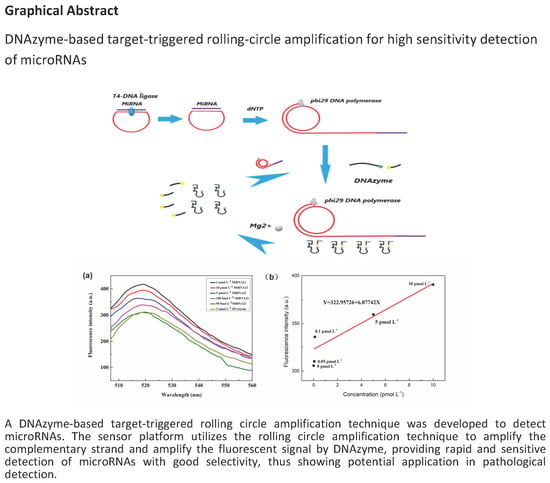
DNAzyme-Based Target-Triggered Rolling-Circle Amplification for High Sensitivity Detection of microRNAs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.1.1. Materials
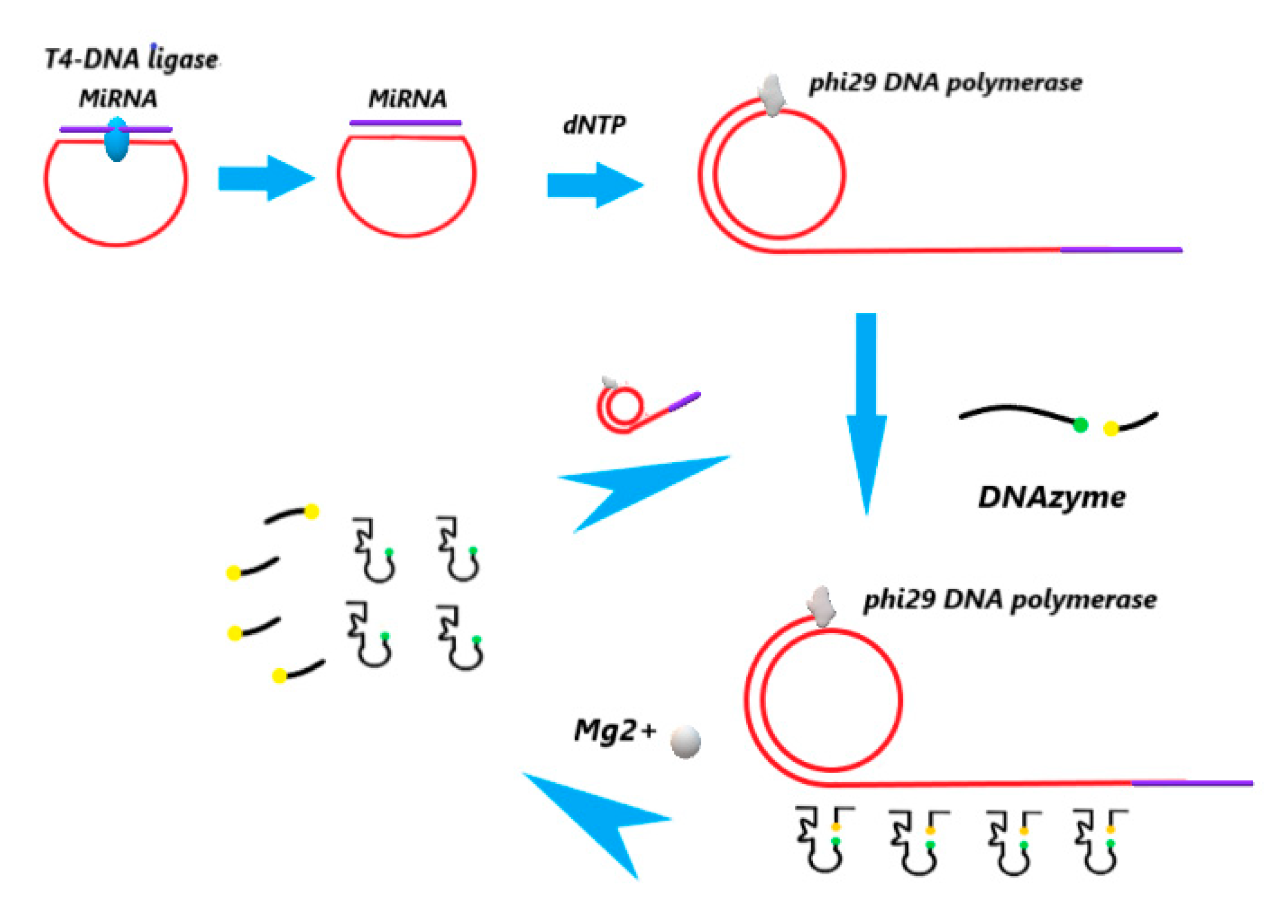
2.1.2. Construction of the Circulating Detection Platform
2.1.3. Instrumentation
2.2. Method
2.2.1. Design Strategy for miR-21 Detection
2.2.2. Ligation Reactions
2.2.3. The RCA Reactions
2.2.4. Fluorescence Signal Amplification based on DNAzyme
3. Results and Discussion
3.1. Feasibility Study
3.2. Optimization Studies
3.2.1. Padlock Probe
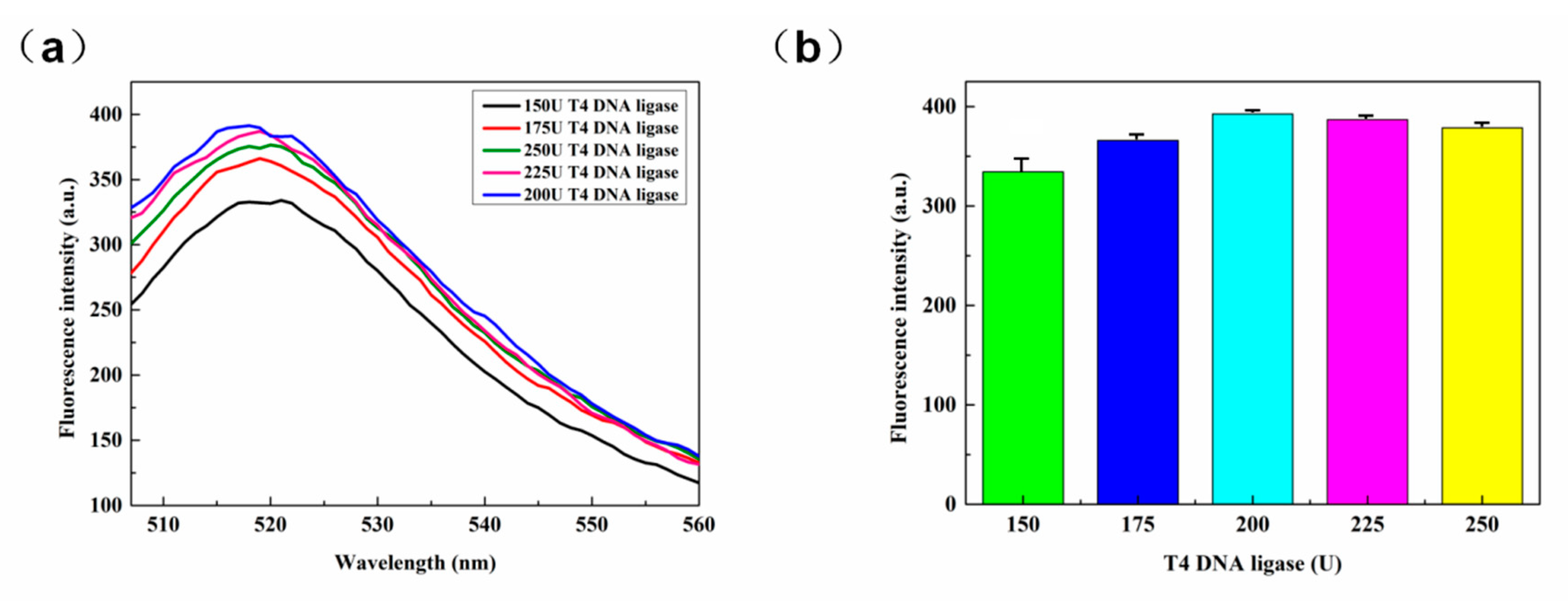
3.2.2. T4 DNA Ligase
3.2.3. Phi29 DNA Polymerase
3.2.4. The Concentration of dNTPs
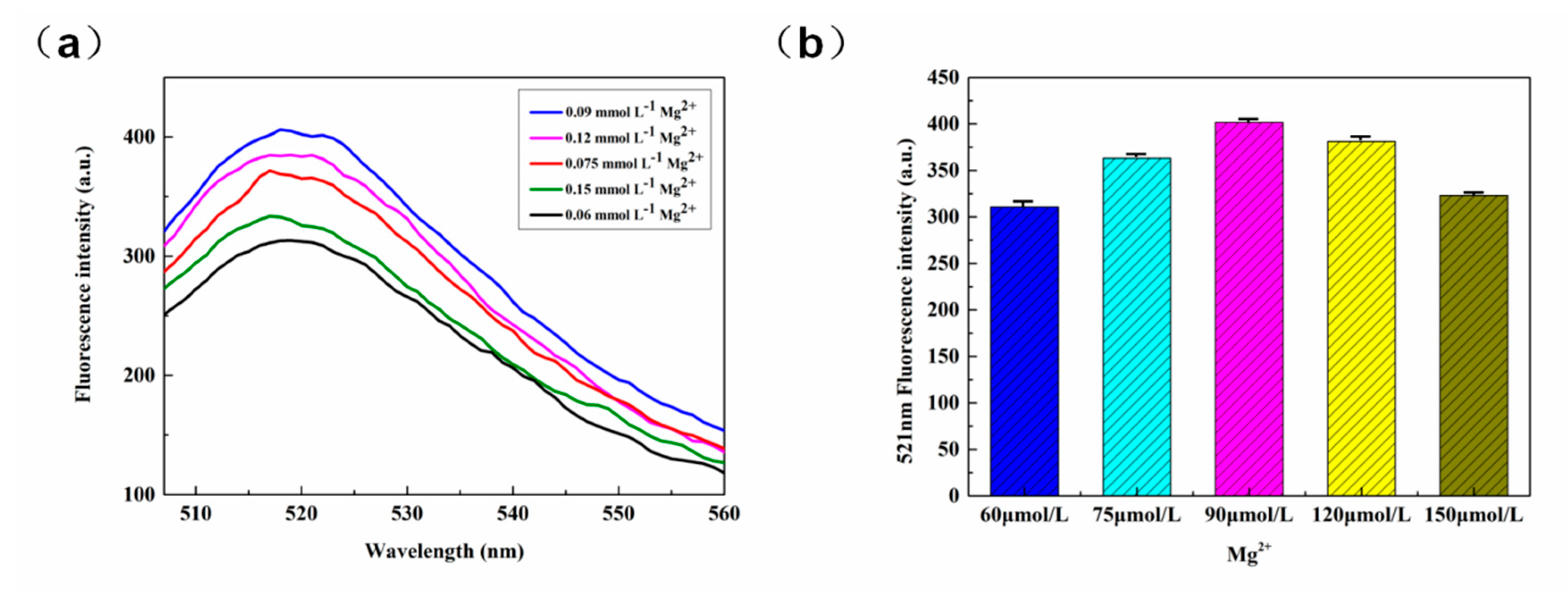
3.2.5. The Concentration of Mg2+
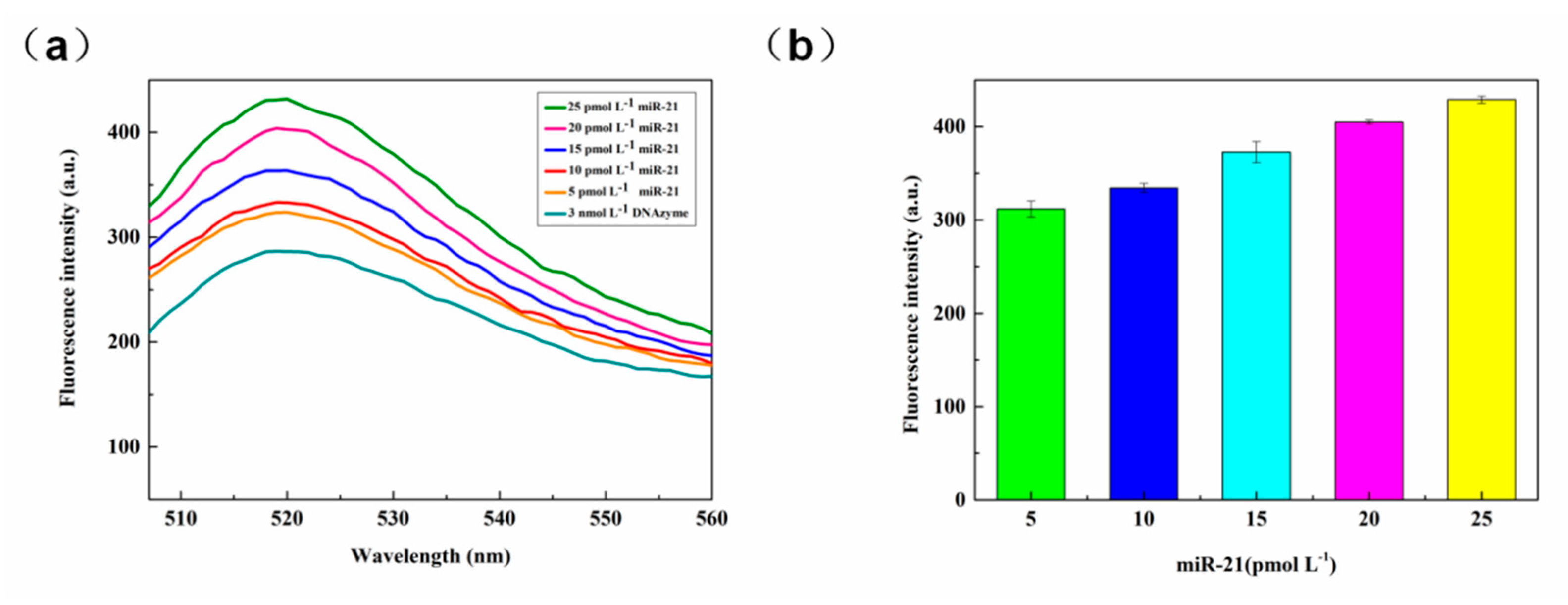
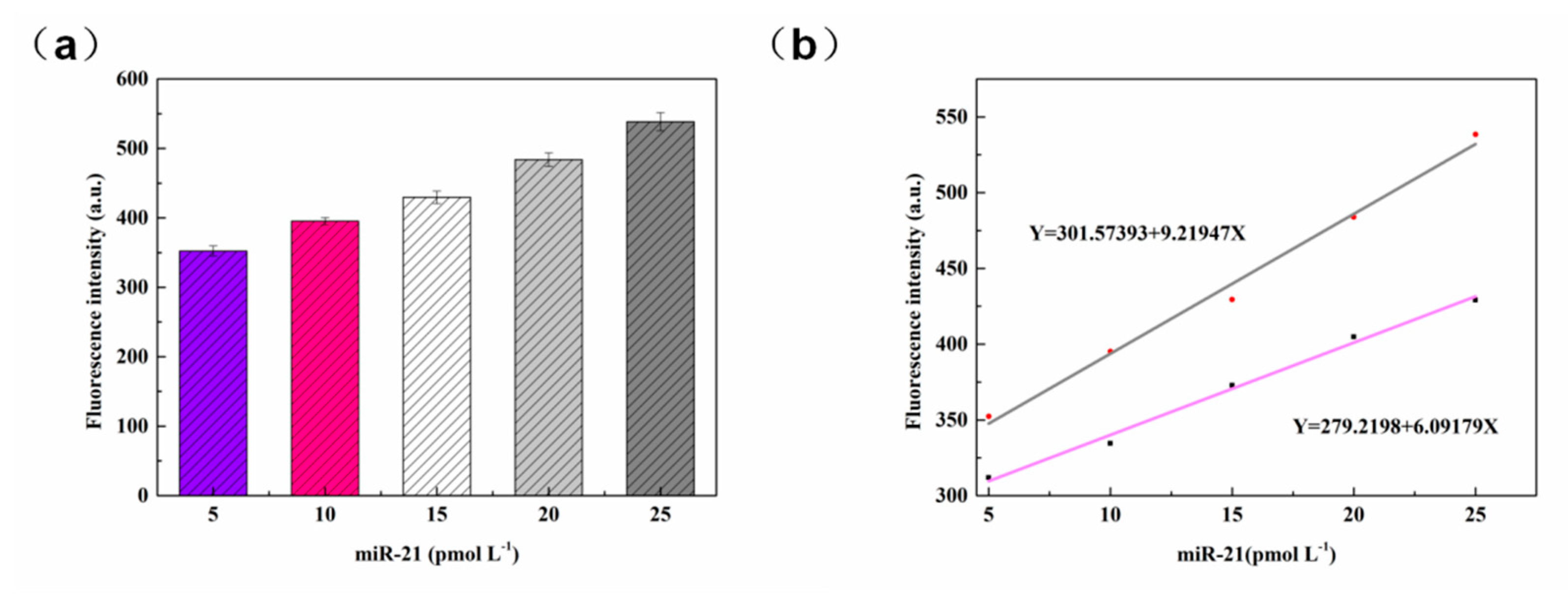
3.2.6. miR-21 Detection
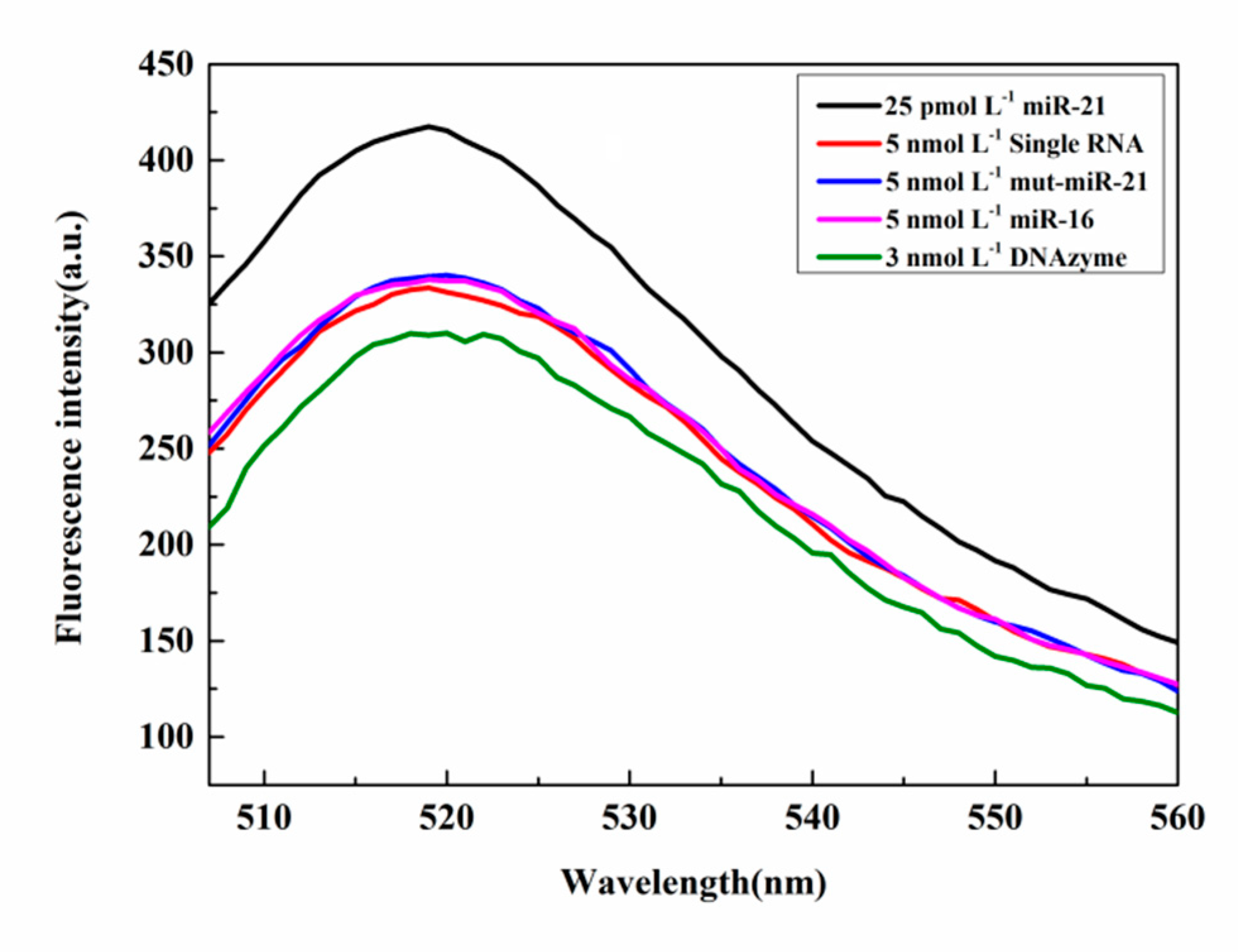
3.2.7. Selectivity of microRNA Detection
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Liu, Q.; Zhou, N.; Mo, Y.-Y. Oncogenic microRNAs in Cancer. In MicroRNA in Cancer; Springer: Dordrecht, The Netherlands, 2013. [Google Scholar]
- Lujambio, A.; Lowe, S.W. The microcosmos of cancer. Nature 2012, 482, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Thum, T.; Gross, C.; Fiedler, J.; Fischer, T.; Kissler, S.; Bussen, M.; Galuppo, P.; Just, S.; Rottbauer, W.; Frantz, S.; et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008, 456, 980–984. [Google Scholar] [CrossRef] [PubMed]
- Koscianska, E.; Starega-Roslan, J.; Czubala, K.; Krzyzosiak, W.J. High-resolution northern blot for a reliable analysis of microRNAs and their precursors. Sci. World J. 2011, 11, 102–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Small, E.M.; Olson, E.N. Pervasive roles of microRNAs in cardiovascular biology. Nature 2011, 469, 336–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, R.; Tang, L.; Tian, Q. Toehold-initiated rolling circle amplification for visualizing individual microRNAs in situ in single cell. Angew. Chem. Int. Ed. Engl. 2014, 53, 2389–2393. [Google Scholar] [CrossRef]
- Ardekani, A.M.; Naeini, M.M. The Role of MicroRNAs in Human Diseases. Avicenna J. Med. Biotechnol. 2010, 2, 161. [Google Scholar]
- Zhou, Y.; Wang, M.; Meng, X.; Yin, H.; Ai, S. Amplified electrochemical microRNA biosensor using a hemin-G-quadruplex complex as the sensing element. RSC Adv. 2012, 2, 7140. [Google Scholar] [CrossRef]
- Tian, T.; Li, L.; Zhang, Y.; Liu, H.; Zhang, L.; Yan, M.; Yu, J. Dual-mode fluorescence biosensor platform based on T-shaped duplex structure for detection of microRNA and folate receptor. Sens. Actuators B Chem. 2018, 261, 44–50. [Google Scholar] [CrossRef]
- Jiang, H.-X.; Zhao, M.-Y.; Niu, C.-D.; Kong, D.-M. Real-time monitoring of rolling circle amplification using aggregation-induced emission: Applications in biological detection. Chem. Commun. 2015, 51, 16518–16521. [Google Scholar] [CrossRef]
- Wan, Y.-H.; Zhou, Y.-J.; Xiao, K.-J.; Nie, C.-P. Target-assisted self-cleavage DNAzyme probes for multicolor simultaneous imaging of tumor-related microRNAs with signal amplification. Chem. Commun. 2019, 55, 3278–3281. [Google Scholar] [CrossRef]
- Chen, Y.-R.; Han, Y.; Liu, R.; Xue, C. Intracellular self-enhanced rolling circle amplification to image specific miRNAs within tumor cells. Sens. Actuators B Chem. 2019, 282, 507–514. [Google Scholar] [CrossRef]
- Zhuang, J.; Lai, W.; Chen, G.; Tang, D. A rolling circle amplification-based DNA machine for miRNA screening coupling catalytic hairpin assembly with DNAzyme formation. Chem. Commun. 2014, 50, 2935–2938. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.X.; Liang, Z.Z.; Ma, Y.H.; Kong, D.M.; Hong, Z.Y. G-quadruplex fluorescent probe-mediated real-time rolling circle amplification strategy for highly sensitive microRNA detection. Anal. Chim. Acta 2016, 943, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.S.; Zhang, S.; Zhou, H.; Shen, G.L.; Yu, R. Universal Aptameric System for Highly Sensitive Detection of Protein Based on Structure-Switching-Triggered Rolling Circle Amplification. Anal. Chem. 2010, 82, 2221–2227. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Li, F.; Zhang, Z.; Zhang, K. Rolling circle amplification: A versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014, 43, 3324–3341. [Google Scholar] [CrossRef]
- Ge, J.; Zhang, L.L.; Liu, S.J.; Yu, R.Q.; Chu, X. A highly sensitive target-primed rolling circle amplification (TPRCA) method for fluorescent in situ hybridization detection of microRNA in tumor cells. Anal. Chem. 2014, 86, 1808–1815. [Google Scholar] [CrossRef]
- Liu, M.; Yin, Q.; McConnell, E.M.; Chang, Y.; Brennan, J.D.; Li, Y. DNAzyme Feedback Amplification: Relaying Molecular Recognition to Exponential DNA Amplification. Chemistry 2018, 24, 4473–4479. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, B.; Wang, M.; Wang, J.; Yin, H.; Ai, S. Fluorometric determination of microRNA based on strand displacement amplification and rolling circle amplification. Microchim. Acta 2017, 184, 4359–4365. [Google Scholar] [CrossRef]
- Kim, H.K.; Liu, J.; Li, J.; Nagraj, N.; Li, M.; Pavot, C.M.B.; Lu, Y. Metal-Dependent Global Folding and Activity of the 8-17 DNAzyme Studied by Fluorescence Resonance Energy Transfer. J. Am. Chem. Soc. 2007, 129, 6896–6902. [Google Scholar] [CrossRef] [PubMed]
- Hollenstein, M.; Hipolito, C.; Lam, C.; Dietrich, D.; Perrin, D.M. A highly selective DNAzyme sensor for mercuric ions. Angew. Chem. Int. Ed. Engl. 2008, 47, 4346–4350. [Google Scholar] [CrossRef]
- Wen, Y.; Xu, Y.; Mao, X.; Wei, Y.; Song, H.; Chen, N.; Huang, Q.; Fan, C.; Li, D. DNAzyme-based rolling-circle amplification DNA machine for ultrasensitive analysis of microRNA in Drosophila larva. Anal. Chem. 2012, 84, 7664–7669. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhang, X.; Li, Z.; Jiao, X.; Wang, Y.; Zhang, Y. Highly sensitive determination of microRNA using target-primed and branched rolling-circle amplification. Angew. Chem. Int. Ed. Engl. 2009, 48, 3268–3272. [Google Scholar] [CrossRef]
- Jiang, H.; Xu, Y.; Dai, L.; Liu, X.; Kong, D. Ultrasensitive, label-free detection of T4 ligase and T4 polynucleotide kinase based on target-triggered hyper-branched rolling circle amplification. Sens. Actuators B Chem. 2018, 260, 70–77. [Google Scholar] [CrossRef]
- Gong, X.; Zhou, W.; Li, D.; Chai, Y.; Xiang, Y.; Yuan, R. RNA-regulated molecular tweezers for sensitive fluorescent detection of microRNA from cancer cells. Biosens. Bioelectron. 2015, 71, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Zhang, L.; Deng, Y.; Li, S.; He, N. Graphene oxide for rapid microRNA detection. Nanoscale 2012, 4, 5840. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y.; Li, W.; Wu, P.; Zhang, H.; Cai, C. Fluorescence Quenching of Graphene Oxide Integrating with the Site-Specific Cleavage of the Endonuclease for Sensitive and Selective MicroRNA Detection. Anal. Chem. 2013, 85, 2536–2542. [Google Scholar] [CrossRef]
- Tian, T.; Xiao, H.; Zhang, Z.; Long, Y.; Peng, S.; Wang, S.; Zhou, X.; Liu, S.; Zhou, X. Sensitive and Convenient Detection of microRNAs Based on Cascade Amplification by Catalytic DNAzymes. Chem. A Eur. J. 2013, 19, 92–95. [Google Scholar] [CrossRef]
- Wang, W.; Kong, T.; Zhang, D.; Zhang, J.; Cheng, G. Label-Free MicroRNA Detection Based on Fluorescence Quenching of Gold Nanoparticles with a Competitive Hybridization. Anal. Chem. 2015, 21, 10822–10829. [Google Scholar] [CrossRef]




| Name | Sequence(5’–3’) |
|---|---|
| Padlock probe | pCTGATAAGCTATTTATTTCCTCAATGCTGCTGCTGTACTACTAGTGATTTACTTGGATGTCTTCAACATCAGT |
| DNAzyme | TTTATTTCAAACT(Dabcyl)rAGGT(FAM)CTTTTTTTTTGACTCCGAGCCGGACGAAGTTAATGCTG |
| MiR-21 | UAGCUUAUCAGACUGAUGUUGA |
| MiR-16 | UAGCAGCACGUAAAUAUUGGCG |
| mut-miR-21 | UAGCUUAACAGACUGAUGUUGA |
| Single RNA | UUGUACUACACAAAAGUACUG |
| Methods | LOD | Linear Range | Correlation Coefficient (R2) | Reference |
|---|---|---|---|---|
| DNA self-assembled molecular tweezers | 0.6 pM | 1–500 nM | 0.9929 | [25] |
| Graphene oxide for rapid microRNA detection | / | 50–400 nM | 0.9562 | [26] |
| Fluorescence quenching of graphene oxide integrating | 3.0fM | 0.02–100 pM | / | [27] |
| A novel DNA nanomachine based on the linear rolling circle amplification strategy | 87fM | 0.1 pM–0.1 nM | 0.9908 | [13] |
| Highly sensitive determination of microRNA using target-primed and branched rolling-circle amplification | 0.25pM | 0.025 pM–2.5 nM | 0.9994 | [23] |
| Fluorometric determination of microRNA based on strand displacement amplification and rolling circle amplification | 1.04fM | 10 fM–0.1 nM | 0.9957 | [19] |
| Cascade amplification by catalytic DNAzymes | 10pM | 10 pM–100 nM | / | [28] |
| Fluorescence quenching of gold nanoparticles with a competitive hybridization | 33.4fM | 100 fM–1.0 nM | / | [29] |
| DNAzyme-based target-triggered rolling-circle amplification | 0.49pM | 0–25 pM | 0.99106 | This work |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Han, J.; Zhou, L.; Zhang, J.; Du, J. DNAzyme-Based Target-Triggered Rolling-Circle Amplification for High Sensitivity Detection of microRNAs. Sensors 2020, 20, 2017. https://doi.org/10.3390/s20072017
Liu C, Han J, Zhou L, Zhang J, Du J. DNAzyme-Based Target-Triggered Rolling-Circle Amplification for High Sensitivity Detection of microRNAs. Sensors. 2020; 20(7):2017. https://doi.org/10.3390/s20072017
Chicago/Turabian StyleLiu, Chen, Jialun Han, Lujian Zhou, Jingjing Zhang, and Jie Du. 2020. "DNAzyme-Based Target-Triggered Rolling-Circle Amplification for High Sensitivity Detection of microRNAs" Sensors 20, no. 7: 2017. https://doi.org/10.3390/s20072017
APA StyleLiu, C., Han, J., Zhou, L., Zhang, J., & Du, J. (2020). DNAzyme-Based Target-Triggered Rolling-Circle Amplification for High Sensitivity Detection of microRNAs. Sensors, 20(7), 2017. https://doi.org/10.3390/s20072017