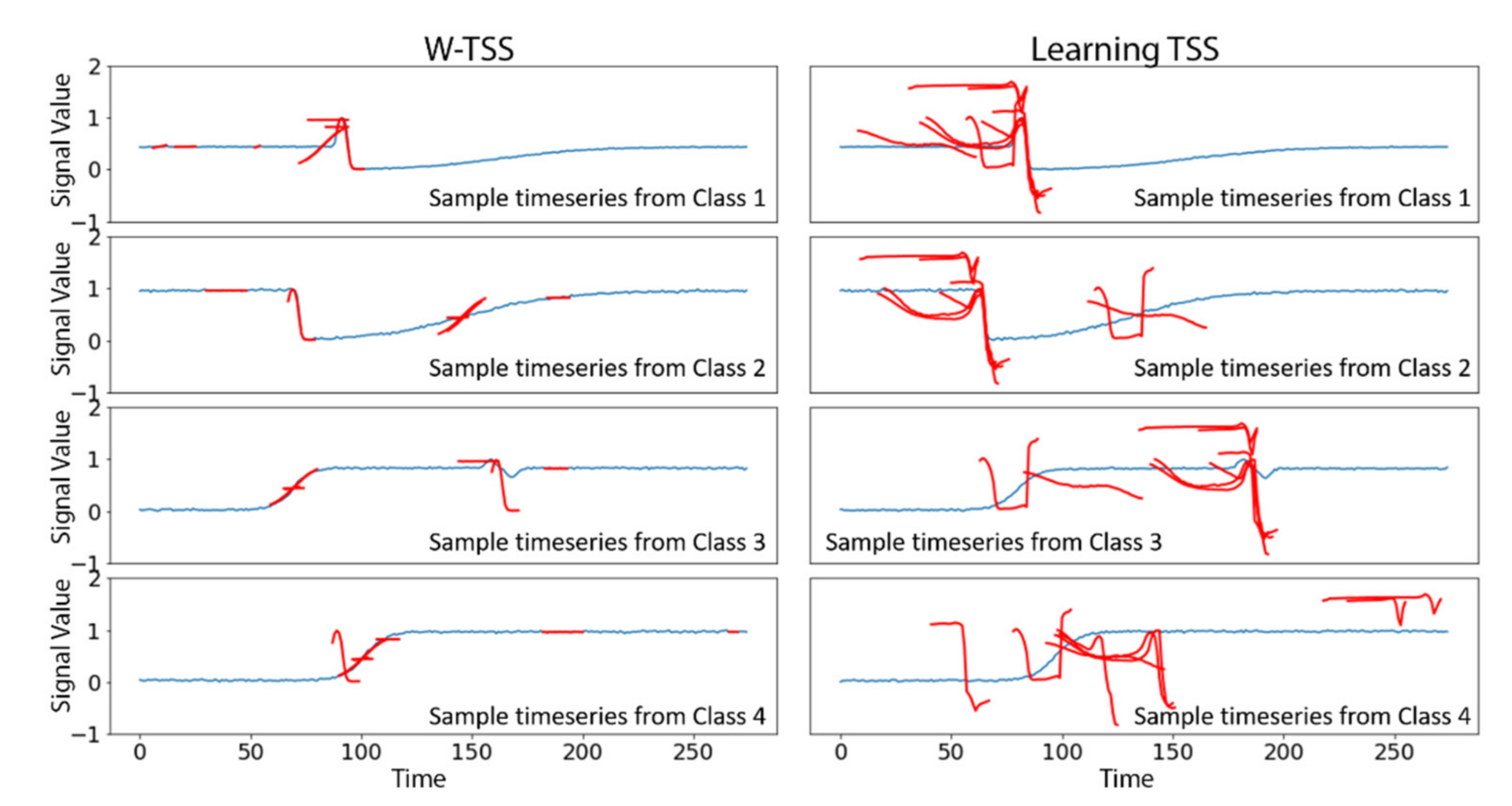
2.1. Learning TSS
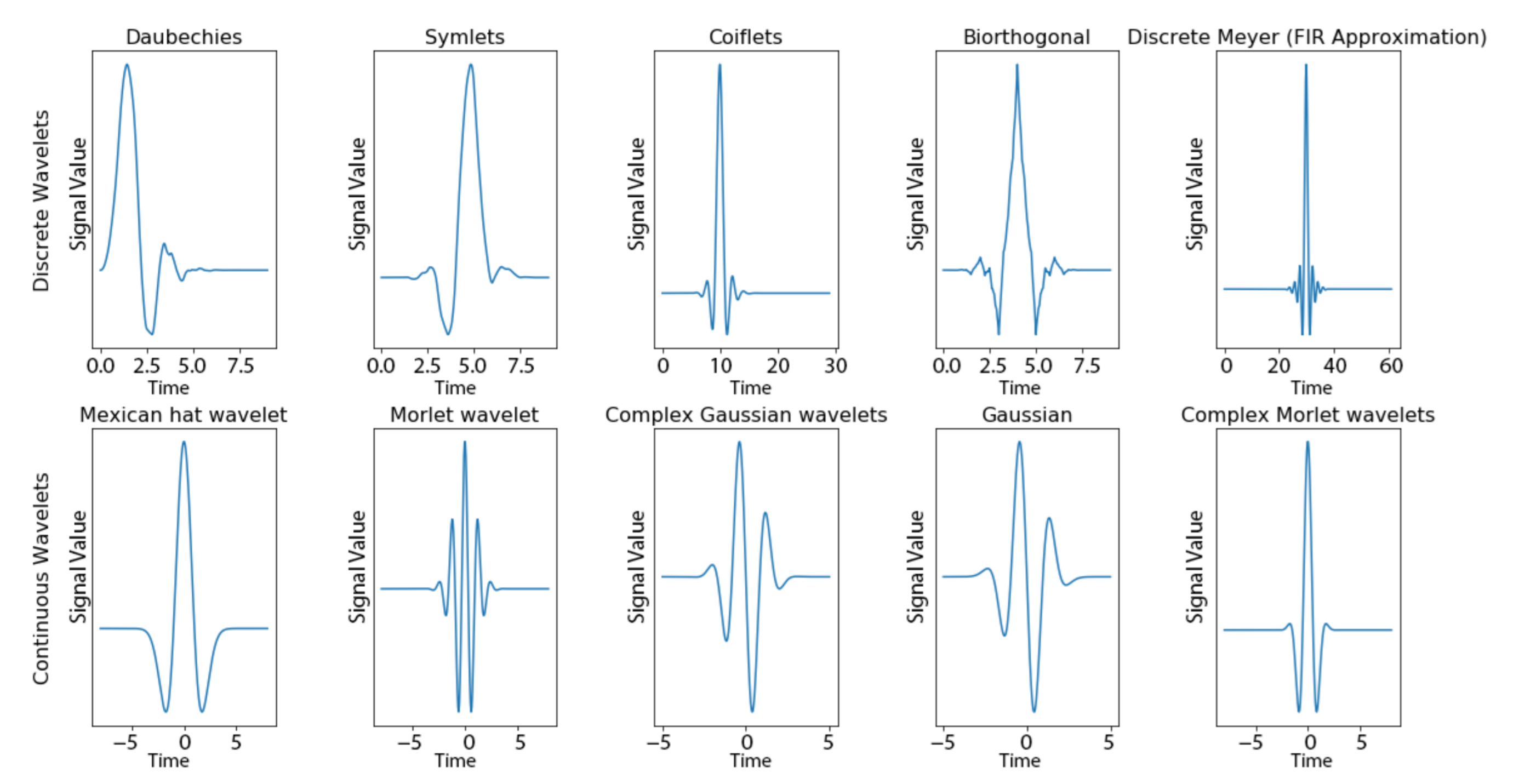
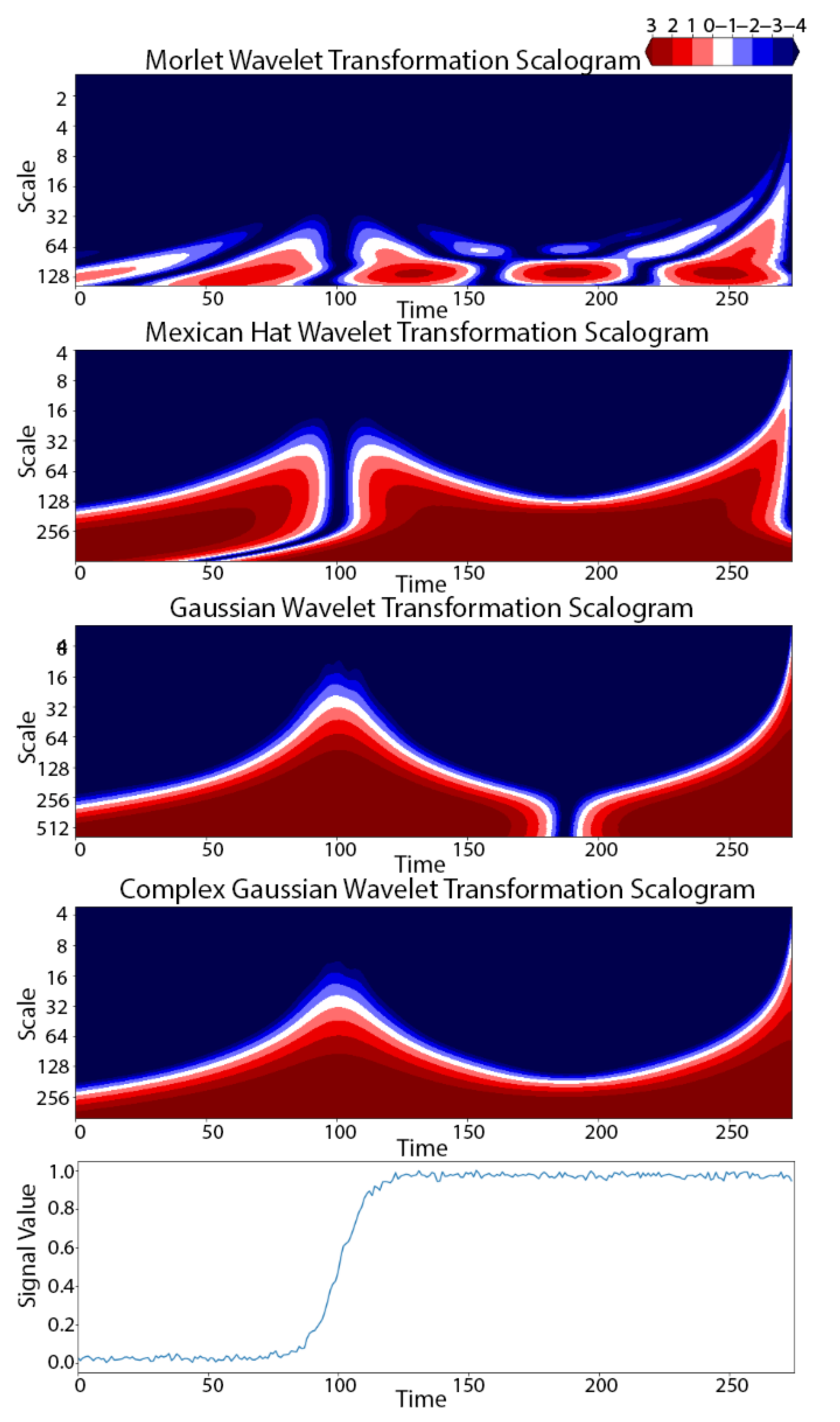
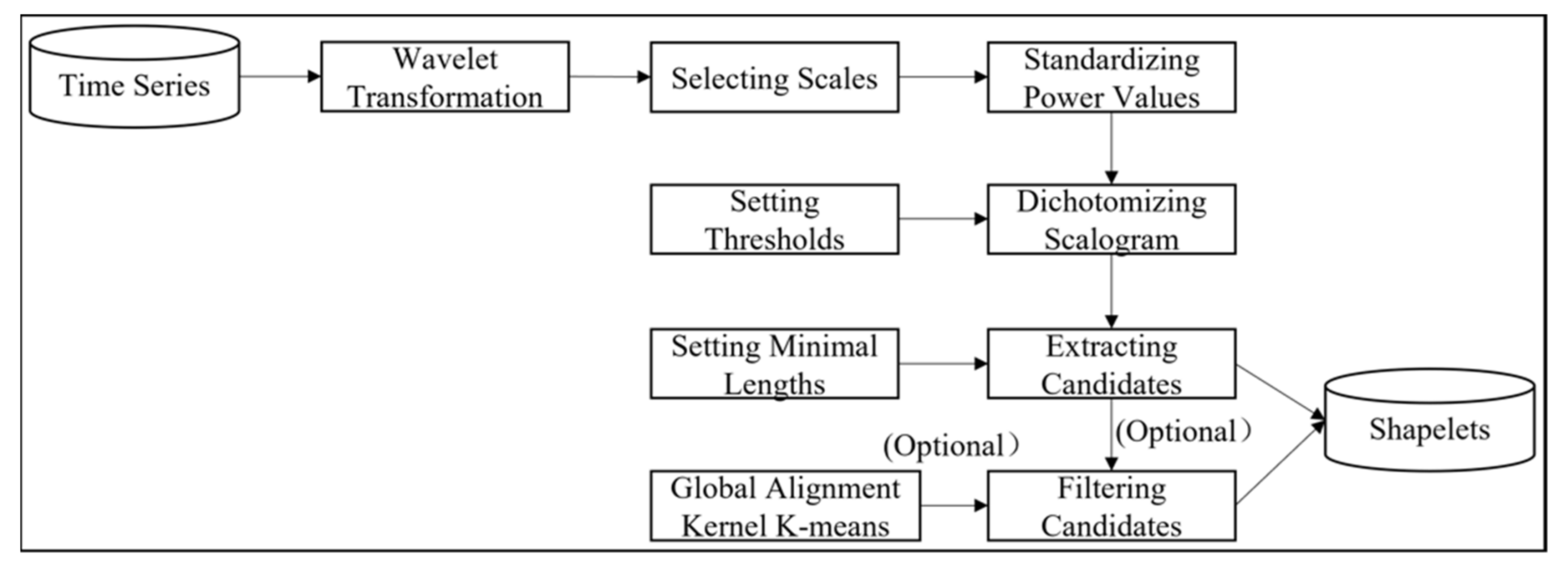
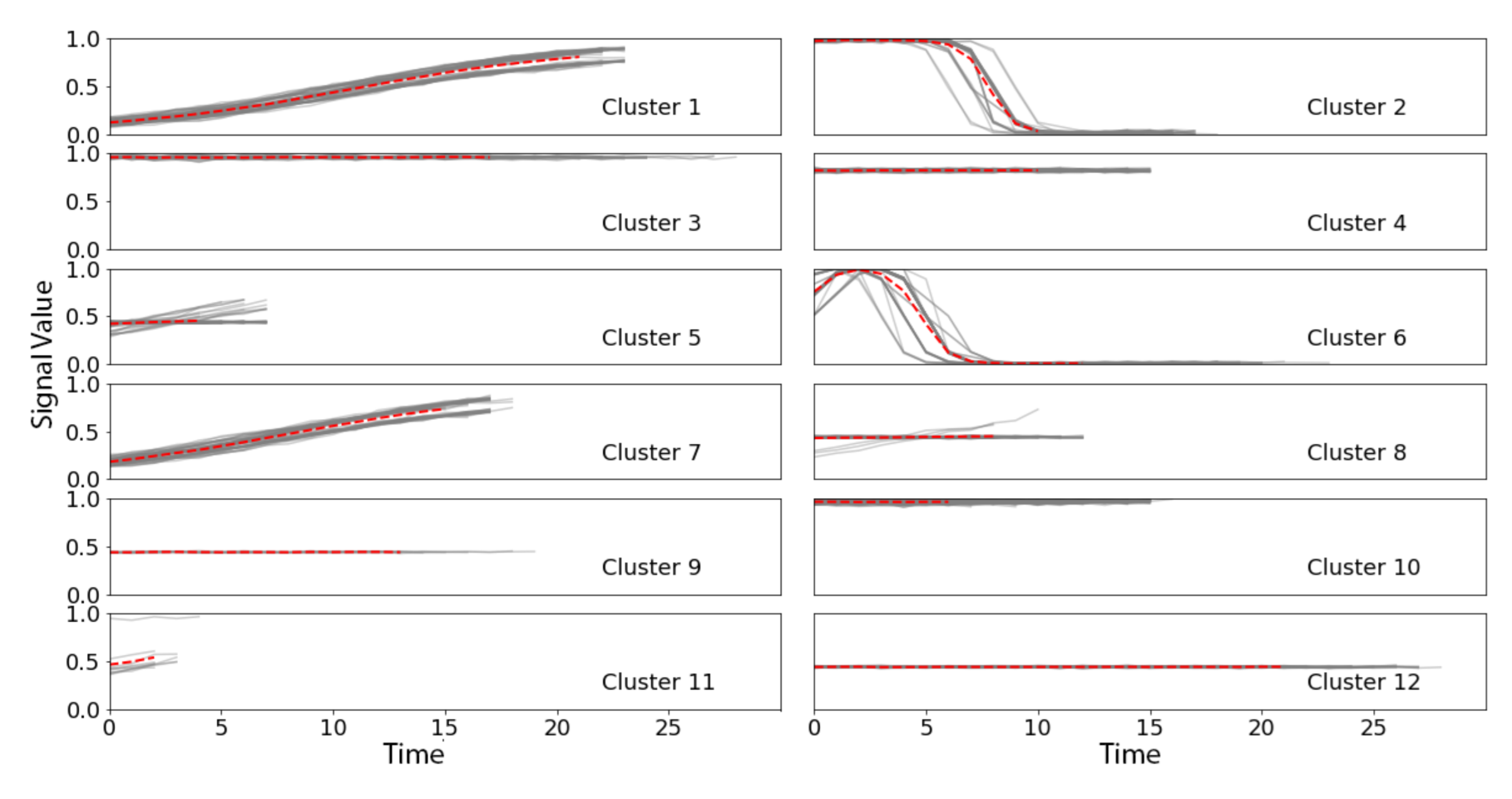
2.2. Wavelet-Based Discovery for TSS (W-TSS)
2.3. Datasets
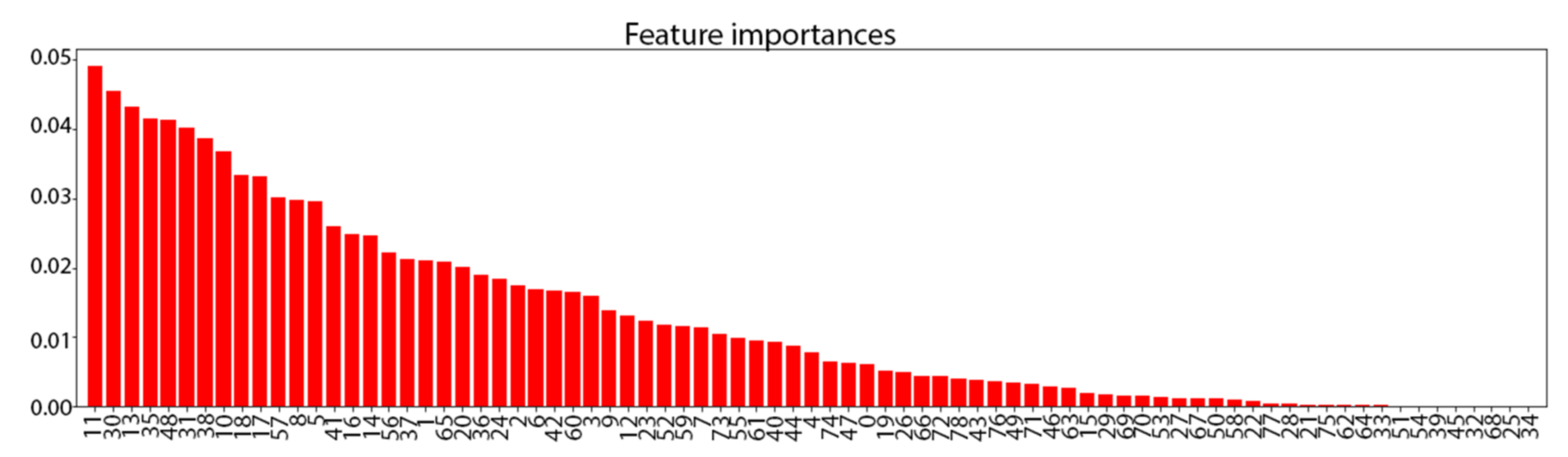
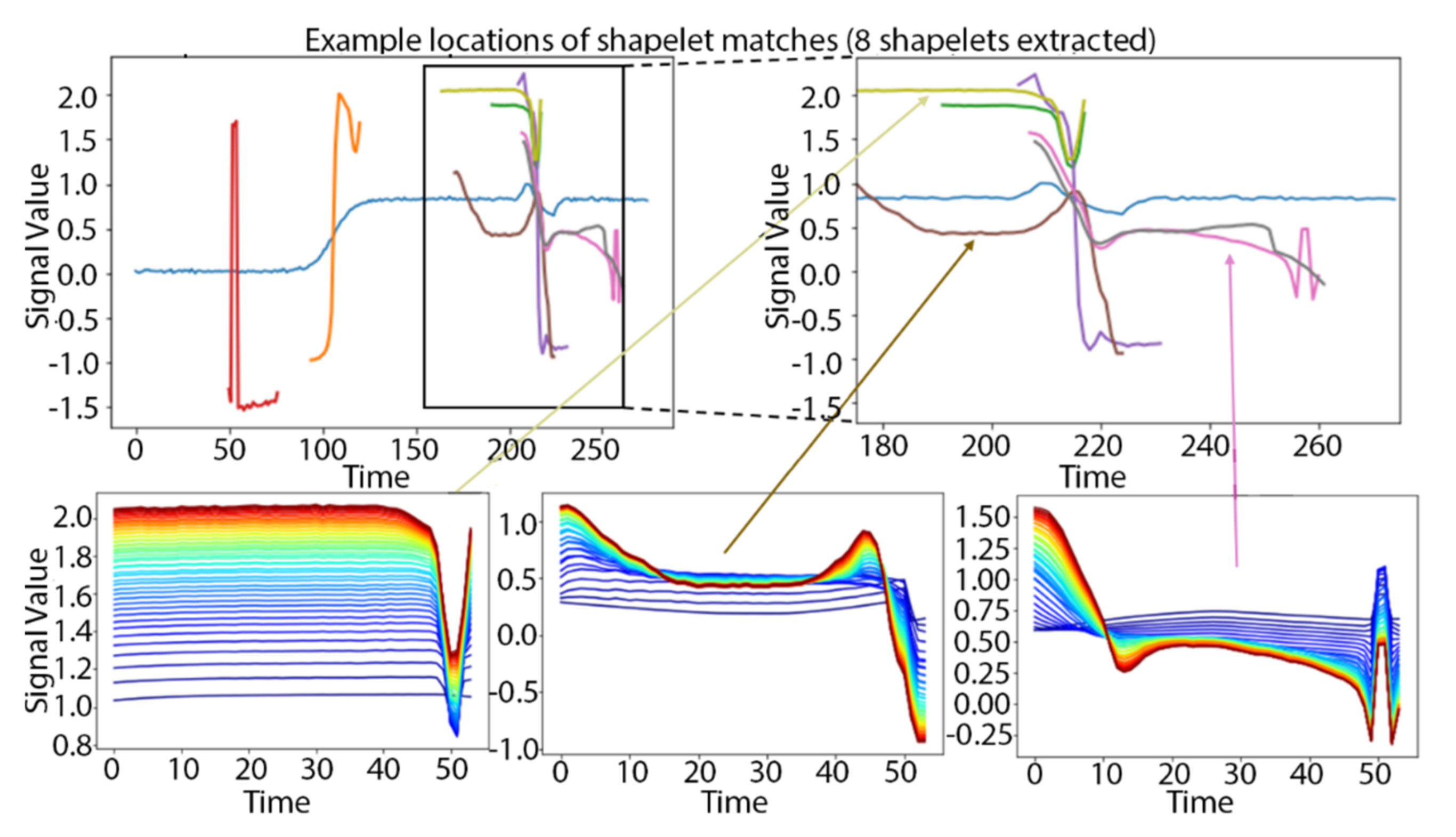
3. Results
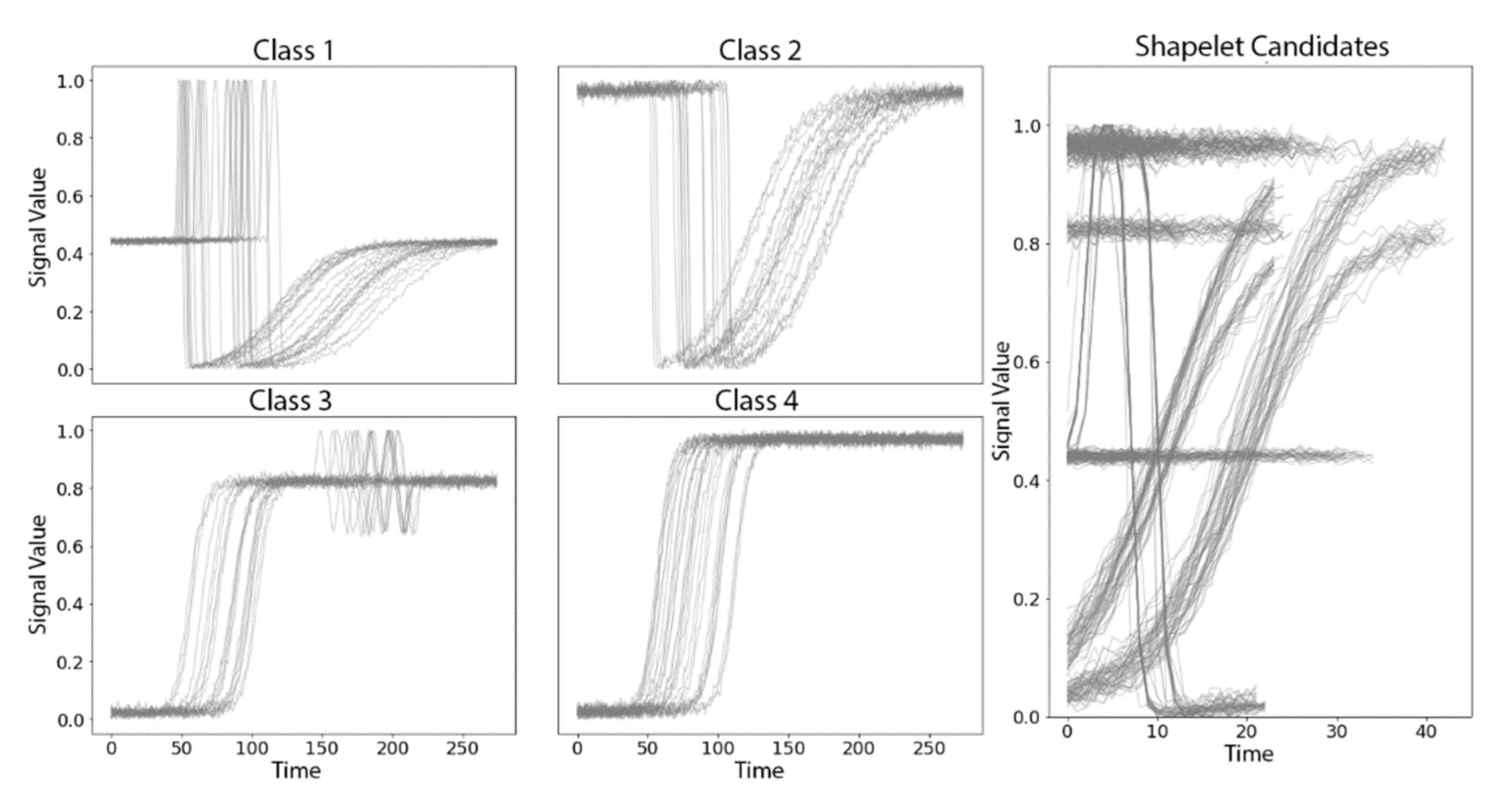
3.1. Synthetic TRACE Dataset
3.2. PRISMS Dataset from Pediatric Asthma Study
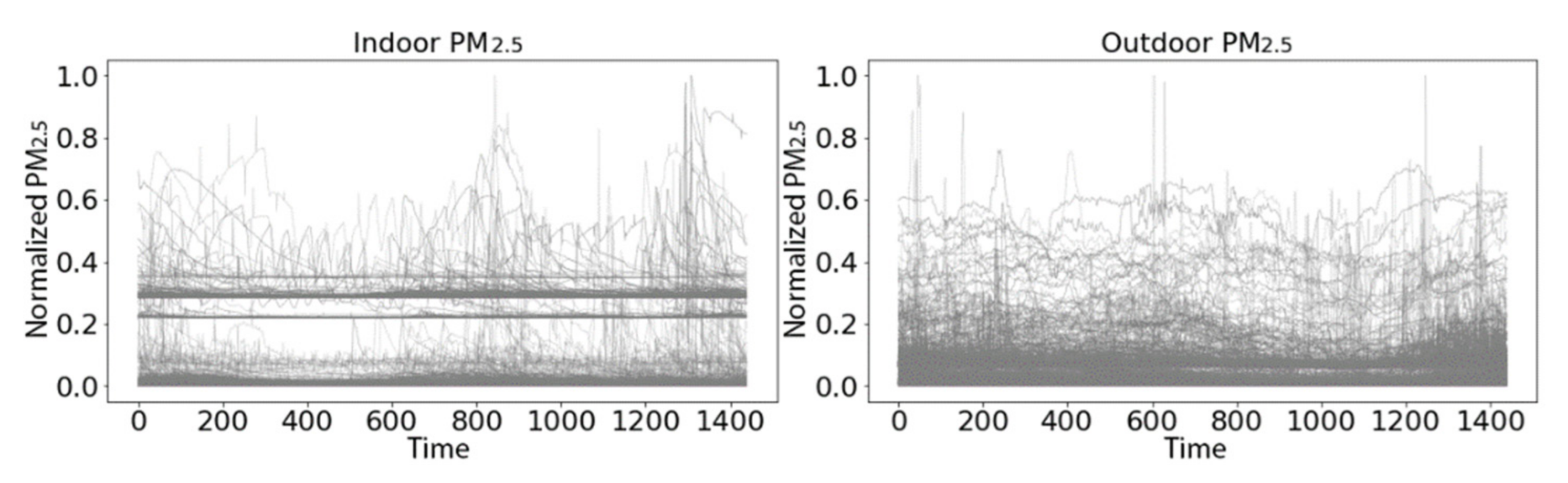
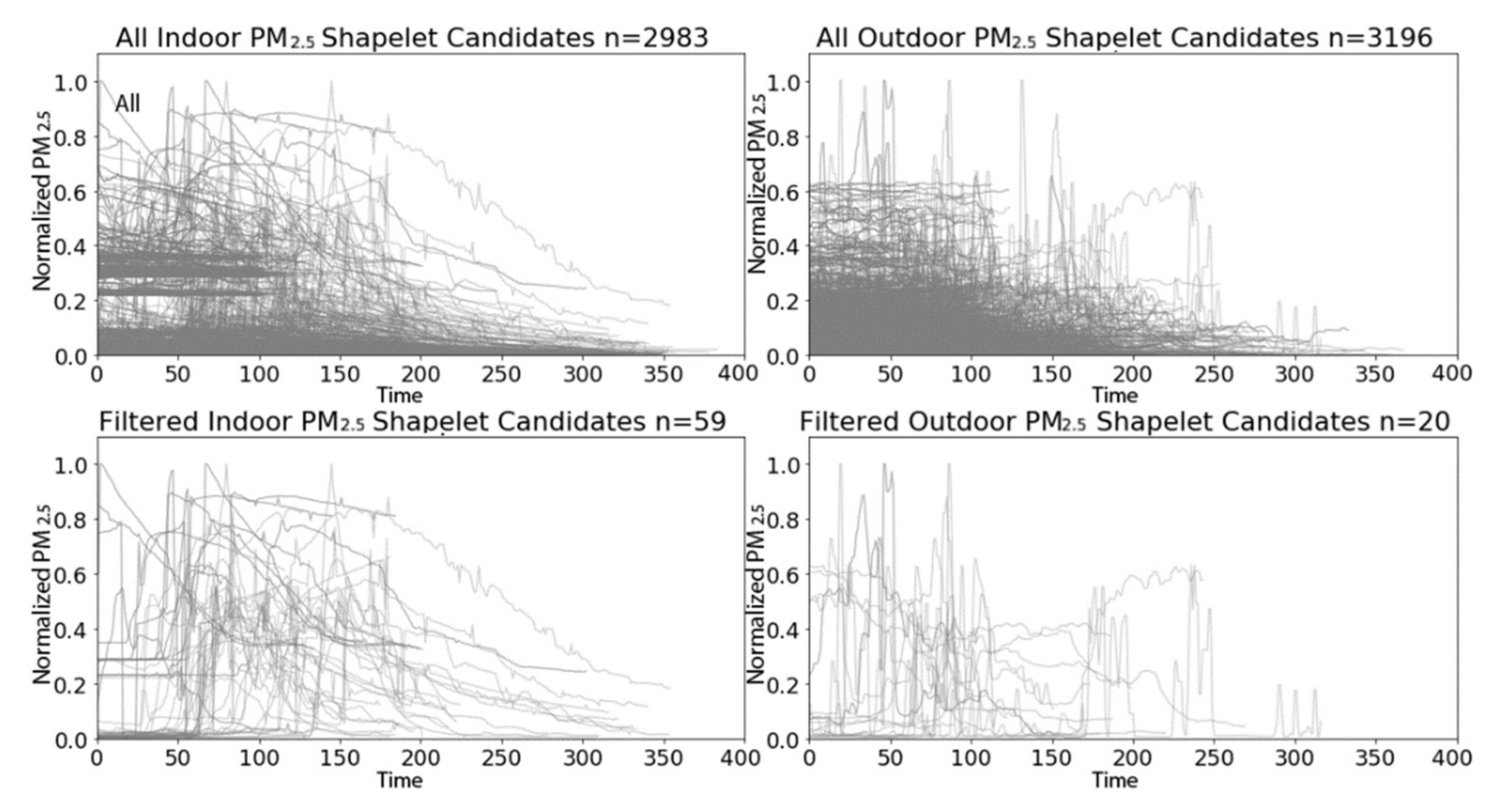
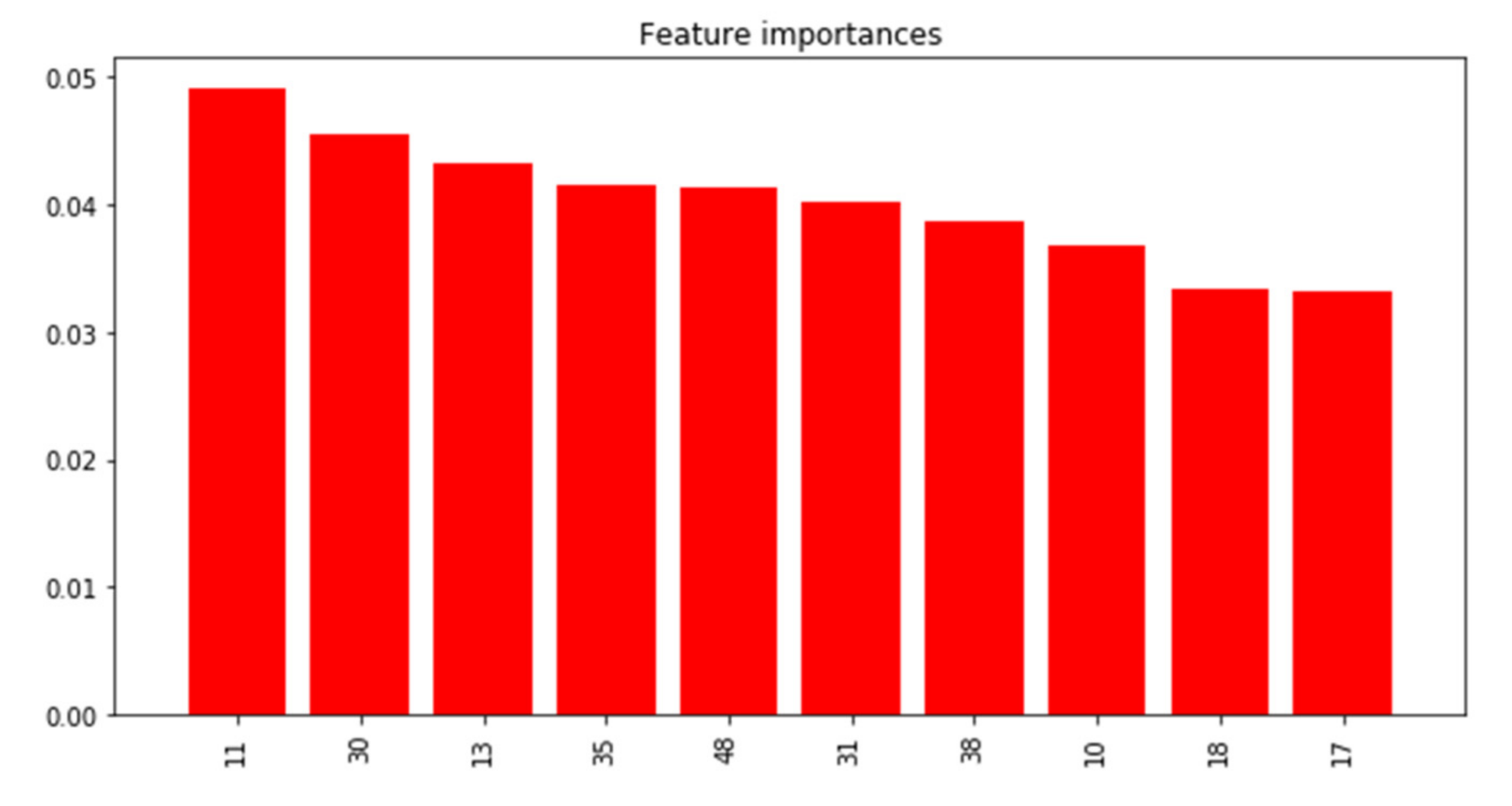
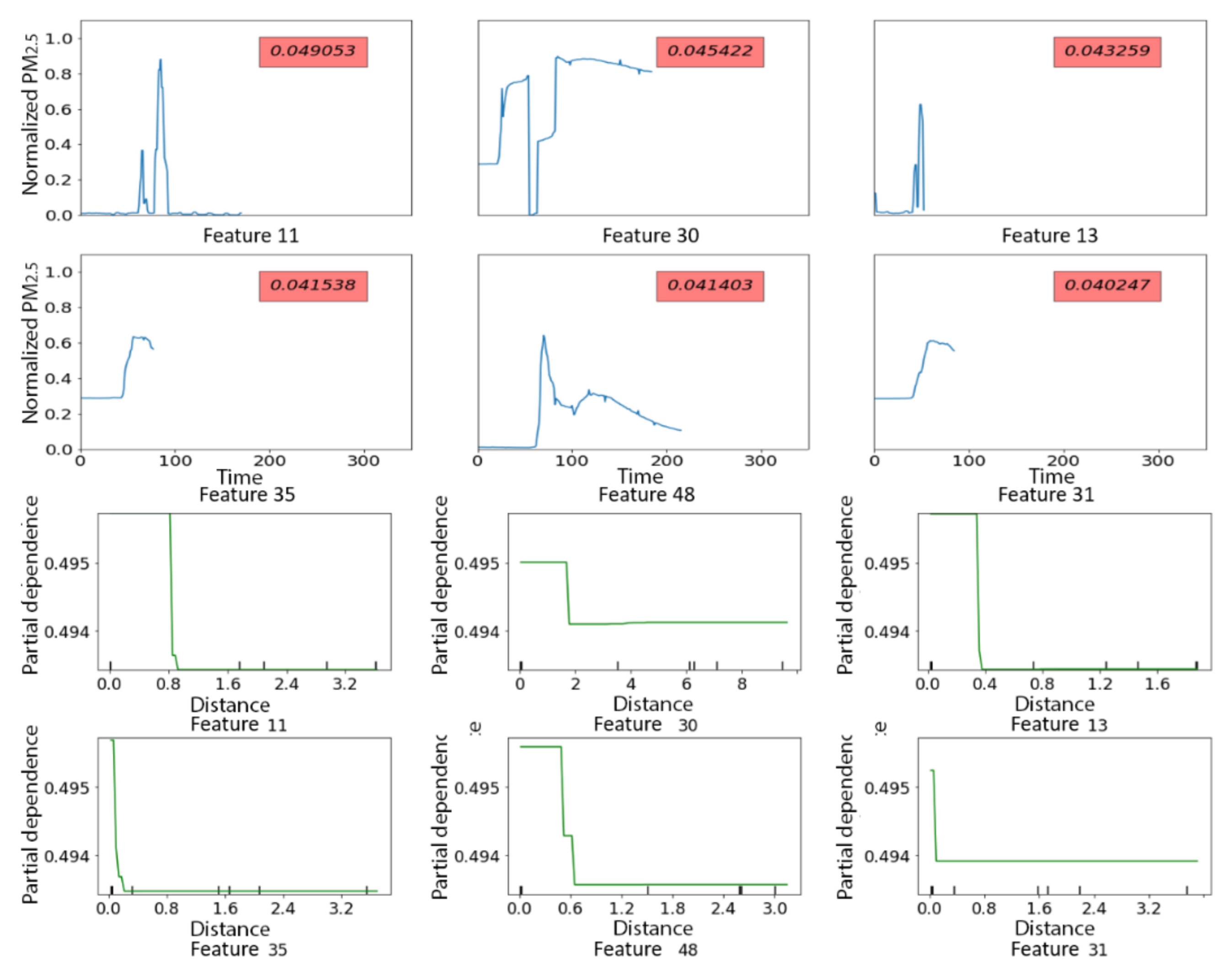
3.2.1. PRISMS: Daily Indoor PM2.5 vs. Outdoor PM2.5 Time Series
3.2.2. PRISMS: Daily Indoor PM2.5 Time Series with and without Rescue Medication Usage
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Hyperparameter Name | Description | Tuning Range |
|---|---|---|
| n_estimators | Number of gradient boosted trees. Equivalent to number of boosting rounds. | Randomly picked from 150 to 500 in each run. |
| learning_rate | Boosting learning rate. | Uniformly picked from 0.01 to 0.07 in each run. |
| subsample | Subsample ratio of the training instance. | Uniformly picked from 0.3 to 0.7 in each run. |
| max_depth | Maximum tree depth for base learners. | 3;4;5;6;7;8;9. |
| colsample_bytree | Subsample ratio of columns when constructing each tree. | Uniformly picked from 0.45 to 0.5 in each run. |
| min_child_weight | Minimum sum of instance weights needed in a child. | 1;2;3. |
| scale_pos_weight | Balancing of positive and negative weights. | 1 for Section 3.2.1; 12.05 for Section 3.2.2. |
| Abbreviation | Description |
|---|---|
| TSS | Time series shapelets |
| W-TSS | Wavelet-based time series shapelets discovery algorithms |
| PRISMS | Pediatric Research Using Integrated Sensor Monitoring Systems |
| TRACE | A Synthetic dataset from University of California Riverside Time Series Archive |
| Xgboost | An implementation of gradient boosted decision trees named eXtreme Gradient Boosting |
| SAX | Symbolic aggregate approximation |
| IDP | Important data points |

References
- Jeong, Y.-S.; Jeong, M.K.; Omitaomu, O.A. Weighted dynamic time warping for time series classification. Pattern Recognit. 2011, 44, 2231–2240. [Google Scholar] [CrossRef]
- Bagnall, A.; Lines, J.; Bostrom, A.; Large, J.; Keogh, E. The great time series classification bake off: A review and experimental evaluation of recent algorithmic advances. Data Min. Knowl. Discov. 2017, 31, 606–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, L.X.; Keogh, E. Time Series Shapelets: A New Primitive for Data Mining. In Proceedings of the 15th ACM SIGKDD Conference on Knowledge Discovery and Data Mining (KDD09), Paris, France, 28 June–1 July 2009; pp. 947–955. [Google Scholar]
- Hills, J.; Lines, J.; Baranauskas, E.; Mapp, J.; Bagnall, A. Classification of time series by shapelet transformation. Data Min. Knowl. Discov. 2014, 28, 851–881. [Google Scholar] [CrossRef] [Green Version]
- Yoon, H.-J.; Xu, S.; Tourassi, G. Predicting Lung Cancer Incidence from Air Pollution Exposures Using Shapelet-based Time Series Analysis. In Proceedings of the 2018 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), Las Vegas, NV, USA, 24–27 February 2016; Volume 2016, pp. 565–568. [Google Scholar]
- Bock, C.; Gumbsch, T.; Moor, M.; Rieck, B.; Roqueiro, D.; Borgwardt, K.M. Association mapping in biomedical time series via statistically significant shapelet mining. Bioinformatics 2018, 34, i438–i446. [Google Scholar] [CrossRef] [Green Version]
- Gumbsch, T.; Bock, C.; Moor, M.; Rieck, B.; Borgwardt, K. Enhancing statistical power in temporal biomarker discovery through representative shapelet mining. Bioinformatics 2020, 36, i840–i848. [Google Scholar] [CrossRef] [PubMed]
- Rakthanmanon, T.; Keogh, E. Fast Shapelets: A Scalable Algorithm for Discovering Time Series Shapelets. In Proceedings of the 2013 SIAM International Conference on Data Mining, Austin, TX, USA, 2–4 May 2013; pp. 668–676. [Google Scholar]
- Ji, C.; Zhao, C.; Pan, L.; Liu, S.; Yang, C.; Wu, L. A Fast Shapelet Discovery Algorithm Based on Important Data Points. Int. J. Web Serv. Res. 2017, 14, 67–80. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, A.; Nishikawa, T. One-Class Learning Time-Series Shapelets. In Proceedings of the 2018 IEEE International Conference on Big Data, Seattle, WA, USA, 10–13 December 2018; pp. 2365–2372. [Google Scholar]
- Shah, M.; Grabocka, J.; Schilling, N.; Wistuba, M.; Schmidt-Thieme, L. Learning DTW-Shapelets for Time-Series Classification. In Proceedings of the 3rd IKDD Conference on Data Science, Pune, India, 13–16 March 2016. [Google Scholar]
- Fang, Z.; Wang, P.; Wang, W. Efficient Learning Interpretable Shapelets for Accurate Time Series Classification. In Proceedings of the 2018 IEEE 34th International Conference on Data Engineering (ICDE), Paris, France, 16–19 April 2018; pp. 497–508. [Google Scholar]
- Grabocka, J.; Schilling, N.; Wistuba, M.; Schmidt-Thieme, L. Learning time-series shapelets. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; pp. 392–401. [Google Scholar]
- Chen, Y.; Keogh, E.; Hu, B.; Begum, N.; Bagnall, A.; Mueen, A.; Batista, G. The UCR Time Series Classification Archive. 2015. Available online: https://www.cs.ucr.edu/~eamonn/time_series_data_2018/ (accessed on 21 May 2021).
- Christensen, O. An Introduction to Wavelet Analysis. In Functions, Spaces, and Expansions: Mathematical Tools in Physics and Engineering; Springer: Boston, MA, USA, 2010; pp. 159–180. [Google Scholar] [CrossRef]
- Addison, P.S. Wavelet transforms and the ECG: A review. Physiol. Meas. 2005, 26, R155–R199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. arXiv 2016, arXiv:1603.02754. [Google Scholar]
- Roverso, D. Multivariate temporal classification by windowed wavelet decomposition and recurrent neural networks. In In Proceedings of the International Topical Meeting on Nuclear Plant Instrumentation, Controls, and Human-Machine Interface Technologies (NPIC&HMIT 2000), Washington, DC, USA, 13–17 November 2000. [Google Scholar]
- Vercellino, R.J.; Sleeth, D.K.; Handy, R.G.; Min, K.T.; Collingwood, S.C. Laboratory evaluation of a low-cost, real-time, aerosol multi-sensor. J. Occup. Environ. Hyg. 2018, 15, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Tzortzis, G.; Likas, A. The global kernel k-means clustering algorithm. In Proceedings of the 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), Hong Kong, China, 1–8 June 2008; pp. 1977–1984. [Google Scholar]
- Goutte, C.; Tofta, P.; Rostrup, E.; Nielsen, F.; Hansen, L.K. On Clustering fMRI Time Series. NeuroImage 1999, 9, 298–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghaderpour, E.; Pagiatakis, S.; Hassan, Q. A Survey on Change Detection and Time Series Analysis with Applications. Appl. Sci. 2021, 11, 6141. [Google Scholar] [CrossRef]
- Ghalwash, M.F.; Radosavljević, V.; Obradovic, Z. Extraction of Interpretable Multivariate Patterns for Early Diagnostics. In Proceedings of the 2013 IEEE 13th International Conference on Data Mining, Dallas, TX, USA, 7–10 December 2013; pp. 201–210. [Google Scholar]

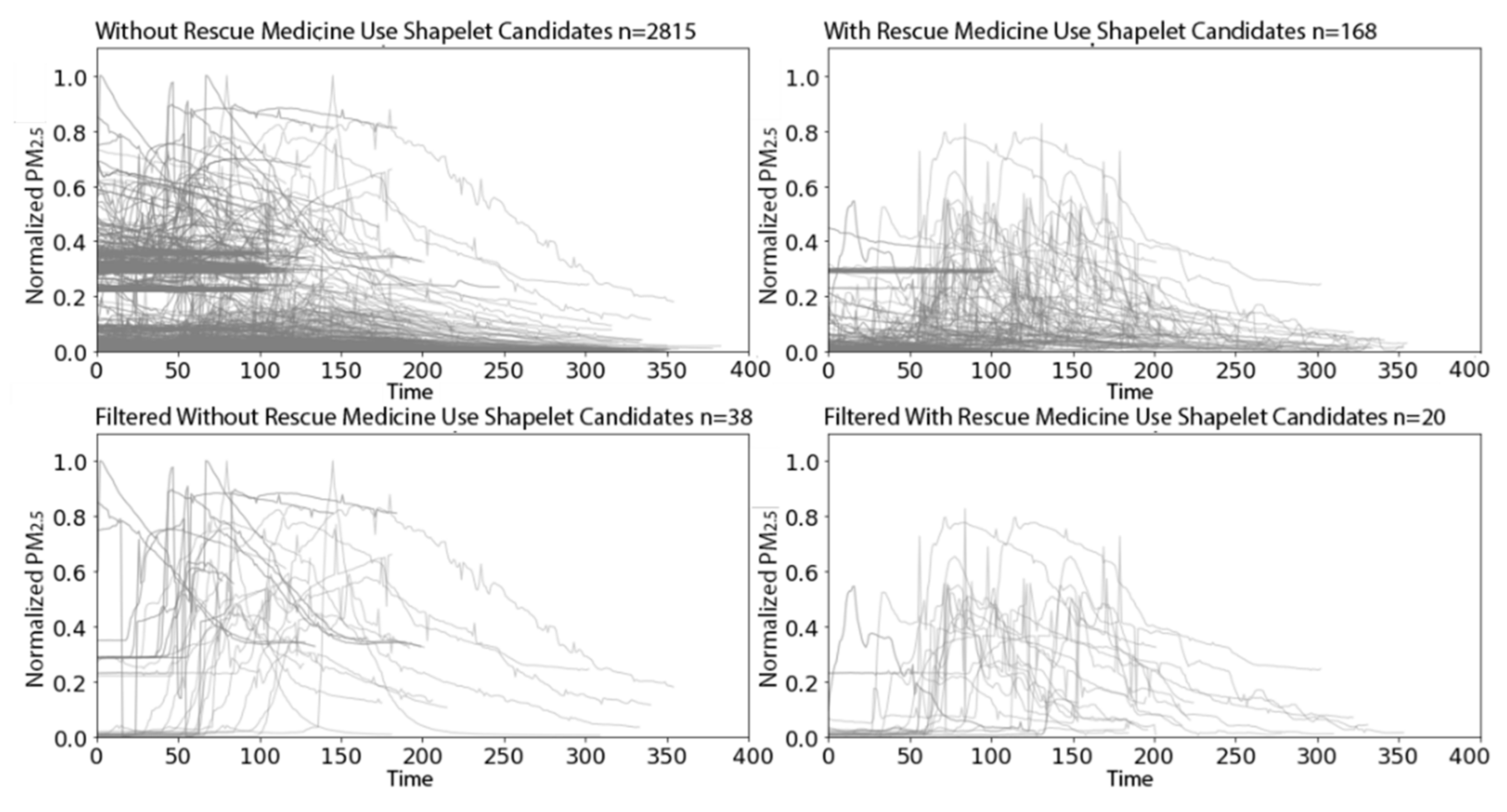
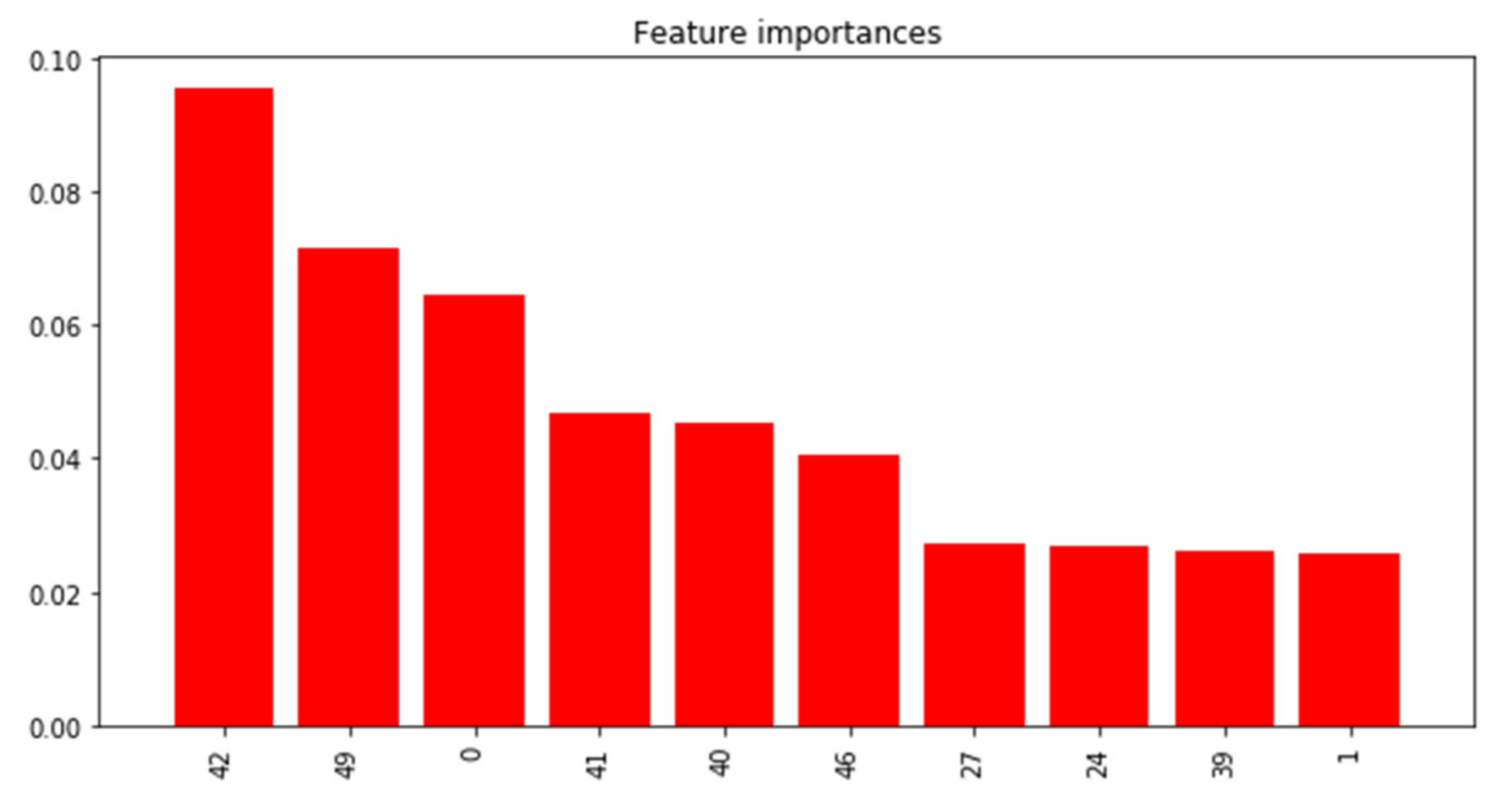
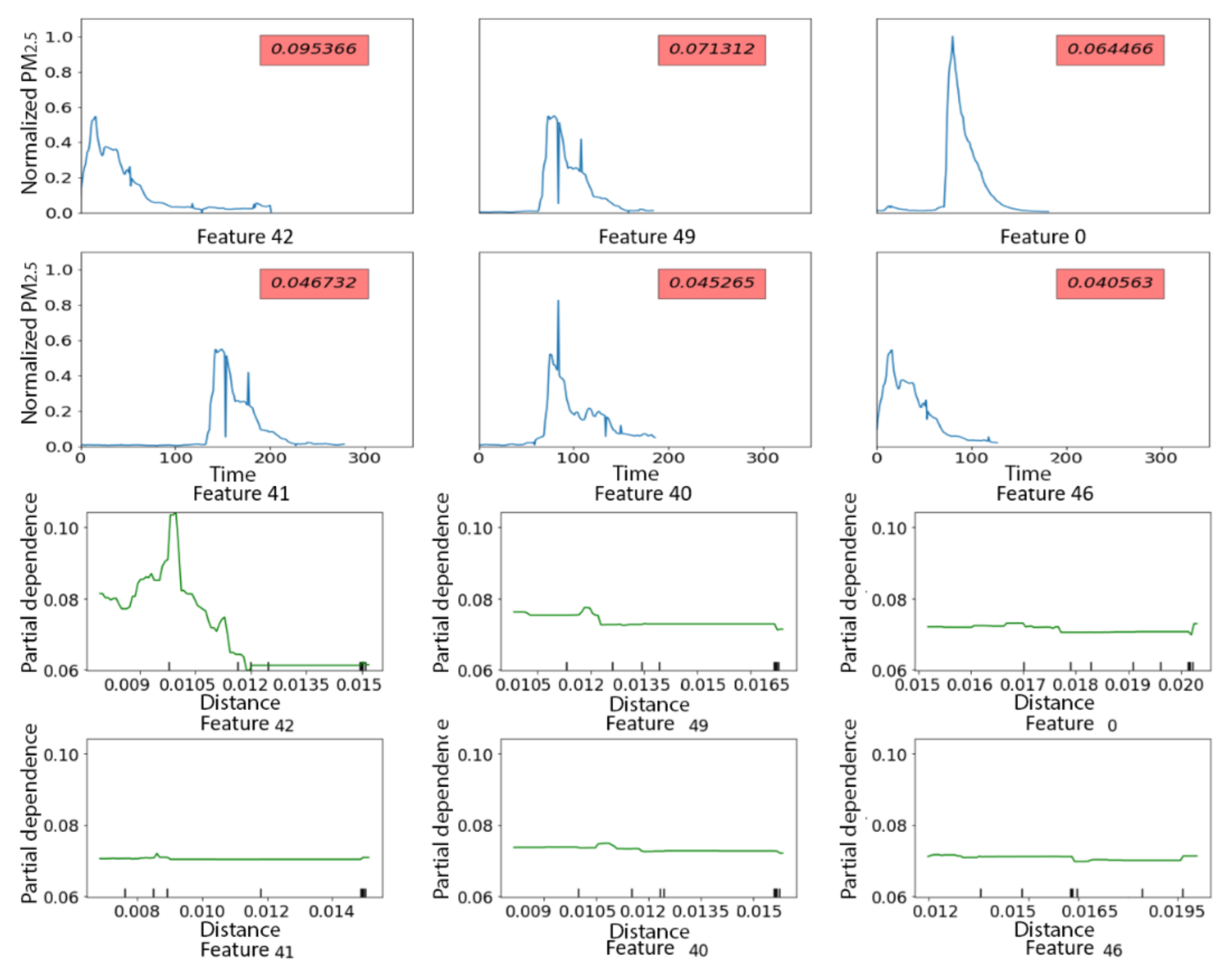
| Test N = 100 | True Class 1 | True Class 2 | True Class 3 | True Class 4 | Total |
|---|---|---|---|---|---|
| Predicted class 1 | 24 | 0 | 0 | 0 | 24 |
| Predicted class 2 | 0 | 29 | 0 | 0 | 29 |
| Predicted class 3 | 0 | 0 | 28 | 0 | 28 |
| Predicted class 4 | 0 | 0 | 0 | 19 | 19 |
| Total | 24 | 29 | 28 | 19 | F1 = 1 |
| Total Days = 544 | True Indoor PM2.5 | True Outdoor PM2.5 | Total |
|---|---|---|---|
| Predicted Indoor PM2.5 | 266 | 0 | 266 |
| Predicted Outdoor PM2.5 | 0 | 278 | 278 |
| Total | 266 | 278 | F1 = 1 |
| Total Days = 272 | True Use Days | True no Use Days | Total |
|---|---|---|---|
| Predicted use days | 4 | 15 | 19 |
| Predicted no-use days | 8 | 245 | 253 |
| Total | 12 | 260 | F1 = 0.26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, K.; Deng, H.; Morrison, J.; Habre, R.; Franklin, M.; Chiang, Y.-Y.; Sward, K.; Gilliland, F.D.; Ambite, J.L.; Eckel, S.P. W-TSS: A Wavelet-Based Algorithm for Discovering Time Series Shapelets. Sensors 2021, 21, 5801. https://doi.org/10.3390/s21175801
Li K, Deng H, Morrison J, Habre R, Franklin M, Chiang Y-Y, Sward K, Gilliland FD, Ambite JL, Eckel SP. W-TSS: A Wavelet-Based Algorithm for Discovering Time Series Shapelets. Sensors. 2021; 21(17):5801. https://doi.org/10.3390/s21175801
Chicago/Turabian StyleLi, Kenan, Huiyu Deng, John Morrison, Rima Habre, Meredith Franklin, Yao-Yi Chiang, Katherine Sward, Frank D. Gilliland, José Luis Ambite, and Sandrah P. Eckel. 2021. "W-TSS: A Wavelet-Based Algorithm for Discovering Time Series Shapelets" Sensors 21, no. 17: 5801. https://doi.org/10.3390/s21175801
APA StyleLi, K., Deng, H., Morrison, J., Habre, R., Franklin, M., Chiang, Y.-Y., Sward, K., Gilliland, F. D., Ambite, J. L., & Eckel, S. P. (2021). W-TSS: A Wavelet-Based Algorithm for Discovering Time Series Shapelets. Sensors, 21(17), 5801. https://doi.org/10.3390/s21175801






