Optical Technologies for the Improvement of Skin Cancer Diagnosis: A Review
Abstract
1. Introduction
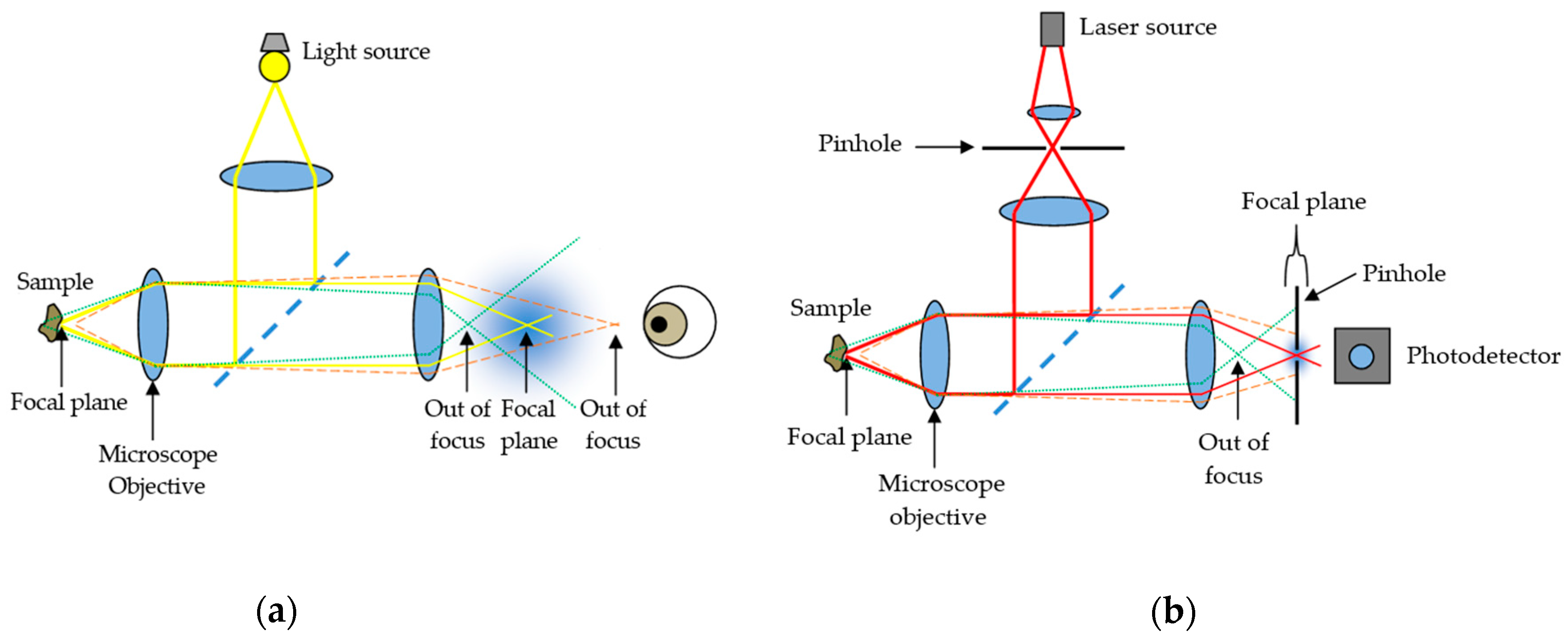
2. Confocal Microscopy
2.1. Reflectance CLSM
2.2. Fluorescence CLSM
2.3. Contributions of CLSM Imaging
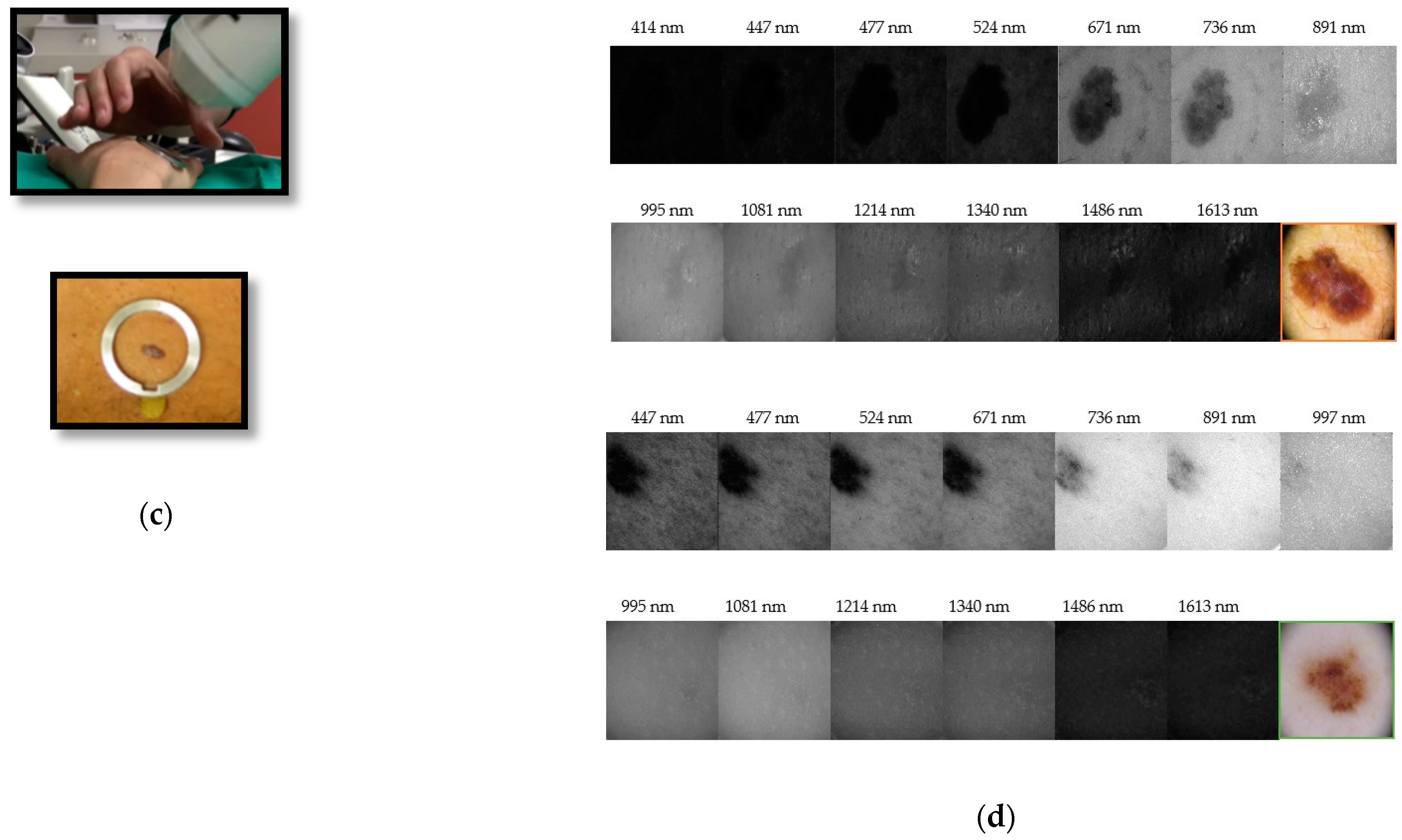
3. Multispectral Imaging
3.1. Area Scanning MS Systems
3.2. Snapshot MS Systems
3.3. Contributions of MS Imaging
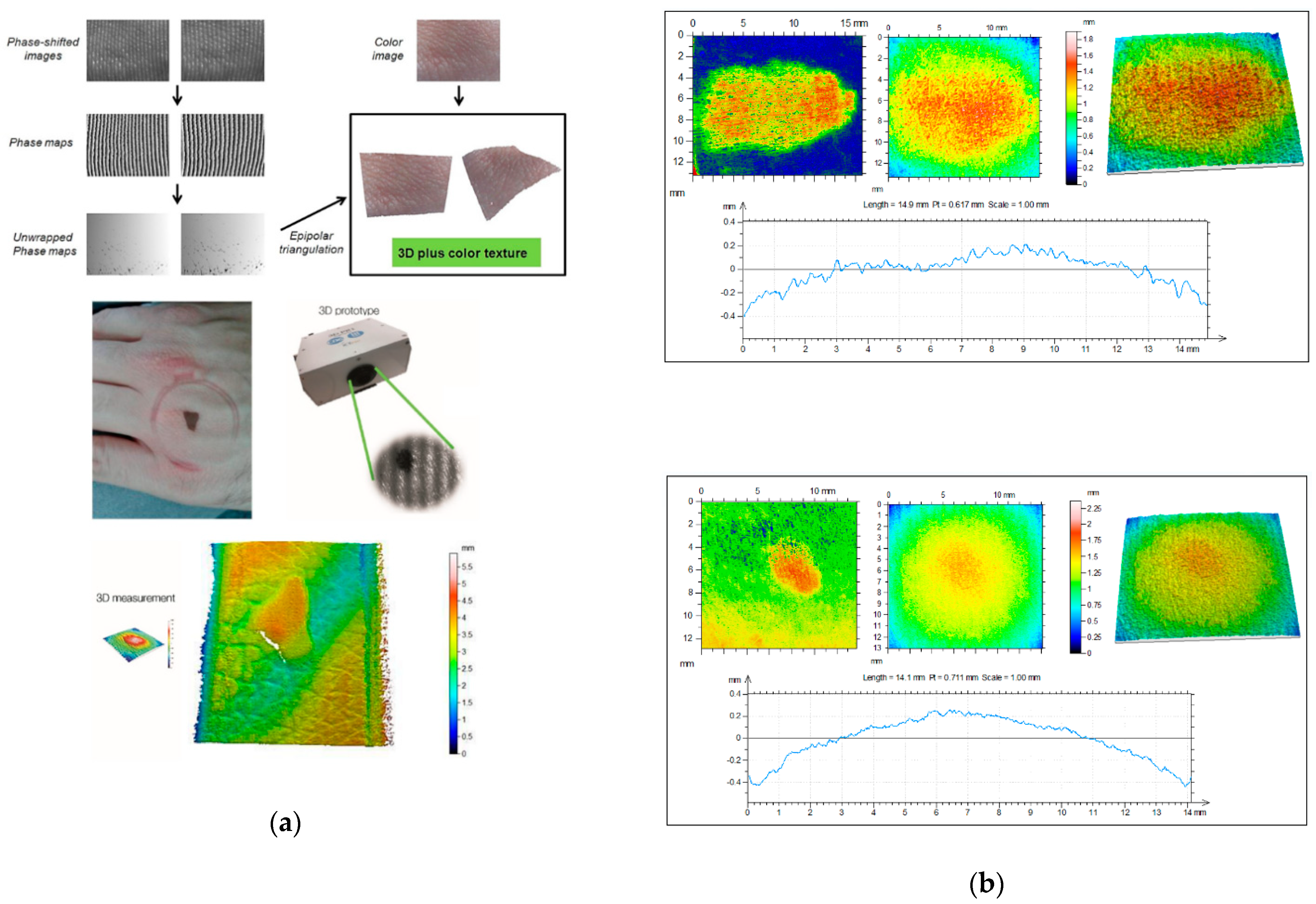
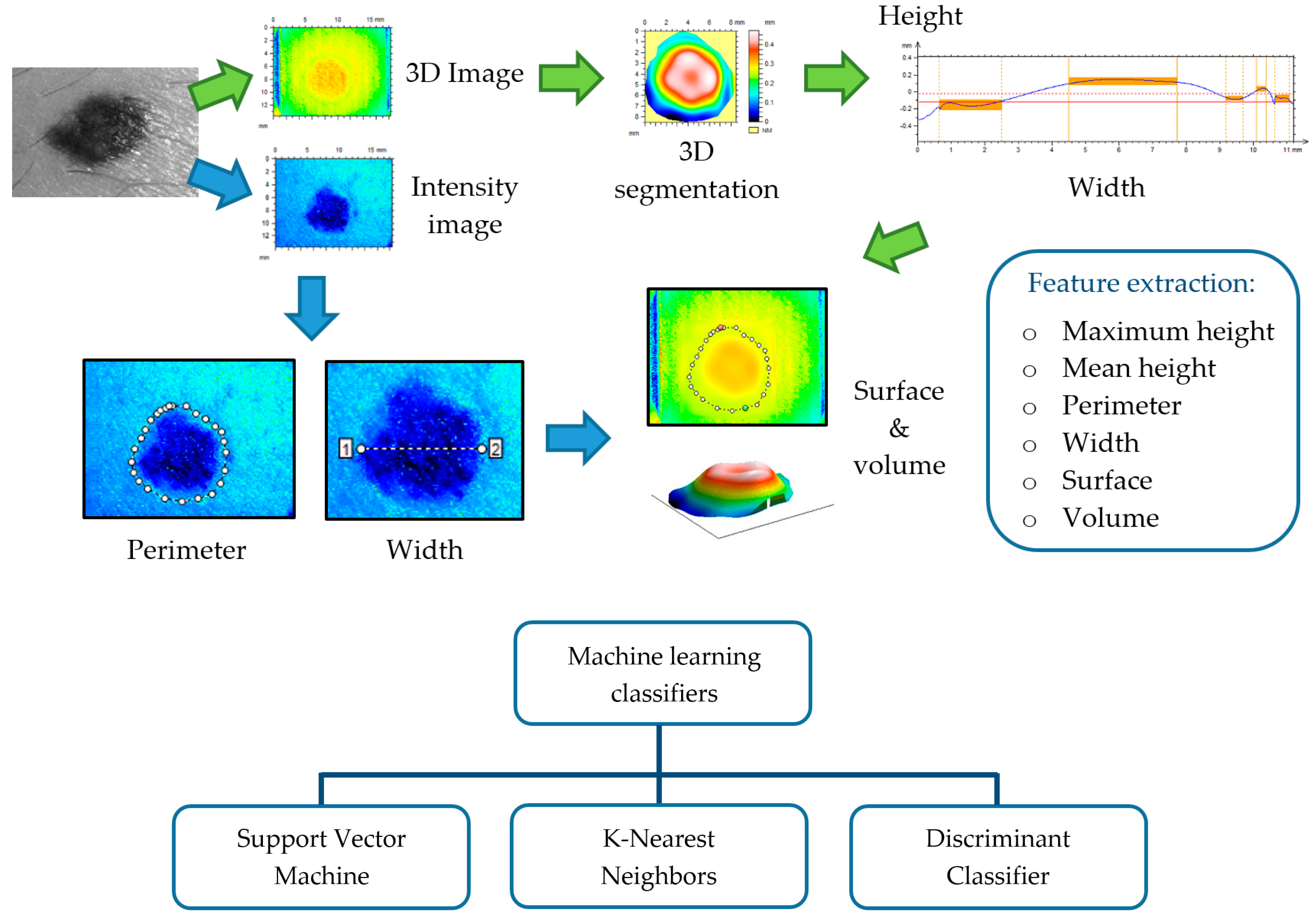
4. Three-Dimensional Topography
4.1. Replica-Based Methods for Skin 3D Imaging
4.1.1. Microtopography
4.1.2. Optical Profilometry
4.2. 3D Systems for In Vivo Skin Analysis
4.3. Contributions of 3D Topography
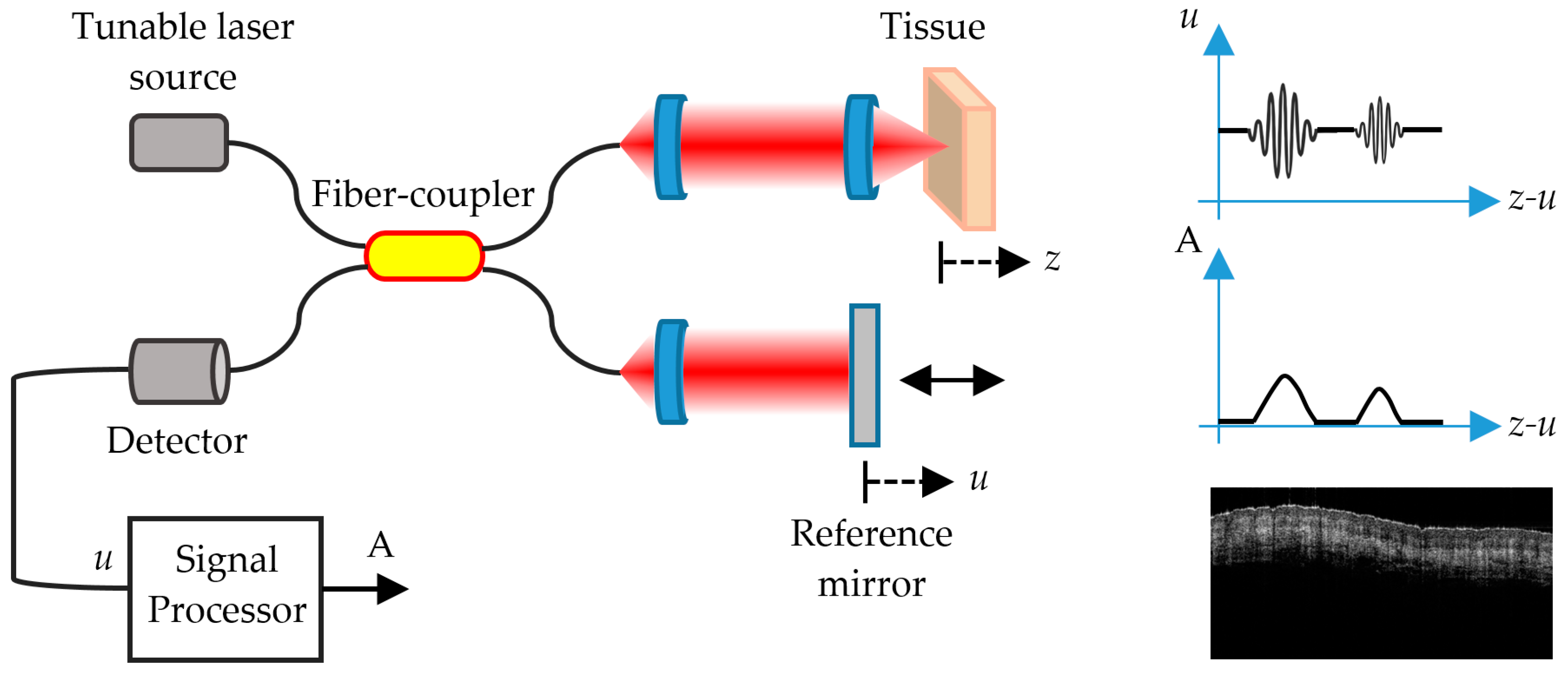
5. Optical Coherence Tomography
5.1. FD-OCT
5.2. Contributions of OCT
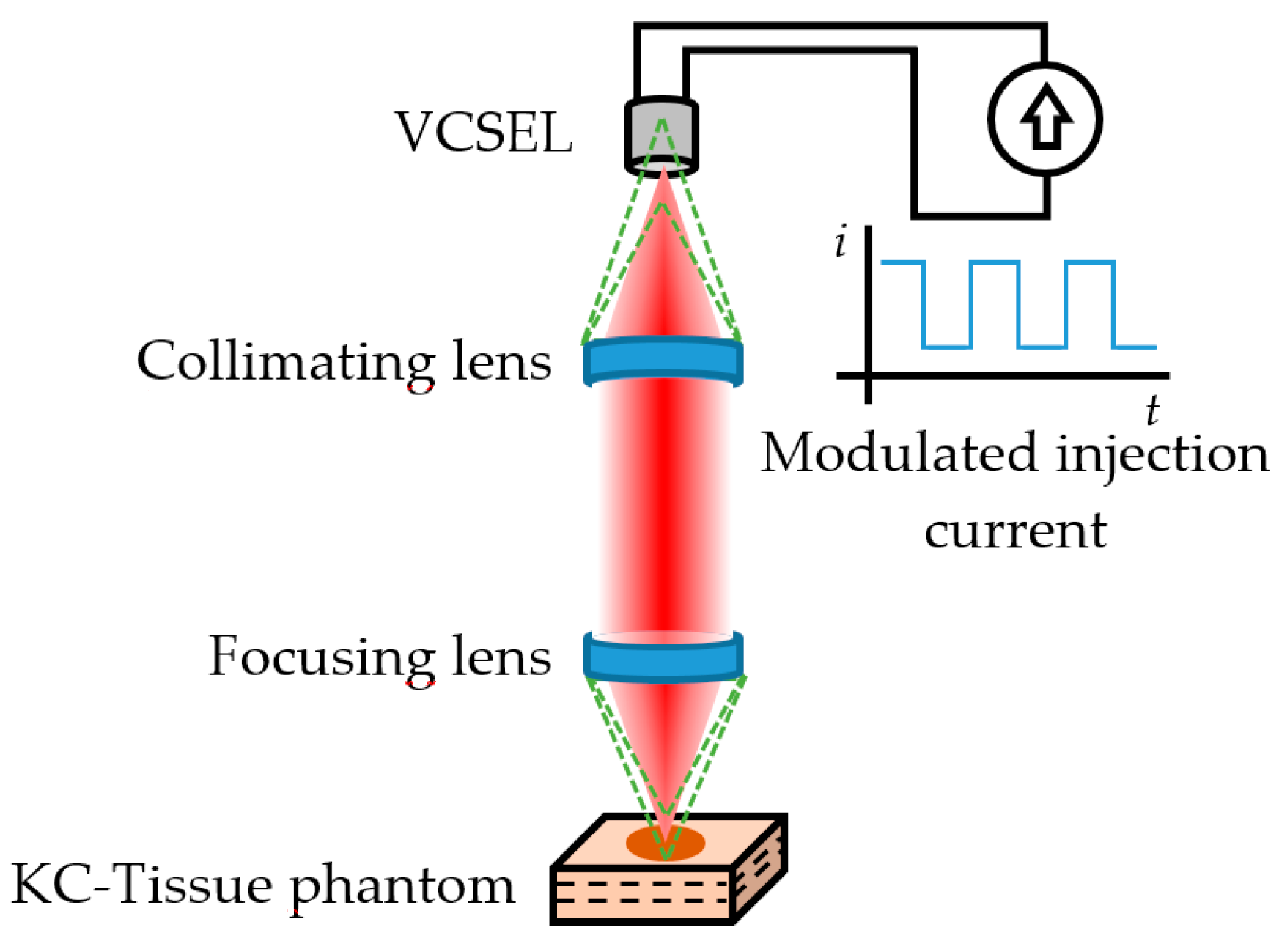
6. Self-Mixing Interferometry
6.1. SMI Imaging
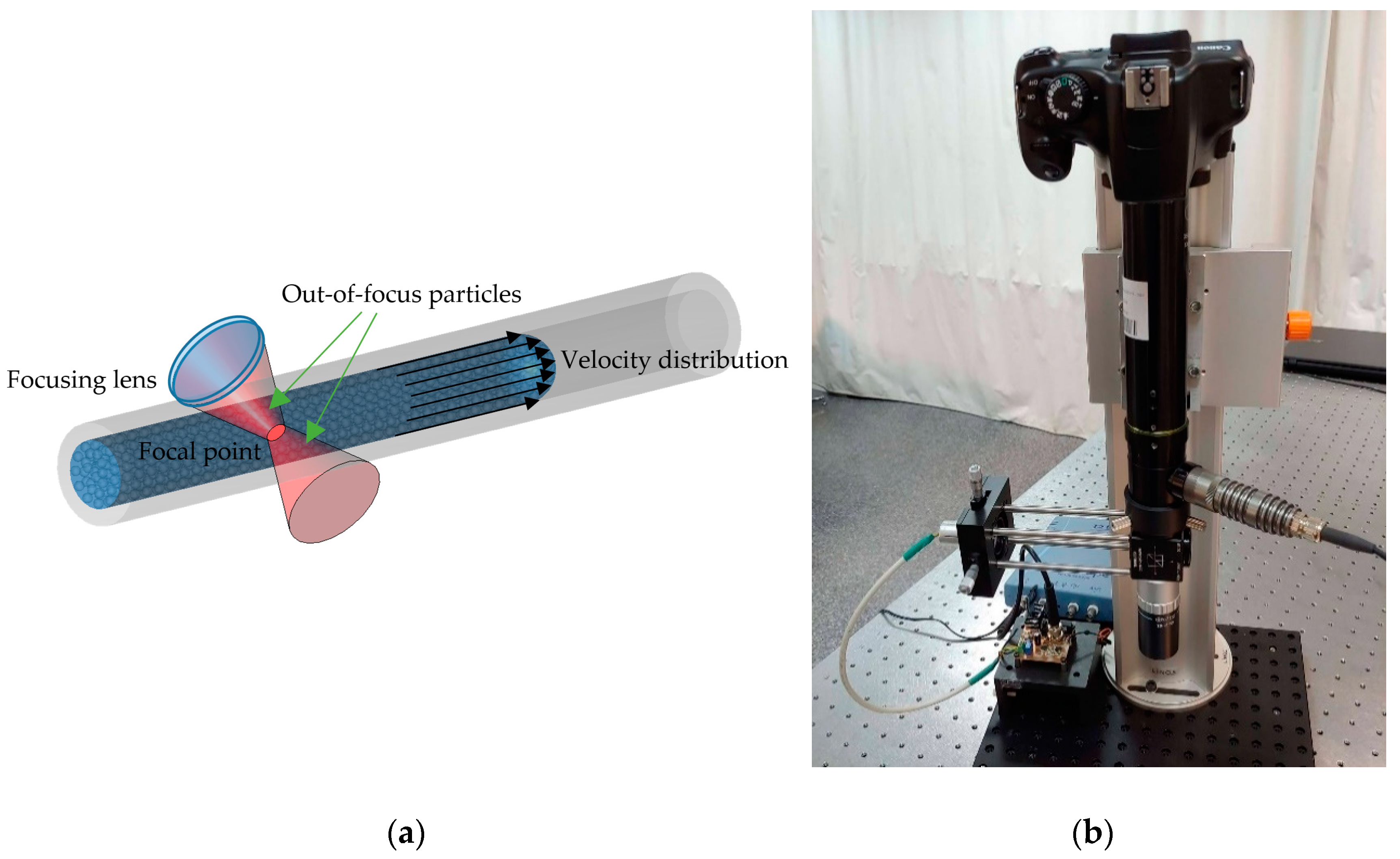
6.2. SMI Flow Sensing
6.3. SMI Flow Cytometry
6.4. Possible Contributions of SMI
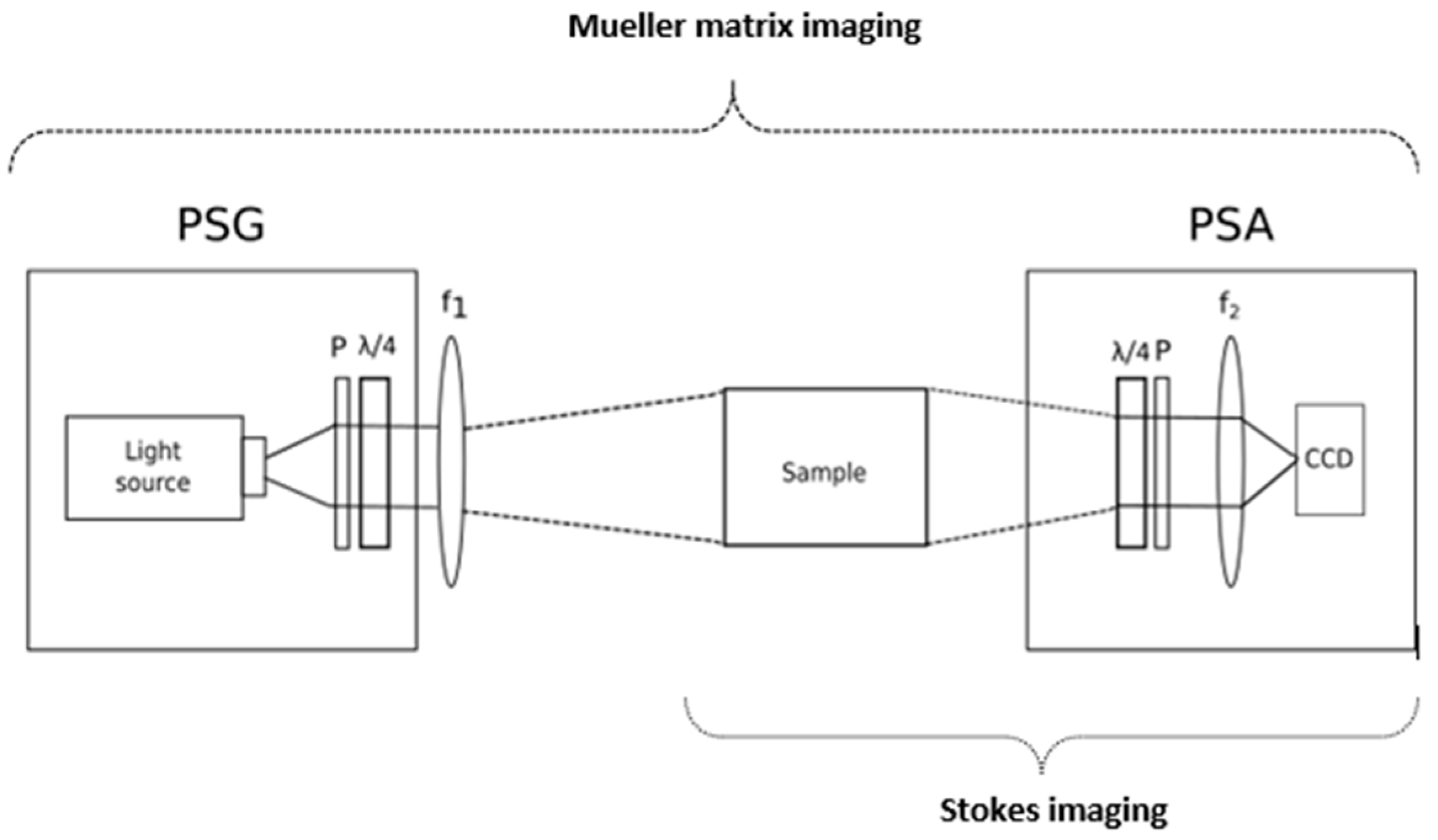
7. Polarimetric Imaging
7.1. Stokes Imaging
7.2. MMI
7.3. Contributions of Polarimetric Imaging
8. Learning Algorithms for Skin Cancer Diagnosis
8.1. Machine Learning
8.2. Deep Learning
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The Skin Cancer Foundation. Available online: https://www.skincancer.org/ (accessed on 19 October 2020).
- Wolff, K.; Allen, J.R. Fitzpatrick’s Color Atlas and Synopsis of Clinical Dermatology; McGraw-Hill Professional: New York, NY, USA, 2009; ISBN 007179302X. [Google Scholar]
- American Cancer Society. Available online: https://www.cancer.org (accessed on 19 October 2020).
- World Health Organization. Available online: https://www.who.int/en/ (accessed on 19 October 2020).
- Gadeliya Goodson, A.; Grossman, D. Strategies for early melanoma detection: Approaches to the patient with nevi. J. Am. Acad. Dermatol. 2009, 60, 719–735. [Google Scholar] [CrossRef] [PubMed]
- Guy, G.P.; Ekwueme, D.U.; Tangka, F.K.; Richardson, L.C. Melanoma Treatment Costs. Am. J. Prev. Med. 2012, 43, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Cancer Research, UK. Available online: http://www.cancerresearchuk.org (accessed on 19 October 2020).
- Ahlgrimm-Siess, V.; Laimer, M.; Rabinovitz, H.S.; Oliviero, M.; Hofmann-Wellenhof, R.; Marghoob, A.A.; Scope, A. Confocal Microscopy in Skin Cancer. Curr. Derm. Rep. 2018, 7, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Fellers, T.J.; Davidson, M.W. OLYMPUS Microscopy Resource Center. Available online: https://www.olympus-lifescience.com/es/microscope-resource/primer/techniques/confocal/confocalintro/ (accessed on 24 October 2020).
- Star, P.; Guitera, P. Advances in the use of reflectance confocal microscopy in melanoma. Melanoma Manag. 2018, 5, 1. [Google Scholar]
- Longo, C.; Ragazzi, M.; Rajadhyaksha, M.; Nehal, K.; Bennassar, A.; Pellacani, G.; Malvehy Guilera, J. In Vivo and Ex Vivo Confocal Microscopy for Dermatologic and Mohs Surgeons. Dermatol. Clin. 2016, 34, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, M.; Lange-Asschenfeldt, S. In vivo confocal microscopy in dermatology: From research to clinical application. J. Biomed. Opt. 2013, 18, 061212. [Google Scholar] [CrossRef] [PubMed]
- Biel, S.S.; Wepf, R.; Wessel, S. Confocal microscopy of skin in vitro and ex vivo. In Bioengineering of the Skin: Skin Imaging and Analysis; Wilhelm, K.P., Elsner, P., Berardesca, E., Maibach, H.I., Eds.; CRC Press Taylor & Francis Group: Boca Raton, FL, USA, 2006; pp. 149–164. ISBN 9781420005516. [Google Scholar]
- Pellacani, G.; Cesinaro, A.M.; Seidenari, S. In vivo assessment of melanocytic nests in nevi and melanomas by reflectance confocal microscopy. Mod. Pathol. 2005, 18, 469–474. [Google Scholar] [CrossRef]
- Guitera, P.; Pellacani, G.; Longo, C.; Seidenari, S.; Avramidis, M.; Menzies, S.W. In Vivo Reflectance Confocal Microscopy Enhances Secondary Evaluation of Melanocytic Lesions. J. Investig. Dermatol. 2009, 129, 131–138. [Google Scholar] [CrossRef]
- Guitera, P.; Menzies, S.W.; Longo, C.; Cesinaro, A.M.; Scolyer, R.A.; Pellacani, G. In Vivo Confocal Microscopy for Diagnosis of Melanoma and Basal Cell Carcinoma Using a Two-Step Method: Analysis of 710 Consecutive Clinically Equivocal Cases. J. Investig. Dermatol. 2012, 132, 2386–2394. [Google Scholar] [CrossRef]
- Segura, S.; Puig, S.; Carrera, C.; Palou, J.; Malvehy, J. Development of a two-step method for the diagnosis of melanoma by reflectance confocal microscopy. J. Am. Acad. Dermatol. 2009, 61, 216–229. [Google Scholar] [CrossRef]
- Ulrich, M.; Maltusch, A.; Rius-Diaz, F.; Röwert-Huber, J.; González, S.; Sterry, W.; Stockfleth, E.; Astner, S. Clinical applicability of in vivo reflectance confocal microscopy for the diagnosis of actinic keratoses. Dermatol. Surg. 2008, 34, 610–619. [Google Scholar] [PubMed]
- Horn, M.; Gerger, A.; Ahlgrimm-Siess, V.; Weger, W.; Koller, S.; Kerl, H.; Samonigg, H.; Smolle, J.; Hofmann-Wellenhof, R. Discrimination of actinic keratoses from normal skin with reflectance mode confocal microscopy. Dermatol. Surg. 2008, 34, 620–625. [Google Scholar] [PubMed]
- Gareau, D.S.; Li, Y.; Huang, B.; Eastman, Z.; Nehal, K.S.; Rajadhyaksha, M. Confocal mosaicing microscopy in Mohs skin excisions: Feasibility of rapid surgical pathology. J. Biomed. Opt. 2008, 13, 054001. [Google Scholar] [CrossRef] [PubMed]
- Gareau, D.S.; Karen, J.K.; Dusza, S.W.; Tudisco, M.; Nehal, K.S.; Rajadhyaksha, M. Sensitivity and specificity for detecting basal cell carcinomas in Mohs excisions with confocal fluorescence mosaicing microscopy. J. Biomed. Opt. 2009, 14, 034012. [Google Scholar] [CrossRef]
- Abeytunge, S.; Li, Y.; Larson, B.; Toledo-Crow, R.; Rajadhyaksha, M. Rapid confocal imaging of large areas of excised tissue with strip mosaicing. J. Biomed. Opt. 2011, 16, 050504. [Google Scholar] [CrossRef]
- Li, Q.; He, X.; Wang, Y.; Liu, H.; Xu, D.; Guo, F. Review of spectral imaging technology in biomedical engineering: Achievements and challenges. J. Biomed. Opt. 2013, 18, 100901. [Google Scholar] [CrossRef]
- Grahn, H.F.; Geladi, P. Techniques and Applications of Hyperspectral Image Analysis; Grahn, H.F., Geladi, P., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2007; ISBN 9780470010884. [Google Scholar]
- Kang, H.R. Computational Color Technology; Society of Photo Optical, Ed.; SPIE: Bellingham, WA, USA, 2006; ISBN 9780819461193. [Google Scholar]
- Godoy, S.E.; Ramirez, D.A.; Myers, S.A.; von Winckel, G.; Krishna, S.; Berwick, M.; Padilla, R.S.; Sen, P.; Krishna, S. Dynamic infrared imaging for skin cancer screening. Infrared Phys. Technol. 2015, 70, 147–152. [Google Scholar] [CrossRef]
- Emery, J.D.; Hunter, J.; Hall, P.N.; Watson, A.J.; Moncrieff, M.; Walter, F.M. Accuracy of SIAscopy for pigmented skin lesions encountered in primary care: Development and validation of a new diagnostic algorithm. BMC Dermatol. 2010, 10, 9. [Google Scholar] [CrossRef]
- Delpueyo, X.; Vilaseca, M.; Royo, S.; Ares, M.; Rey-Barroso, L.; Sanabria, F.; Puig, S.; Malvehy, J.; Pellacani, G.; Noguero, F.; et al. Multispectral imaging system based on light-emitting diodes for the detection of melanomas and basal cell carcinomas: A pilot study. J. Biomed. Opt. 2017, 22, 065006. [Google Scholar] [CrossRef][Green Version]
- Monheit, G.; Cognetta, A.B.; Ferris, L.; Rabinovitz, H.; Gross, K.; Martini, M.; Grichnik, J.M.; Mihm, M.; Prieto, V.G.; Googe, P.; et al. The performance of MelaFind: A prospective multicenter study. Arch. Dermatol. 2011, 147, 188–194. [Google Scholar] [CrossRef]
- Bekina, A.; Diebele, I.; Rubins, U.; Zaharans, J.; Derjabo, A.; Spigulis, J. Multispectral assessment of skin malformations using a modified video-microscope. Latv. J. Phys. Tech. Sci. 2012, 49, 4–8. [Google Scholar]
- Kapsokalyvas, D.; Bruscino, N.; Alfieri, D.; de Giorgi, V.; Cannarozzo, G.; Cicchi, R.; Massi, D.; Pimpinelli, N.; Pavone, F.S. Spectral morphological analysis of skin lesions with a polarization multispectral dermoscope. Opt. Express 2013, 21, 4826. [Google Scholar] [CrossRef] [PubMed]
- Stamnes, J.J.; Ryzhikov, G.; Biryulina, M.; Hamre, B.; Zhao, L.; Stamnes, K. Optical detection and monitoring of pigmented skin lesions. Biomed. Opt. Express 2017, 8, 2946. [Google Scholar] [CrossRef] [PubMed]
- Kuzmina, I.; Diebele, I.; Jakovels, D.; Spigulis, J.; Valeine, L.; Kapostinsh, J.; Berzina, A. Towards noncontact skin melanoma selection by multispectral imaging analysis. J. Biomed. Opt. 2011, 16, 060502. [Google Scholar] [CrossRef] [PubMed]
- Jolivot, R.; Benezeth, Y.; Marzani, F. Skin parameter map retrieval from a dedicated multispectral imaging system applied to dermatology/cosmetology. Int. J. Biomed. Imaging 2013, 2013, 978289. [Google Scholar] [CrossRef] [PubMed]
- Farina, B.; Bartoli, C.; Bono, A.; Colombo, A.; Lualdi, M.; Tragni, G.; Marchesini, R. Multispectral imaging approach in the diagnosis of cutaneous melanoma: Potentiality and limits. Phys. Med. Biol. 2000, 45, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Tomatis, S.; Bono, A.; Bartoli, C.; Carrara, M.; Lualdi, M.; Tragni, G.; Marchesini, R. Automated melanoma detection: Multispectral imaging and neural network approach for classification. Med. Phys. 2003, 30, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Fioravanti, V.; Brandhoff, L.; van den Driesche, S.; Breiteneder, H.; Kitzwögerer, M.; Hafner, C.; Vellekoop, M. An Infrared Absorbance Sensor for the Detection of Melanoma in Skin Biopsies. Sensors 2016, 16, 1659. [Google Scholar] [CrossRef]
- Carrara, M.; Bono, A.; Bartoli, C.; Colombo, A.; Lualdi, M.; Moglia, D.; Santoro, N.; Tolomio, E.; Tomatis, S.; Tragni, G.; et al. Multispectral imaging and artificial neural network: Mimicking the management decision of the clinician facing pigmented skin lesions. Phys. Med. Biol. 2007, 52, 2599–2613. [Google Scholar] [CrossRef]
- Diebele, I.; Kuzmina, I.; Lihachev, A.; Kapostinsh, J.; Derjabo, A.; Valeine, L.; Spigulis, J. Clinical evaluation of melanomas and common nevi by spectral imaging. Biomed. Opt. Express 2012, 3, 467. [Google Scholar] [CrossRef]
- Jakovels, D.; Lihacova, I.; Kuzmina, I.; Spigulis, J. Evaluation of skin melanoma in spectral range 450–950 nm using principal component analysis. In Medical Laser Applications and Laser-Tissue Interactions VI; Lilge, L.D., Sroka, R., Eds.; SPIE: Bellingham, WA, USA, 2013; Volume 8803, p. 88030C. [Google Scholar]
- Kim, S.; Cho, D.; Kim, J.; Kim, M.; Youn, S.; Jang, J.E.; Je, M.; Lee, D.H.; Lee, B.; Farkas, D.L.; et al. Smartphone-based multispectral imaging: System development and potential for mobile skin diagnosis. Biomed. Opt. Express 2016, 7, 5294–5307. [Google Scholar] [CrossRef] [PubMed]
- Vasaturo, A.; Di Blasio, S.; Verweij, D.; Blokx, W.A.M.; van Krieken, J.H.; de Vries, I.J.M.; Figdor, C.G. Multispectral imaging for highly accurate analysis of tumour-infiltrating lymphocytes in primary melanoma. Histopathology 2017, 70, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Rey-Barroso, L.; Burgos-Fernández, F.; Delpueyo, X.; Ares, M.; Royo, S.; Malvehy, J.; Puig, S.; Vilaseca, M. Visible and Extended Near-Infrared Multispectral Imaging for Skin Cancer Diagnosis. Sensors 2018, 18, 1441. [Google Scholar] [CrossRef]
- Hashimoto, K. New methods for surface ultrastructure: Comparative studies of scanning electron microscopy, transmission electron microscopy and replica method. Int. J. Dermatol. 1974, 13, 357–381. [Google Scholar] [CrossRef] [PubMed]
- Hocken, R.J.; Creath, K.; Schmit, J.; Wyant, J.C. Optical Metrology of Diffuse Surfaces. In Optical Shop Testing; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006; Volume 1, pp. 756–807. ISBN 9780470135976. [Google Scholar]
- Tchvialeva, L.; Zeng, H.; Markhvida, I.; McLean, I.D.; Lui, H.; Lee, K.T. Skin Roughness Assessment. In New Developments in Biomedical Engineering; Campolo, D., Ed.; InTech: Makati, Philippines, 2010; pp. 341–358. ISBN 978-953-7619-57-2. [Google Scholar]
- Kottner, J.; Schario, M.; Bartels, N.G.; Pantchechnikova, E.; Hillmann, K.; Blume-Peytavi, U. Comparison of two in vivo measurements for skin surface topography. Skin Res. Technol. 2013, 19, 84–90. [Google Scholar] [CrossRef]
- Egawa, M.; Oguri, M.; Kuwahara, T.; Takahashi, M. Effect of exposure of human skin to a dry environment. Skin Res. Technol. 2002, 8, 212–218. [Google Scholar] [CrossRef]
- Setaro, M.; Sparavigna, A. Irregularity skin index (ISI): A tool to evaluate skin surface texture. Skin Res. Technol. 2001, 7, 159–163. [Google Scholar] [CrossRef]
- Humbert, P.G.; Haftek, M.; Creidi, P.; Lapière, C.; Nusgens, B.; Richard, A.; Schmitt, D.; Rougier, A.; Zahouani, H. Topical ascorbic acid on photoaged skin. Clinical, topographical and ultrastructural evaluation: Double-blind study vs. placebo. Exp. Dermatol. 2003, 12, 237–244. [Google Scholar] [CrossRef]
- Connemann, B.J.; Busche, H.; Kreusch, J.; Teichert, H.-M.; Wolff, H.H. Quantitative surface topography as a tool in the differential diagnosis between melanoma and naevus. Skin Res. Technol. 1995, 1, 180–186. [Google Scholar] [CrossRef]
- Kim, E.; Nam, G.W.; Kim, S.; Lee, H.; Moon, S.; Chang, I. Influence of polyol and oil concentration in cosmetic products on skin moisturization and skin surface roughness. Skin Res. Technol. 2007, 13, 417–424. [Google Scholar] [CrossRef]
- Bielfeldt, S.; Buttgereit, P.; Brandt, M.; Springmann, G.; Wilhelm, K.P. Non-invasive evaluation techniques to quantify the efficacy of cosmetic anti-cellulite products. Skin Res. Technol. 2008, 14, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Lévêque, J.L.; Querleux, B. SkinChip®, a new tool for investigating the skin surface in vivo. Skin Res. Technol. 2003, 9, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Seo, Y.K.; Baek, J.H.; Koh, J.S. Comparison between ultrasonography (Dermascan C version 3) and transparency profilometry (Skin Visiometer SV600). Skin Res. Technol. 2008, 14, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Lotay, A.S.; Carvalho, M.T.; Girkin, J.M. Non-invasive assessment of skin roughness through speckle pattern analysis. In Proceedings of the Biomedical Optics 2016, Hollywood, FL, USA, 3–6 April 2016; Optics InfoBase Conference Papers; OSA: Washington, DC, USA, 2016; p. JTu3A.6. [Google Scholar]
- Tchvialeva, L.; Zeng, H.; Markhvida, I.; Dhadwal, G.; McLean, L.; McLean, D.I.; Lui, H.; Lee, T.K. Optical discrimination of surface reflection from volume backscattering in speckle contrast for skin roughness measurements. In Proceedings of the Photonic Therapeutics and Diagnostics V, San Jose, CA, USA, 19 February 2009; Kollias, N., Choi, B., Zeng, H., Malek, R.S., Wong, B.J., Ilgner, J.F.R., Gregory, K.W., Tearney, G.J., Marcu, L., Hirschberg, H., et al., Eds.; SPIE: San Jose, CA, USA, 2009; p. 71610I. [Google Scholar]
- Jaspers, S.; Hopermann, H.; Sauermann, G.; Hoppe, U.; Lunderstädt, R.; Ennen, J. Rapid in vivo measurement of the topography of human skin by active image triangulation using a digital micromirror device. Skin Res. Technol. 1999, 5, 195–207. [Google Scholar] [CrossRef]
- Gorpas, D.S.; Politopoulos, K.; Alexandratou, E.; Yova, D. A binocular machine vision system for non-melanoma skin cancer 3D reconstruction. In Multimodal Biomedical Imaging; Azar, F.S., Metaxas, D.N., Eds.; IEEE: San Jose, CA, USA, 2006; Volume 6081, p. 60810D. [Google Scholar]
- Lee, K.-S.; Zhao, H.; Ibrahim, S.F.; Meemon, N.; Khoudeir, L.; Rolland, J.P. Three-dimensional imaging of normal skin and nonmelanoma skin cancer with cellular resolution using Gabor domain optical coherence microscopy. J. Biomed. Opt. 2012, 17, 1. [Google Scholar] [CrossRef]
- Lagarde, J.M.; Rouvrais, C.; Black, D.; Diridollou, S.; Gall, Y. Skin topography measurement by interference fringe projection: A technical validation. Skin Res. Technol. 2001, 7, 112–121. [Google Scholar] [CrossRef]
- Lagarde, J.M.; Rouvrais, C.; Black, D. Topography and anisotropy of the skin surface with ageing. Skin Res. Technol. 2005, 11, 110–119. [Google Scholar] [CrossRef]
- Moore, C.J.; Burton, D.R.; Skydan, O.; Sharrock, P.J.; Lalor, M. 3D Body Surface Measurement and Display in Radiotherapy Part I: Technology of Structured Light Surface Sensing. In Proceedings of the International Conference on Medical Information Visualisation—BioMedical Visualisation (MedVis’06), London, UK, 5–7 July 2006; IEEE: London, UK, 2006; pp. 97–102. [Google Scholar]
- Ares, M.; Royo, S.; Vilaseca, M.; Herrera, J.A.; Delpueyo, X.; Sanabria, F. Handheld 3D Scanning System for In-Vivo Imaging of Skin Cancer. In Proceedings of the 5th International Conference on 3D Body Scanning Technologies, Lugano, Switzerland, 21–22 October 2014; Hometrica Consulting-Dr. Nicola D’Apuzzo: Ascona, Switzerland, 2014; pp. 231–236. [Google Scholar]
- Rey-Barroso, L.; Burgos-Fernández, F.J.; Ares, M.; Royo, S.; Puig, S.; Malvehy, J.; Pellacani, G.; Espinar, D.; Sicilia, N.; Ricart, M.V. Morphological study of skin cancer lesions through a 3D scanner based on fringe projection and machine learning. Biomed. Opt. Express 2019, 10, 3404. [Google Scholar] [CrossRef]
- Korn, V.; Surber, C.; Imanidis, G. Skin Surface Topography and Texture Analysis of Sun-Exposed Body Sites in View of Sunscreen Application. Skin Pharmacol. Physiol. 2017, 29, 291–299. [Google Scholar] [CrossRef]
- Kim, S.-W.; Park, H.-G. Moiré topography by slit beam scanning. Appl. Opt. 1992, 31, 6157–6161. [Google Scholar] [CrossRef]
- Weissman, J.; Hancewicz, T.; Kaplan, P. Optical coherence tomography of skin for measurement of epidermal thickness by shapelet-based image analysis. Opt. Express 2004, 12, 5760. [Google Scholar] [CrossRef] [PubMed]
- Villiger, M.L.; Bouma, B.E. Center for Biomedical Oct Research & Translation-Working Principle of OCT. Available online: https://octresearch.org/the-center/oct-and-biomedical-optics/working-principle-of-oct/ (accessed on 24 October 2020).
- Mogensen, M.; Thrane, L.; Joergensen, T.M.M.; Andersen, P.E.; Jemec, G.B.E. Optical coherence tomography for imaging of skin and skin diseases. Semin. Cutan. Med. Surg. 2009, 28, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.; Wang, K.; Markowitz, O. Optical Coherence Tomography in the Diagnosis of Skin Cancer. Dermatol. Clin. 2017, 35, 465–488. [Google Scholar] [CrossRef] [PubMed]
- Ferrante di Ruffano, L.; Dinnes, J.; Deeks, J.J.; Chuchu, N.; Bayliss, S.E.; Davenport, C.; Takwoingi, Y.; Godfrey, K.; O’Sullivan, C.; Matin, R.N.; et al. Optical coherence tomography for diagnosing skin cancer in adults. Cochrane Database Syst. Rev. 2018. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.R.; Roisman, L.; Zhang, Q.; Zheng, F.; Rafael de Oliveira Dias, J.; Yehoshua, Z.; Schaal, K.B.; Feuer, W.; Gregori, G.; Chu, Z.; et al. Comparison between spectral-domain and swept-source optical coherence tomography angiographic imaging of choroidal neovascularization. Investig. Ophthalmol. Vis. Sci. 2017, 58, 1499–1505. [Google Scholar] [CrossRef] [PubMed]
- Abreu-González, R.; Gallego-Pinazo, R.; Dolz-Marco, R.; Donate López, J.; López Guajardo, L. Swept Source OCT versus Spectral Domain OCT: Myths and realities. Arch. la Soc. Española Oftalmol. 2016, 91, 459–460. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, M.; Themstrup, L.; de Carvalho, N.; Manfredi, M.; Grana, C.; Ciardo, S.; Kästle, R.; Holmes, J.; Whitehead, R.; Jemec, G.B.E.; et al. Dynamic Optical Coherence Tomography in Dermatology. Dermatology 2016, 232, 298–311. [Google Scholar] [CrossRef]
- Olsen, J.; Holmes, J.; Jemec, G.B.E. Advances in optical coherence tomography in dermatology—a review. J. Biomed. Opt. 2018, 23, 1. [Google Scholar] [CrossRef]
- Deegan, A.J.; Talebi-Liasi, F.; Song, S.; Li, Y.; Xu, J.; Men, S.; Shinohara, M.M.; Flowers, M.E.; Lee, S.J.; Wang, R.K. Optical coherence tomography angiography of normal skin and inflammatory dermatologic conditions. Lasers Surg. Med. 2018, 50, 183–193. [Google Scholar] [CrossRef]
- Choi, W.J.; Reif, R.; Yousefi, S.; Wang, R.K. Improved microcirculation imaging of human skin in vivo using optical microangiography with a correlation mapping mask. J. Biomed. Opt. 2014, 19, 036010. [Google Scholar] [CrossRef]
- Alex, A.; Weingast, J.; Hofer, B.; Eibl, M.; Binder, M.; Pehamberger, H.; Drexler, W.; Považay, B. 3D optical coherence tomography for clinical diagnosis of nonmelanoma skin cancers. Imaging Med. 2011, 3, 653–674. [Google Scholar] [CrossRef]
- Korde, V.R.; Bonnema, G.T.; Xu, W.; Krishnamurthy, C.; Ranger-Moore, J.; Saboda, K.; Slayton, L.D.; Salasche, S.J.; Warneke, J.A.; Alberts, D.S.; et al. Using optical coherence tomography to evaluate skin sun damage and precancer. Lasers Surg. Med. 2007, 39, 9. [Google Scholar] [CrossRef] [PubMed]
- Themstrup, L.; Welzel, J.; Ciardo, S.; Kaestle, R.; Ulrich, M.; Holmes, J.; Whitehead, R.; Sattler, E.C.; Kindermann, N.; Pellacani, G.; et al. Validation of Dynamic optical coherence tomography for non-invasive, in vivo microcirculation imaging of the skin. Microvasc. Res. 2016, 107, 97–105. [Google Scholar] [CrossRef]
- Xiong, Y.-Q.; Mo, Y.; Wen, Y.-Q.; Cheng, M.-J.; Huo, S.-T.; Chen, X.-J.; Chen, Q. Optical coherence tomography for the diagnosis of malignant skin tumors: A meta-analysis. J. Biomed. Opt. 2018, 23, 1. [Google Scholar] [CrossRef] [PubMed]
- Perchoux, J.; Quotb, A.; Atashkhooei, R.; Azcona, F.; Ramírez-Miquet, E.; Bernal, O.; Jha, A.; Luna-Arriaga, A.; Yanez, C.; Caum, J.; et al. Current Developments on Optical Feedback Interferometry as an All-Optical Sensor for Biomedical Applications. Sensors 2016, 16, 694. [Google Scholar] [CrossRef]
- Rakić, A.D.; Taimre, T.; Bertling, K.; Lim, Y.L.; Wilson, S.J.; Nikolić, M.; Valavanis, A.; Indjin, D.; Linfield, E.H.; Davies, A.G.; et al. THz QCL self-mixing interferometry for biomedical applications. In Terahertz Emitters, Receivers, and Applications V; Razeghi, M., Baranov, A.N., Zavada, J.M., Pavlidis, D., Eds.; SPIE: Bellingham, WA, USA, 2014; Volume 9199, p. 91990M. [Google Scholar]
- de Mul, F.F.M.; Koelink, M.H.; Weijers, A.L.; Greve, J.; Aarnoudse, J.G.; Graaff, R.; Dassel, A.C.M. Self-mixing laser-Doppler velocimetry of liquid flow and of blood perfusion in tissue. Appl. Opt. 1992, 31, 5844. [Google Scholar] [CrossRef]
- Cattini, S.; Norgia, M.; Pesatori, A.; Rovati, L. Blood flow measurement in extracorporeal circulation using self-mixing laser diode. In Optical Diagnostics and Sensing X: Toward Point-of-Care Diagnostics; Coté, G.L., Ed.; SPIE: Bellingham, WA, USA, 2010; Volume 7572, p. 75720A. [Google Scholar]
- Norgia, M.; Pesatori, A.; Rovati, L. Self-Mixing Laser Doppler Spectra of Extracorporeal Blood Flow: A Theoretical and Experimental Study. IEEE Sens. J. 2012, 12, 552–557. [Google Scholar] [CrossRef]
- Zhao, Y.; Shen, X.; Zhang, M.; Yu, J.; Li, J.; Wang, X.; Perchoux, J.; Moreira, R.D.C.; Chen, T. Self-Mixing Interferometry-Based Micro Flow Cytometry System for Label-Free Cells Classification. Appl. Sci. 2020, 10, 478. [Google Scholar] [CrossRef]
- Lacot, E.; Day, R.; Stoeckel, F. Laser optical feedback tomography. Opt. Lett. 1999, 24, 744. [Google Scholar] [CrossRef]
- Mowla, A.; Du, B.W.; Taimre, T.; Bertling, K.; Wilson, S.; Soyer, H.P.; Rakić, A.D. Confocal laser feedback tomography for skin cancer detection. Biomed. Opt. Express 2017, 8, 4037. [Google Scholar] [CrossRef]
- Yáñez, C.; Azcona, F.J.; Royo, S. Confocal flowmeter based on self-mixing interferometry for real-time velocity profiling of turbid liquids flowing in microcapillaries. Opt. Express 2019, 27, 24340. [Google Scholar] [CrossRef] [PubMed]
- Spandana, K.U.; Mahato, K.K.; Mazumder, N. Polarization-resolved Stokes-Mueller imaging: A review of technology and applications. Lasers Med. Sci. 2019, 34, 1283–1293. [Google Scholar]
- Rey-Barroso, L.; Burgos-Fernández, F.J.; Royo, S.; Puig, S.; Malvehy, J.; Pellacani, G.; Delpueyo, X.; Peña, S.; Díaz-Doutón, F.; Vilaseca, M. Polarized multispectral imaging for the diagnosis of skin cancer. Final Progr. Proc.-IS T/SID Color Imaging Conf. 2019, 2019, 381–385. [Google Scholar] [CrossRef]
- Louie, D.C.; Phillips, J.; Tchvialeva, L.; Kalia, S.; Lui, H.; Wang, W.; Lee, T.K. Degree of optical polarization as a tool for detecting melanoma: Proof of principle. J. Biomed. Opt. 2018, 23, 1. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; He, C.; Elson, D.S. Real time complete Stokes polarimetric imager based on a linear polarizer array camera for tissue polarimetric imaging. Biomed. Opt. Express 2017, 8, 4933. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, N.; Kao, F.J. Stokes polarimetry-based second harmonic generation microscopy for collagen and skeletal muscle fiber characterization. Lasers Med. Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.-Y.; Chipman, R.A. Interpretation of Mueller matrices based on polar decomposition. J. Opt. Soc. Am. A 1996, 13, 1106. [Google Scholar] [CrossRef]
- Ortega-Quijano, N.; Haj-Ibrahim, B.; García-Caurel, E.; Arce-Diego, J.L.; Ossikovski, R. Experimental validation of Mueller matrix differential decomposition. Opt. Express 2012, 20, 1151. [Google Scholar] [CrossRef]
- Alali, S.; Vitkin, A. Polarized light imaging in biomedicine: Emerging Mueller matrix methodologies for bulk tissue assessment. J. Biomed. Opt. 2015, 20, 061104. [Google Scholar] [CrossRef]
- Zaffar, M.; Pradhan, A. Assessment of anisotropy of collagen structures through spatial frequencies of Mueller matrix images for cervical pre-cancer detection. Appl. Opt. 2020, 59, 1237–1248. [Google Scholar] [CrossRef]
- Novikova, T.; Pierangelo, A.; Manhas, S.; Benali, A.; Validire, P.; Gayet, B.; De Martino, A. The origins of polarimetric image contrast between healthy and cancerous human colon tissue. Appl. Phys. Lett. 2013, 102, 241103. [Google Scholar] [CrossRef]
- Ahmad, I.; Khaliq, A.; Iqbal, M.; Khan, S. Mueller matrix polarimetry for characterization of skin tissue samples: A review. Photodiagnosis Photodyn. Ther. 2020, 30, 101708. [Google Scholar] [CrossRef] [PubMed]
- Ghassemi, P.; Lemaillet, P.; Germer, T.A.; Shupp, J.W.; Venna, S.S.; Boisvert, M.E.; Flanagan, K.E.; Jordan, M.H.; Ramella-Roman, J.C. Out-of-plane Stokes imaging polarimeter for early skin cancer diagnosis. J. Biomed. Opt. 2012, 17, 0760141. [Google Scholar] [CrossRef] [PubMed]
- Fricke, D.; Maas, S.; Jütte, L.; Wollweber, M.; Roth, B. Non-Contact Fast Mueller Matrix Measurement System for Investigation of Inflammatory Skin Diseases; SPIE: Bellingham, WA, USA, 2019. [Google Scholar]
- Magalhaes, C.; Mendes, J.; Vardasca, R. The role of AI classifiers in skin cancer images. Skin Res. Technol. 2019, 25, 750–757. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Q.; Ding, Y.; Jiang, H.; Thiers, B.H.; Wang, J.Z. Automatic diagnosis of melanoma using machine learning methods on a spectroscopic system. BMC Med. Imaging 2014, 14, 1–12. [Google Scholar] [CrossRef]
- Marvdashti, T.; Duan, L.; Aasi, S.Z.; Tang, J.Y.; Ellerbee Bowden, A.K. Classification of basal cell carcinoma in human skin using machine learning and quantitative features captured by polarization sensitive optical coherence tomography. Biomed. Opt. Express 2016, 7, 3721. [Google Scholar] [CrossRef]
- Suresh, L.P.; Shunmuganathan, K.L.; Veni, S.H.K. Dermoscopic Image Segmentation using Machine Learning Algorithm. Am. J. Appl. Sci. 2011, 8, 1159–1168. [Google Scholar] [CrossRef]
- Janney, J.B.; Roslin, S.E. Classification of melanoma from Dermoscopic data using machine learning techniques. Multimed. Tools Appl. 2020, 79, 3713–3728. [Google Scholar] [CrossRef]
- Dreiseitl, S.; Ohno-Machado, L.; Kittler, H.; Vinterbo, S.; Billhardt, H.; Binder, M. A comparison of machine learning methods for the diagnosis of pigmented skin lesions. J. Biomed. Inform. 2001, 34, 28–36. [Google Scholar] [CrossRef]
- Capdehourat, G.; Corez, A.; Bazzano, A.; Alonso, R.; Musé, P. Toward a combined tool to assist dermatologists in melanoma detection from dermoscopic images of pigmented skin lesions. Pattern Recognit. Lett. 2011, 32, 2187–2196. [Google Scholar] [CrossRef]
- Arshaghi, A.; Ashourian, M.; Ghabeli, L. Detection of skin cancer image by feature selection methods using new buzzard optimization (BUZO) algorithm. Trait. du Signal 2020, 37, 181–194. [Google Scholar] [CrossRef]
- Rastgoo, M.; Garcia, R.; Morel, O.; Marzani, F. Automatic differentiation of melanoma from dysplastic nevi. Comput. Med. Imaging Graph. 2015, 43, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Gautam, D.; Ahmed, M.; Meena, Y.K.; Ul Haq, A. Machine learning–based diagnosis of melanoma using macro images. Int. J. Numer. Method. Biomed. Eng. 2018, 34, e2953. [Google Scholar] [CrossRef] [PubMed]
- Torre, E.L.; Caputo, B.; Tommasi, T. Learning methods for melanoma recognition. Int. J. Imaging Syst. Technol. 2010, 20, 316–322. [Google Scholar] [CrossRef]
- Sáez, A.; Sánchez-Monedero, J.; Gutiérrez, P.A.; Hervás-Martínez, C. Machine Learning Methods for Binary and Multiclass Classification of Melanoma Thickness from Dermoscopic Images. IEEE Trans. Med. Imaging 2016, 35, 1036–1045. [Google Scholar] [CrossRef]
- Lee, H.D.; Mendes, A.I.; Spolaôr, N.; Oliva, J.T.; Sabino Parmezan, A.R.; Wu, F.C.; Fonseca-Pinto, R. Dermoscopic assisted diagnosis in melanoma: Reviewing results, optimizing methodologies and quantifying empirical guidelines. Knowl. -Based Syst. 2018, 158, 9–24. [Google Scholar] [CrossRef]
- Hameed, N.; Hameed, F.; Shabut, A.; Khan, S.; Cirstea, S.; Hossain, A. An Intelligent Computer-Aided Scheme for Classifying Multiple Skin Lesions. Computers 2019, 8, 62. [Google Scholar] [CrossRef]
- Barros, W.K.P.; Morais, D.S.; Lopes, F.F.; Torquato, M.F.; Barbosa, R.d.M.; Fernandes, M.A.C. Proposal of the CAD system for melanoma detection using reconfigurable computing. Sensors 2020, 20, 3168. [Google Scholar] [CrossRef]
- Pujol, J. A device for the color measurement and detection of spots on the skin. 2008, 14, 65–70. Skin Res. Technol. 2008, 14, 65–70. [Google Scholar]
- Vapnik, V.N. An overview of statistical learning theory. IEEE Trans. Neural Networks 1999, 10, 988–999. [Google Scholar] [CrossRef]
- Dasarathy, B. V Nearest Neighbor (NN) Norms: NN Pattern Classification Techniques; IEEE Comput. Soc. Press.: Los Alamitos, CA, USA, 1991. [Google Scholar]
- Bishop, C.M. Neural Networks: A Pattern Recognition Perspective, 2nd ed.; Oxford University Press: Oxford, UK, 1996; ISBN 0-14-853864-2. [Google Scholar]
- Quinlan, J.R. Simplifying decision trees. Int. J. Man. Mach. Stud. 1987, 27, 221–234. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Yuan, Y.; Chao, M.; Lo, Y. Deep Fully Convolutional Networks With Jaccard Distance. IEEE Trans. Med. Imaging 2017, 36, 1876–1886. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shen, L. Skin lesion analysis towards melanoma detection using deep learning network. Sensors 2018, 18, 556. [Google Scholar] [CrossRef]
- Refianti, R.; Mutiara, A.B.; Priyandini, R.P. Classification of melanoma skin cancer using convolutional neural network. Int. J. Adv. Comput. Sci. Appl. 2019, 10, 409–417. [Google Scholar] [CrossRef]
- Maiti, A.; Chatterjee, B. Improving detection of Melanoma and Naevus with deep neural networks. Multimed. Tools Appl. 2020, 79, 15635–15654. [Google Scholar] [CrossRef]
- Premaladha, J.; Ravichandran, K.S. Novel Approaches for Diagnosing Melanoma Skin Lesions through Supervised and Deep Learning Algorithms. J. Med. Syst. 2016, 40, 96. [Google Scholar] [CrossRef]
- Mahbod, A.; Schaefer, G.; Ellinger, I.; Ecker, R.; Pitiot, A.; Wang, C. Fusing fine-tuned deep features for skin lesion classification. Comput. Med. Imaging Graph. 2019, 71, 19–29. [Google Scholar] [CrossRef]
- Saba, T.; Khan, M.A.; Rehman, A.; Marie-Sainte, S.L. Region Extraction and Classification of Skin Cancer: A Heterogeneous framework of Deep CNN Features Fusion and Reduction. J. Med. Syst. 2019, 43, 289. [Google Scholar] [CrossRef]
- Rodrigues, D.d.A.; Ivo, R.F.; Satapathy, S.C.; Wang, S.; Hemanth, J.; Filho, P.P.R. A new approach for classification skin lesion based on transfer learning, deep learning, and IoT system. Pattern Recognit. Lett. 2020, 136, 8–15. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015-Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 17–19 June 2015; pp. 1–9. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; 2016; pp. 770–778. [Google Scholar]
- Mahbod, A.; Schaefer, G.; Wang, C.; Dorffner, G.; Ecker, R.; Ellinger, I. Transfer learning using a multi-scale and multi-network ensemble for skin lesion classification. Comput. Methods Programs Biomed. 2020, 193, 105475. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Jiang, X.; Zhou, F.; Qin, J.; Ni, D.; Chen, S.; Lei, B.; Wang, T. Melanoma Recognition in Dermoscopy Images via Aggregated Deep Convolutional Features. IEEE Trans. Biomed. Eng. 2019, 66, 1006–1016. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.Y.; Zhang, L.; Lim, C.P. Intelligent skin cancer diagnosis using improved particle swarm optimization and deep learning models. Appl. Soft Comput. J. 2019, 84, 105725. [Google Scholar] [CrossRef]
- Harangi, B. Skin lesion classification with ensembles of deep convolutional neural networks. J. Biomed. Inform. 2018, 86, 25–32. [Google Scholar] [CrossRef]
- Olivas, E.S.; Guerrero, J.D.M.; Martinez Sober, M.; Magdalena Benedito, J.R.; Serrano López, A.J. Handbook of Research on Machine Learning Applications and Trends: Algorithms, Methods, and Techniques; IGI Global: Hershey, PA, USA, 2009; ISBN 9781605667669. [Google Scholar]
- Albert, B.A. Deep Learning from Limited Training Data: Novel Segmentation and Ensemble Algorithms Applied to Automatic Melanoma Diagnosis. IEEE Access 2020, 8, 31254–31269. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, S.; Liu, J.; Tao, C. Towards improving diagnosis of skin diseases by combining deep neural network and human knowledge. BMC Med. Inform. Decis. Mak. 2018, 18, 59. [Google Scholar] [CrossRef]
- Qin, Z.; Liu, Z.; Zhu, P.; Xue, Y. A GAN-based image synthesis method for skin lesion classification. Comput. Methods Programs Biomed. 2020, 195, 105568. [Google Scholar] [CrossRef]



| Study | Year | Skin Cancer Type | No. of Lesions | Modality | Illumination Source | No. of Images | Axial Res. | Lateral Res. | SE/SP |
|---|---|---|---|---|---|---|---|---|---|
| Guitera et al. | 2009 | Melanoma, Nevus | >300 | RCM, commercial CLSM | Laser diode, 830 nm | >100 | 3–5 μm | 1 μm | Mel. Light- colored: 85%/84% Pigmented: 92%/65% |
| Guitera et al. | 2012 | Melanoma, BCC, Nevus, Pigmented facial macules, Other tumors | >700 | RCM, commercial CLSM | Laser diode, 830 nm | >100 | 3–5 μm | 1 μm | 100% 88.5% |
| Segura et al. | 2009 | Melanoma, BCC, SCC, Keratosis, Nevus | >150 | RCM, commercial CLSM | Laser diode, 830 nm | >100 | 5 μm | 2 μm | 86.1% 95.3% |
| Ulrich et al. | 2008 | AK, perilesional healthy skin | >20 | RCM, commercial CLSM | Laser diode, 830 nm | 4–6 images | 3–5 μm | 1 μm | - |
| Horn et al. | 2008 | AK, perilesional healthy skin | 30 | RCM, commercial CLSM | Laser diode, 830 nm | >50 | 3–5 μm | 1 μm | 93.34% 88.34% |
| Gareau et al. | 2008 | BCC | - | FCM, mosaicing | Argon-ion laser, 488 nm | 36 × 36 images for a mosaic | 1.1 μm | 0.25 μm | - |
| Gareau et al. | 2009 | BCC, healthy skin | >40 | FCM, mosaicing | Argon-ion laser, 488 nm | 45 confocal mosaics | 1.1 μm | 0.25 μm | 96.6% 89.2% |
| Abeytunge et al. | 2011 | BCC | 1 | FCM, strip mosaicing | Argon-ion laser, 488 nm | 31 strips for a mosaic | 1.61 μm | 0.33 μm | - |
| Study | Year | Skin Cancer Type | No. of Lesions | Modality | Spectral Range | No. of Images | Spectral Res. | Lateral Res. | SE/SP |
|---|---|---|---|---|---|---|---|---|---|
| Tomatis et al. | 2003 | Melanoma, BCC, Keratosis, Nevus, Other tumors | >500 | MS, Pasive Staring imaging | 400–1040 nm | 17 images | 40 nm | 300 μm pixel size | 78% 76% |
| Bekina et al. | 2012 | Papilloma, Melanoma | <10 | MS, Active Staring imaging | 450, 545, 660 and 940 nm | 4 images | - | - | - |
| Jakovels et al. | 2013 | Bening pigmented lesions, Melanoma | >100 | MS, Pasive Staring imaging | 450–950 nm | 51 images | 10 nm | 75 μm pixel size | - |
| Kim et al. | 2016 | Nevus, Acne lesion | 2 | MS, Smartphone-based, Active Staring imaging | 440–690 nm | 9 images | - | 18 μm pixel size | - |
| Stamnes et al. | 2017 | Melanoma, BCC, SCC, Keratosis, Nevus | >500 | MS, Active Staring imaging | 365–1000 nm | 10 images at different illum./acq. angles | - | 25 μm pixel size | 99% 93% |
| Delpueyo et al. | 2017 | Melanoma, BCC, Nevus | >400 | MS, Active Staring imaging | 414–995 nm | 8 images at different pol. angles | - | 18 μm pixel size | Melanoma: 87.2% 54.5% All: 91.3% 54.5% |
| Vasaturo et al. | 2017 | Melanoma | - | MS, Ex-vivo microscopy | 420–720 nm | 15 images | 20 nm | 1392 x 1040 pixels | - |
| Rey- Barroso et al. | 2018 | Melanoma, Nevus | >50 | VIS and exNIR MS, Active Staring imaging | 414–1613 nm | 14 images | - | CCD: 18 μm pixel size InGaAs: 70 μm pixel size | 78.6% 84.6% |
| Godoy et al. | 2015 | Melanoma, BCC, SCC, Benign pigmented lesions | >100 | MS, LWIR Staring imaging | 750–2500 nm | 60 frames/s during 2 min | - | 300 μm pixel size | 95% 83% |
| Fioravanti et al. | 2016 | Primary melanoma, Metastasis, healthy skin | 15 | MS, Ex-vivo IR spectrometry | 3330–3570 nm | None – Integrated spectral information | 12 nm | - | - |
| Study | Year | Skin Cancer Type | No. of Lesions | Modality | Illumination Source | No. of Images | Axial Res. | Lateral Res. | SE/SP |
|---|---|---|---|---|---|---|---|---|---|
| Gorpas et al. | 2006 | Animal model tumor | 1 | Fringe projection | Blue laser diode 440 nm | - | 4.4 μm | - | |
| Moore et al. | 2006 | Multiple cancers | - | Fringe projection | He-Ne laser 633 nm | - | - | - | - |
| Ares et al. | 2014 | - | - | Fringe projection | He-Ne laser 633 nm | - | - | - | - |
| Rey-Barroso et al. | 2014 | Melanoma, BCC, Nevus | >170 | Fringe projection | He-Ne laser 633 nm | - | - | - | 80.0% 76.7% |
| Korn et al. | 2017 | None – body parts exposed to sun | 5 body sites in 20 patients | 3D optical profilometry | Hallogen lamp | 8 images | 0.005 μm | 1.1 μm | - |
| Study | Year | Skin Cancer Type | No. of Lesions | Modality | Laser Source | No. of Images | Axial Res. | Lateral Res. | SE/SP |
|---|---|---|---|---|---|---|---|---|---|
| Korde et al. | 2008 | AK, body parts exposed to sun | 4 body sites in 112 patients | FD-OCT | 1310 nm | 1344 OCT images | 12 μm | 12 μm | 86% 83% |
| Themstrup et al. | 2016 | None–healthy skin subject to different conditions | - | OCTA, commercial OCT | 1305 nm | - | 5 μm | 7.5 μm | - |
| Weissman et al. | 2004 | None–healthy skin to compare with BCC | - | FD-OCT, commercial OCT | LEDs, 1300 nm | 47 OCT images | 5 μm | 3 μm | - |
| Study | Year | Skin Cancer Type | No. of Lesions | Modality | Illuminationsource | No. of Images | Polarization Configurations | Lateral Res. | SE/SP |
|---|---|---|---|---|---|---|---|---|---|
| Rey- Barroso et al. | 2019 | MM, Nevus | 40 | Polarized MS imaging | LEDs, 414–995 nm | 8 images | 0°, 45° and 90° | 18 μm pixel size | - |
| Ghassemi et al. | 2012 | MM, Nevus, healthy skin | >20 | Stokes imaging | Tricolor LED-based, Hemisferical | 16 images at illum. angles 0°, 24°, and 49° | 0°, 45°, 90° and 135° | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rey-Barroso, L.; Peña-Gutiérrez, S.; Yáñez, C.; Burgos-Fernández, F.J.; Vilaseca, M.; Royo, S. Optical Technologies for the Improvement of Skin Cancer Diagnosis: A Review. Sensors 2021, 21, 252. https://doi.org/10.3390/s21010252
Rey-Barroso L, Peña-Gutiérrez S, Yáñez C, Burgos-Fernández FJ, Vilaseca M, Royo S. Optical Technologies for the Improvement of Skin Cancer Diagnosis: A Review. Sensors. 2021; 21(1):252. https://doi.org/10.3390/s21010252
Chicago/Turabian StyleRey-Barroso, Laura, Sara Peña-Gutiérrez, Carlos Yáñez, Francisco J. Burgos-Fernández, Meritxell Vilaseca, and Santiago Royo. 2021. "Optical Technologies for the Improvement of Skin Cancer Diagnosis: A Review" Sensors 21, no. 1: 252. https://doi.org/10.3390/s21010252
APA StyleRey-Barroso, L., Peña-Gutiérrez, S., Yáñez, C., Burgos-Fernández, F. J., Vilaseca, M., & Royo, S. (2021). Optical Technologies for the Improvement of Skin Cancer Diagnosis: A Review. Sensors, 21(1), 252. https://doi.org/10.3390/s21010252







