This Section presents the details of performed classification experiments: the dataset, the procedure, evaluation method, results and discussion.
3.1. Evaluation Dataset
Hyperspectral images used in our experiments come from the dataset described in [
23], which is publicly available under an open licence (the dataset with snippet code for data loading:
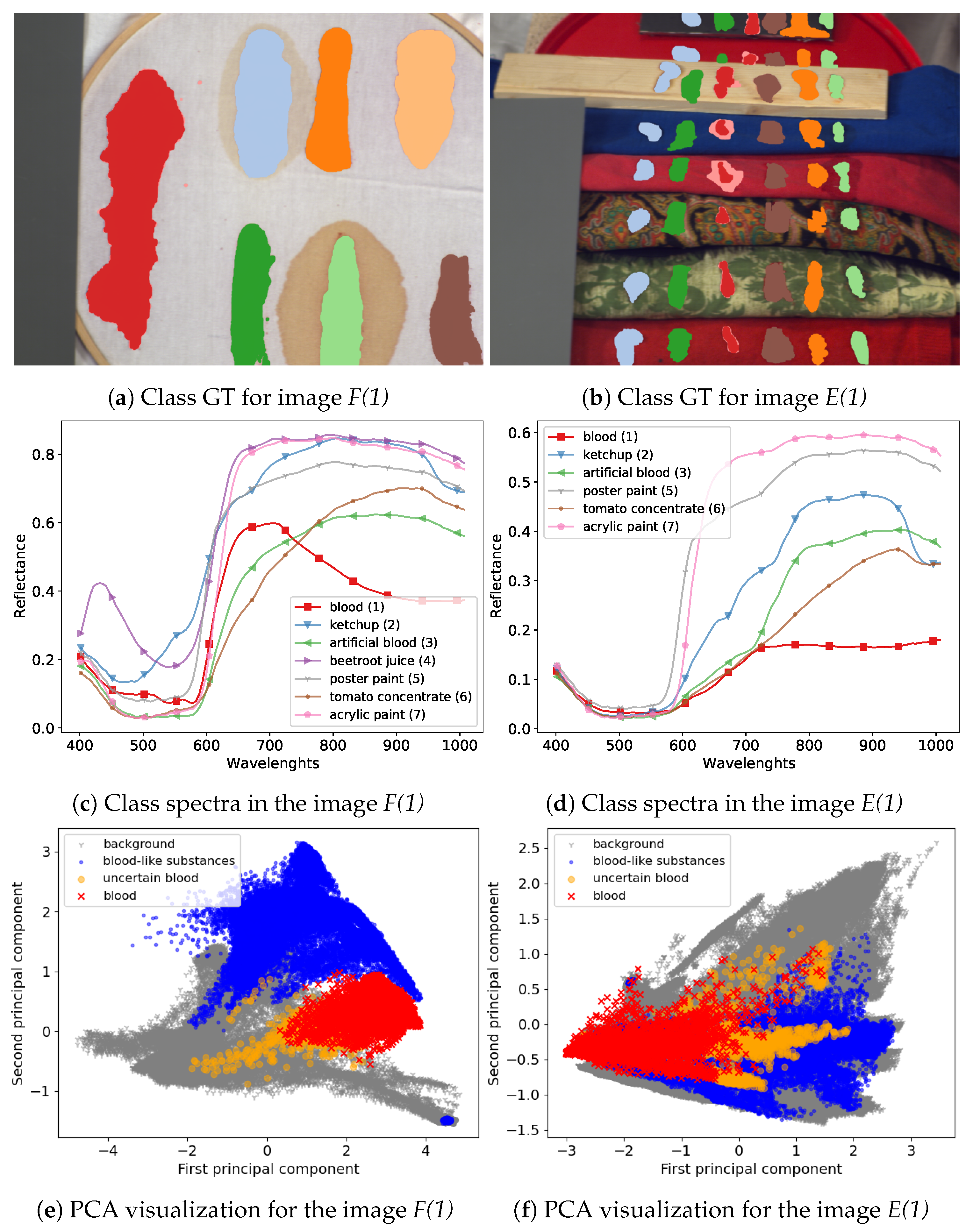
https://zenodo.org/record/3984905). The dataset consists of annotated hyperspectral images of blood and six other visually similar substances:
artificial blood,
tomato concentrate,
ketchup,
beetroot juice,
poster and
acrylic paint. Images in the dataset were recorded on the course of several days, to capture changes in spectra related to the process of the time-related substance decay. On every image, hyperspectral pixels where classes are visible were annotated by authors, therefore they can be treated as labelled examples i.e., pairs
from a set of labelled examples
, where vectors
are hyperspectral pixels,
denotes the number of bands and
are class labels.
The majority of images in the dataset were captured using an SOC710 hyperspectral camera operating in spectral range 377–1046 nm with 128 bands. Following suggestions in [
23] we removed bands [0–4], [48–50] and [122–128], leaving 113 bands. From six image types in the dataset we decided to use two that are most useful for our experimental setting. First, the
frame scenes, denoted as
F in [
23], present class examples on a uniform, white fabric background; this image has the advantage of distinct separation and clear spectral characteristic of the substances which makes it a good benchmark dataset for tested models. The other type of images that was used is the
comparison scenes, denoted as
E. These present the same substances on diverse backgrounds consisting of multiple materials and fabrics types. This means that substance spectra are mixed with the background spectra, which provides a more challenging classification setting. Since one of the classes, the
beetroot juice was not present in all images, it was removed in our experiments, leaving six classes.
Additionally, we used images captured in different days to take into account changes in the blood spectrum over time. For both scene types we used images from days
(6 images in total), denoted
F(1), F(7), F(21) and
E(1), E(7), E(21), respectively. For scenes
F we additionally used an image denoted
F(1a), taken sever hours later than the image
F(1) and two images obtained in the second day, denoted
F(2) and
F(2k). The image
F(2k) was captured with a different hyperspectral camera. Spectra in this image were linearly interpolated to match bands from the SOC710 camera. A more detailed description of the dataset can be found in [
23]. Visualization of example images from the dataset and a comparison of their spectra are presented in
Figure 8.
3.2. Experimental Procedure
We performed two types of classification experiments (see
Section 1 for motivation and discussion):
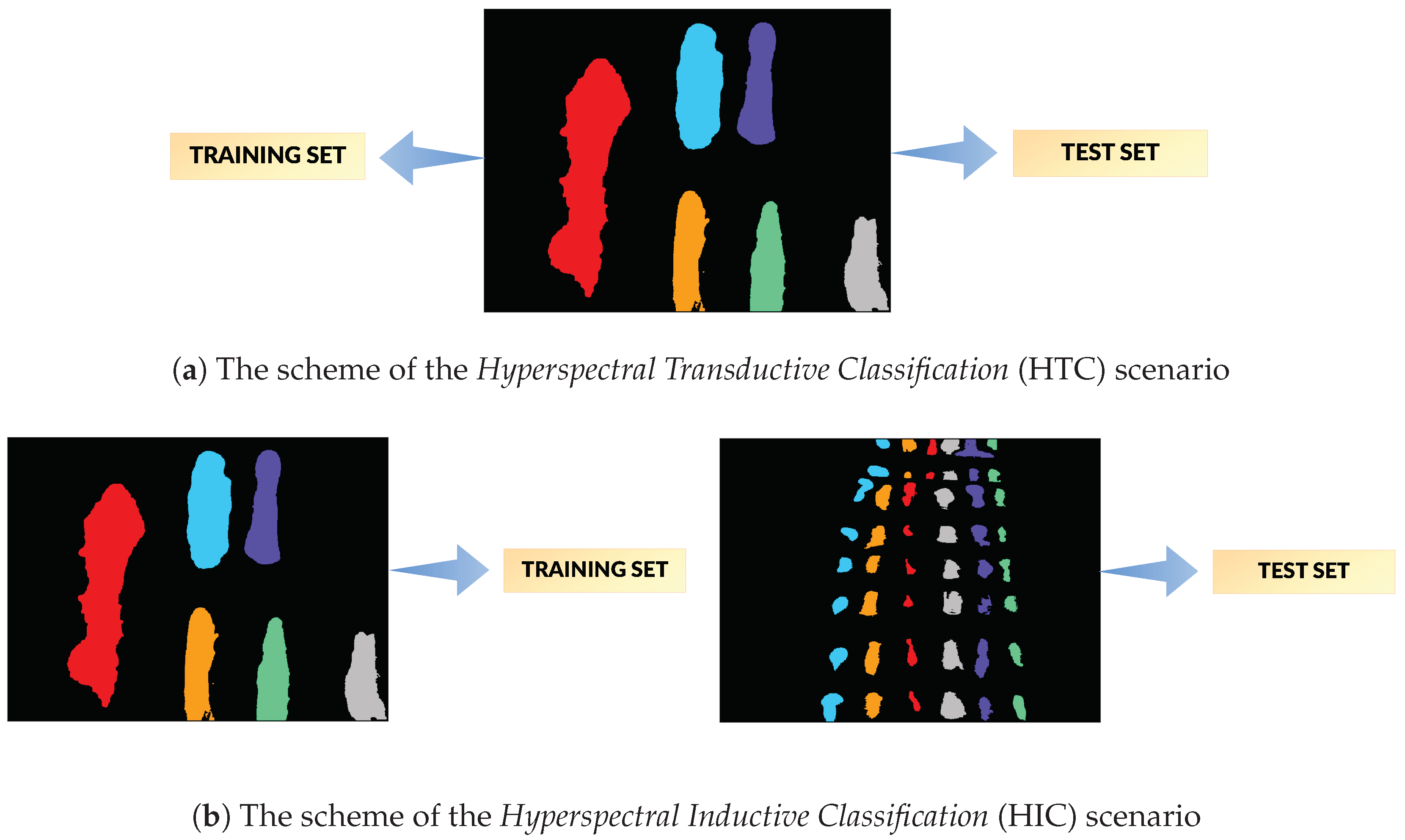
Hyperspectral Transductive Classification (HTC) scenario treats every image in the dataset separately. In every experiment, the set of labelled pixels in the image is divided between the training and the test set, i.e., .
Hyperspectral Inductive Classification (HIC) uses a pair of images: a set of labelled pixels from one image is used to train the classifier which is later tested on full set of labelled pixels from the second image.
Based on the discussion in [
23], pairs of images for training and testing the classifier were chosen in the HIC experiment: a
F/frame is used for training, as it simulates the lab acquired reference data, while a
E/comparison scene image is used as test, as it simulates a real-life crime scene. The complexity of HIC is enhanced by the fact that not every
F image has its
E counterpart acquired at the same time. Among pairs are ones with corresponding acquisition times (e.g.,
F(7)→E(7)), acquisition times differing by hours (e.g.,
F(1a)→E(1)) or days (e.g.,
F(2)→E(7)). Those differences are introduced on purpose, to allow examination of the effect of time-related change of spectral shape on the classification performance. For similar reasons, we have included two pairs
F(2)↔F(2k); while they share a similar scene, they were acquired with different cameras and thus allow to investigate the effect of the equipment change.
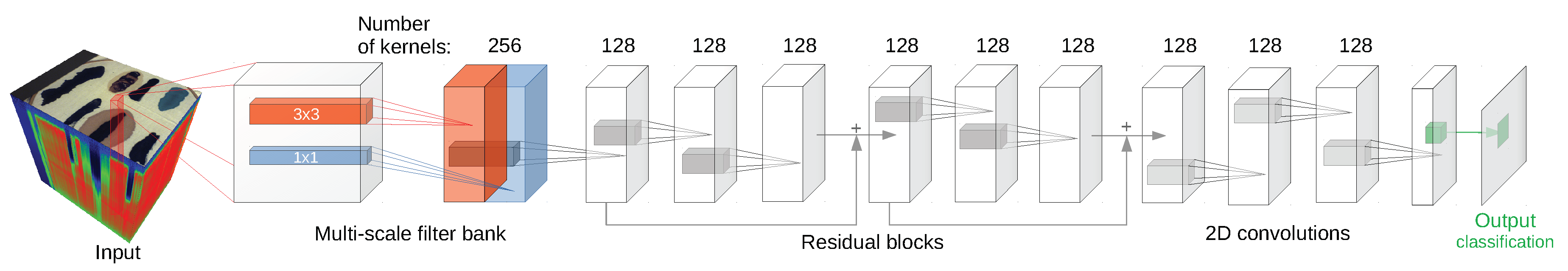
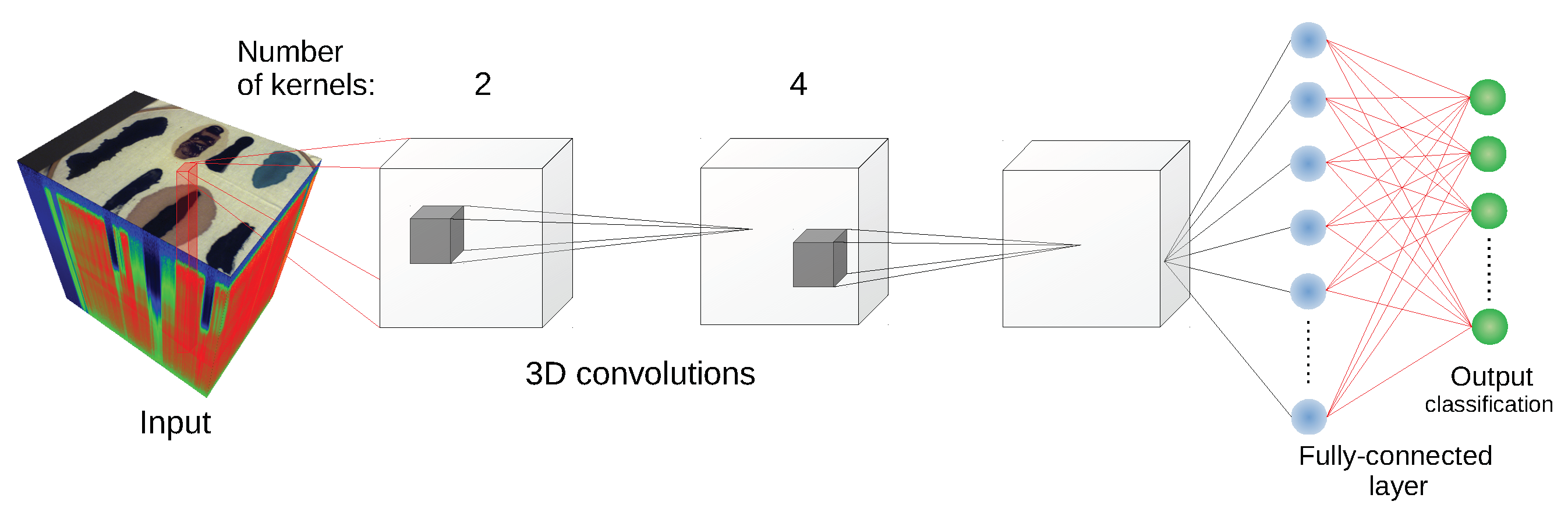
Since some of the architectures considered (e.g., 2D CNN [
24] or 3D CNN [
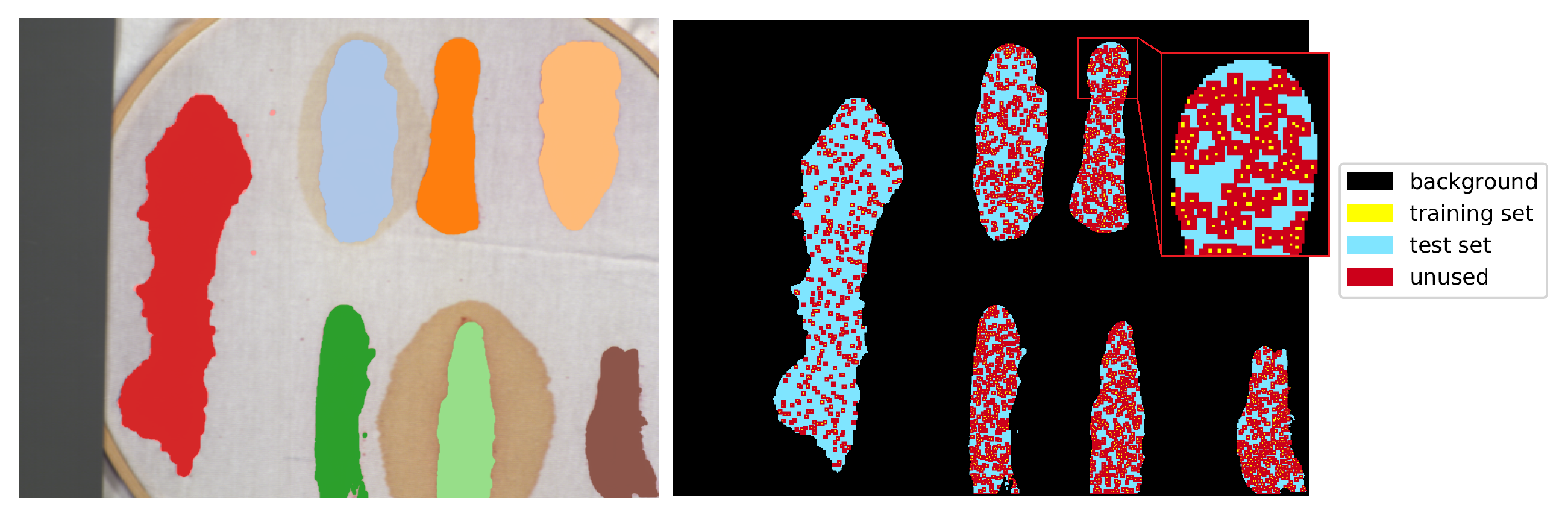
11]) use a block of pixels as an input, there was a need to create a uniform training/testing picture split for every image. The objective was to avoid patches from the training set having non-empty intersections with patches from the test set. For each class, a set of
samples, where
is the number of pixels of the least numerous class, was randomly, uniformly selected as training pixels. Each class was thus represented by the same number of samples. The 2-pixel neighbourhood of the training set—a maximal size of the neighbourhood used by tested architectures—was marked as unusable, along with the margin at the image borders of the same dimension. All of the remaining labelled pixels formed a test set. An example illustration of the effect of this procedure is presented in
Figure 9. For every image
different sets of randomized training pixels were prepared; the same collection of training sets was used for each network, ensuring comparability of results between individual architectures. The results for each scenario of network type/image configuration were averaged over 10 runs with the above specified fixed training datasets. Given 17 image scenarios, seven methods and ten runs for each image/method configuration, the total number of runs was
.
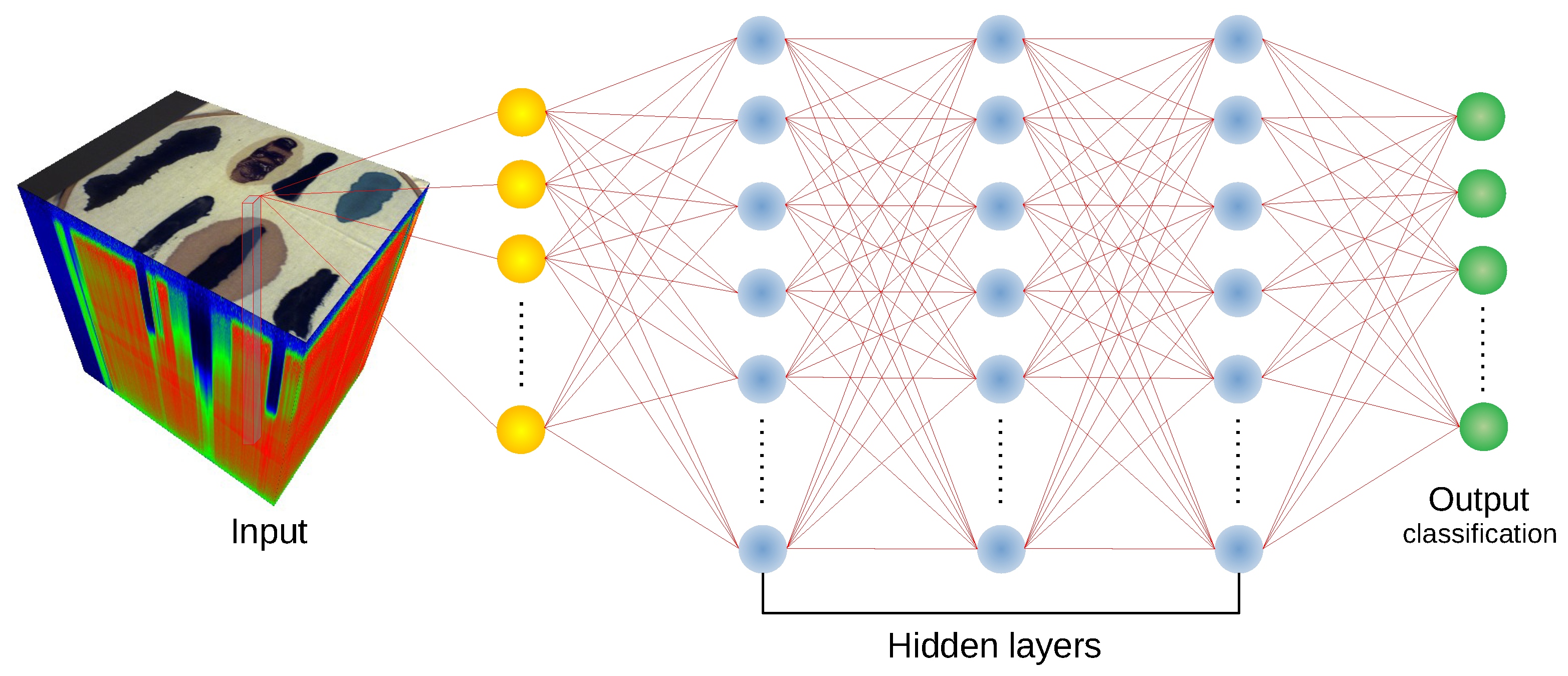
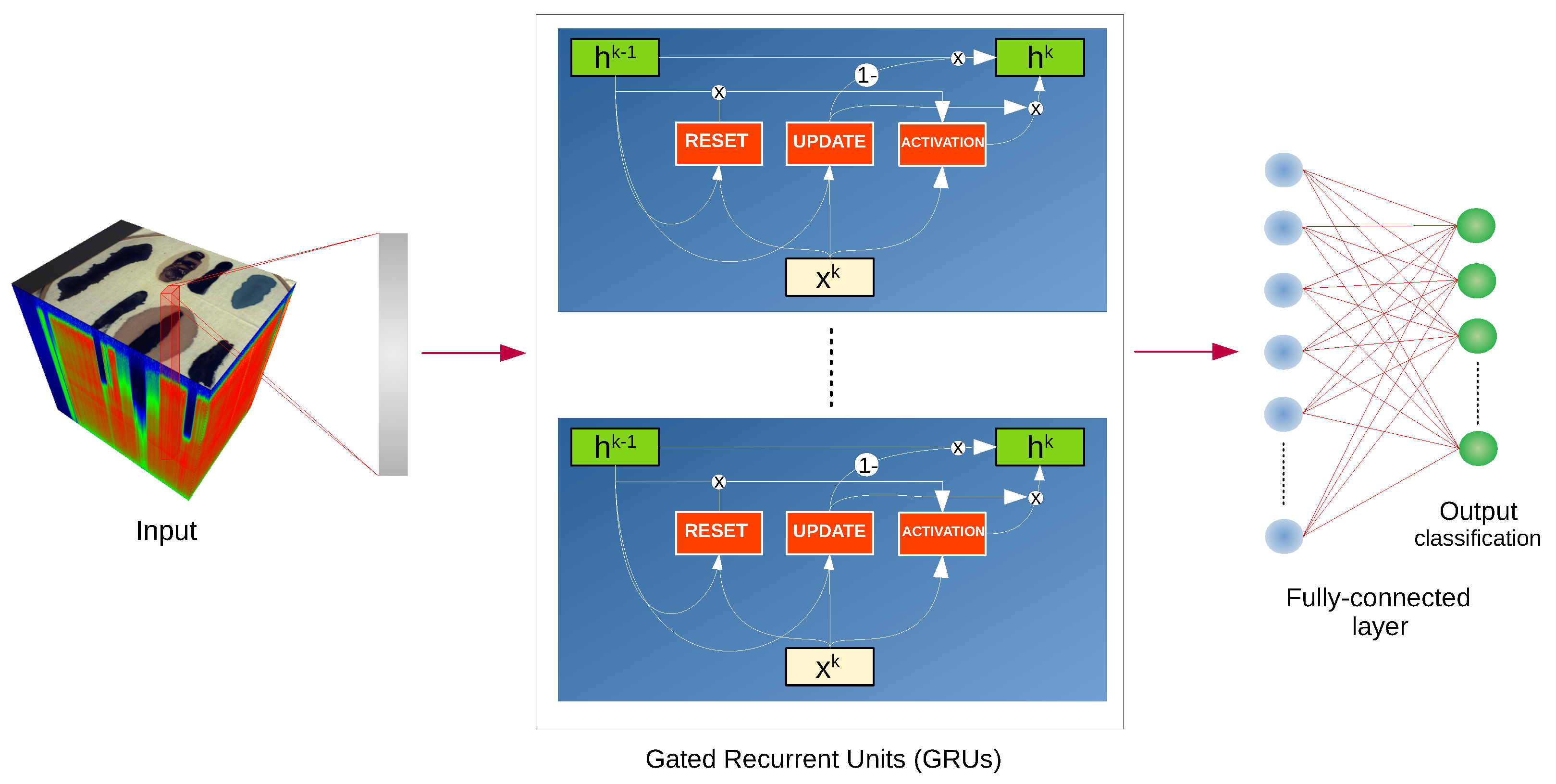
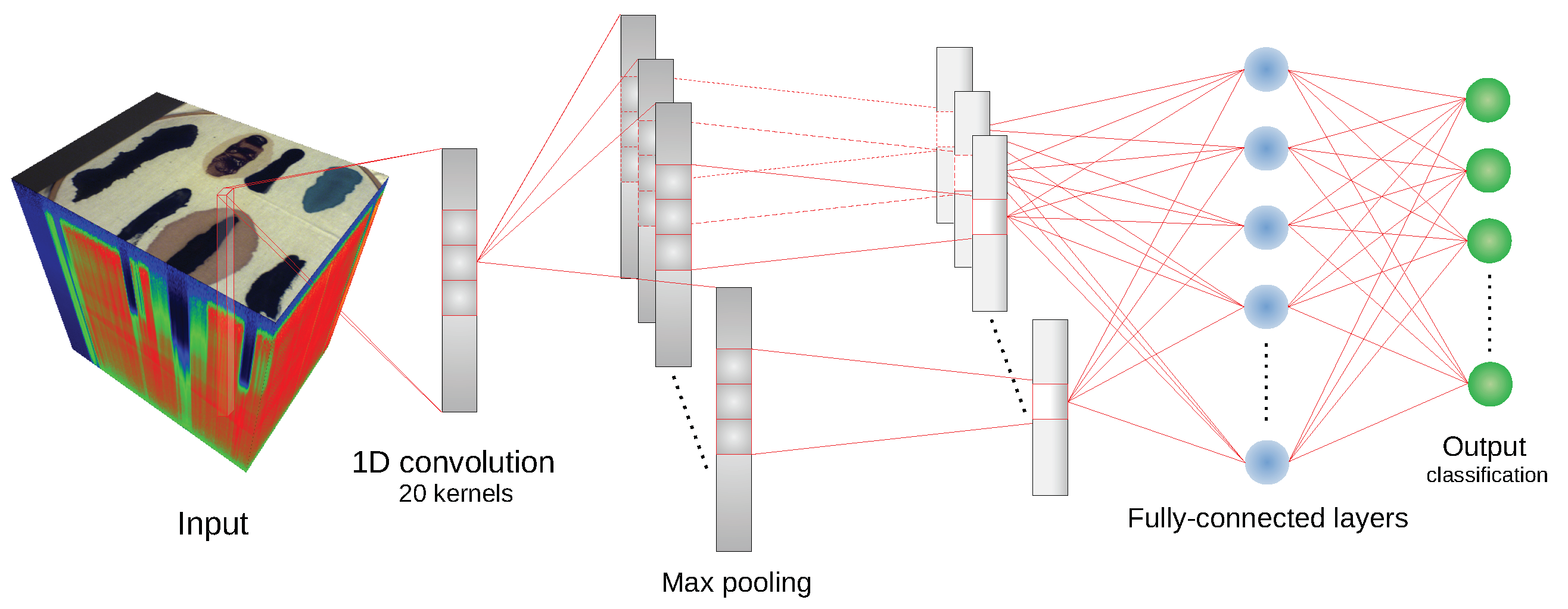
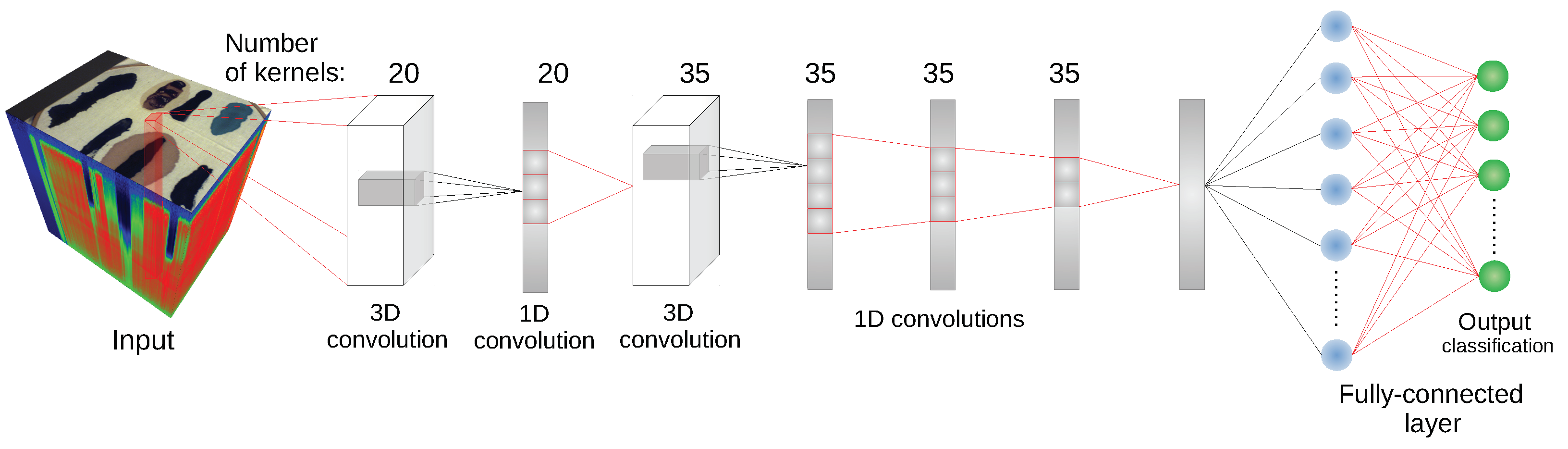
We have used the implementation of network architectures provided with DeepHyperX library [
19]. The models were initialized with the default values of hyperparameters as distributed with the library. Initial trial runs were performed to verify the general performance and assess the stability and suitability of the hyperparameter choice. In all but one case the default parameters were used for the main experiment. For the one case—1D CNN [
10] architecture—a minor adjustment was made: number of epochs was increased to
and batch size was decreased to
. While there exists a possibility that extensive hyperparameter tuning would improve performance of the models, it would also make it more difficult to compare our results with [
19] and assess generalization capability of each network. By applying the architectures to our data with similar hyperparameters as in [
19], we broaden the perspective while at the same time testing the robustness of the default hyperparameter choice. Additionally, hyperparameter optimisation requires a high degree of intimacy with a given architecture; this poses a risk to produce more effective optimisations for some models than the others, thus introducing a bias in the results. Too much optimisation may overtrain a model to a given dataset, which would introduce additional bias, and would go against our objective of ‘stress testing’ the selected architectures.
For the reasons discussed in the previous paragraph, we have not used any preprocessing that would significantly transform the spectra. The only preprocessing performed, as suggested in [
23], was spectra normalisation: for each image, every hyperspectral pixel vector was divided by its median. This procedure is intended to compensate for non-uniform lightning exposure.
As a reference for the tested deep learning architectures, we tested the SVM classifier [
51]. For its construction, we have used the Radial Basis Function (RBF) kernel and parameters chosen through internal cross-validation from a range of
and
, where
with
is the number of bands and
v is the variance of data.
3.4. Experimental Results
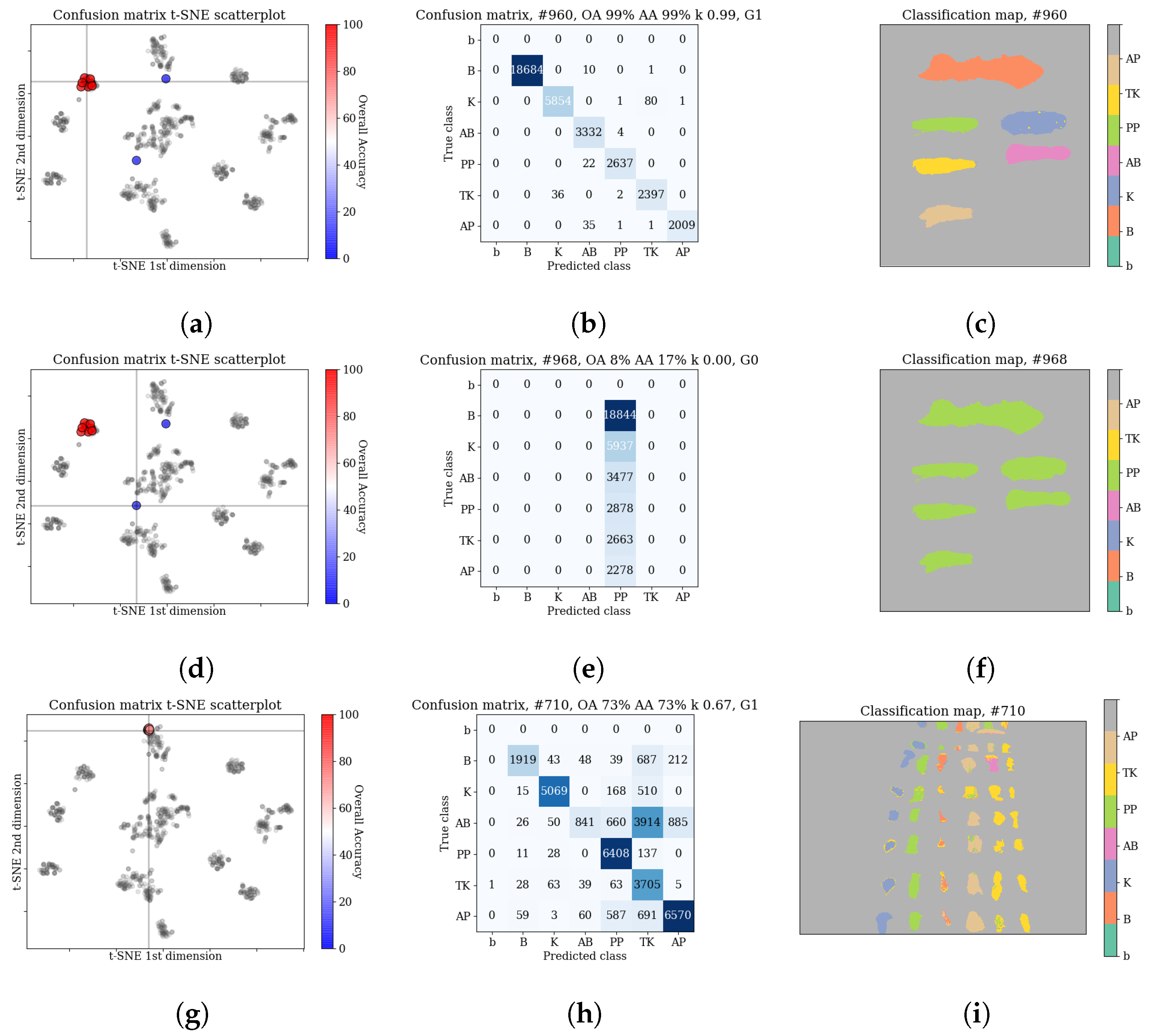
The qualitative analysis of the results, discussed in
Section 3.3.2, identified
cases of failed network training. In all those cases the predicted labels belonged to a single class (In one of those cases, all but one (
samples) were classified as one class, and a single sample was marked as an another class. For the sake of simplicity, we classify this case as in line with the rest of training failures.). We have removed those cases before computing statistics of the results. The scores—means and standard deviations of OA, AA and kappa—for the transductive classification (HTC experiment) are presented in
Table 1, while results for inductive classification (HIC experiment) are presented in
Table 2. Additionally, per-class error percentages are presented in
Table 3,
Table 4,
Table 5 and
Table 6. A graphical presentation of the results in
Table 1 and
Table 2 is in
Figure 11. The detailed information about running times of applied methods are presented in
Appendix A.
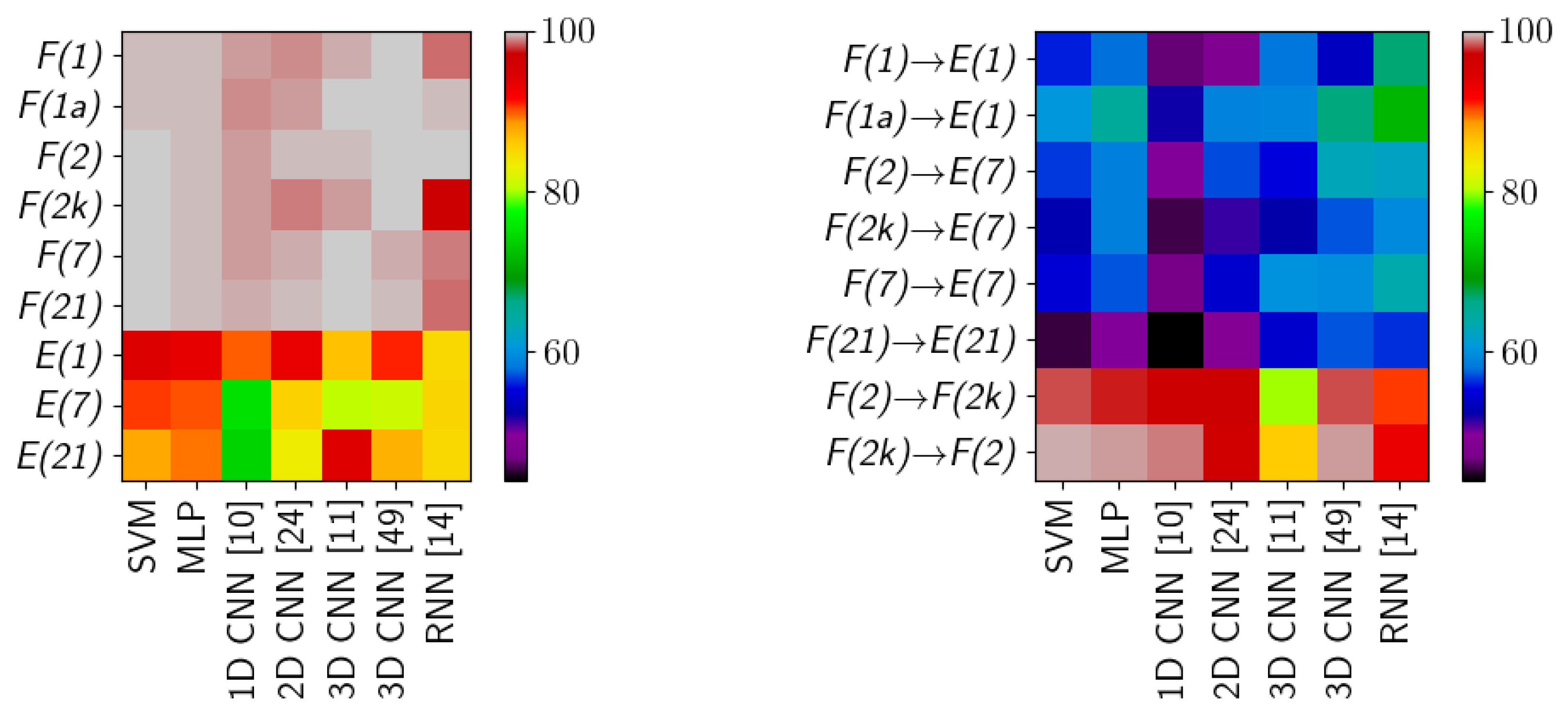
The scores for HTC form two groups, clustered around the
F/frame and
E/comparison scene type images. For
F/frame scenes the results are consistently high for all methods, often above 99%. Those images have a simple, uniform white background which does not interfere with individual class spectra. Left alone, those spectra are easily modelled by methods considered in this study. Inspection of an averaged confusion matrix reveals that the most common errors are misclassifications between
ketchup and
tomato concentrate, which is not unexpected as they represent similar substances. Qualitative inspection of individual results shows that while the scores are similar, the types of errors for individual methods are distinct, i.e., some models ‘favour’ certain classes (see
Table 3). For example, 1D CNN [
10] has the highest error rate for
ketchup (
) and RNN [
14] for
tomato concentrate (
); another example is the range of errors, e.g.,
artificial blood is from
(3D CNN [
49]) to
(RNN [
14]). This suggest that various architectures are able to model different features of the spectra. Quantitative investigation of scatterplots of confusion matrices (see
Section 3.3.2) reveals that in a small number of cases, errors are concentrated mostly within a subset of classes, e.g., ‘bloodlust’ when the model has the tendency to label a sample as
blood, or ‘artistic differences’ when the model tends to misclassify among the
paints classes. We discuss possible reasons for that in the ‘Discussion’ section.
Results are visibly lower for
E/comparison scene images, where the decrease in accuracy is about 5–10%. Quantitative inspection reveals that some architectures (SVM, MLP, 1D CNN [
10], 2D CNN [
24]) have consistent results, i.e., models from different runs have very similar scores and confusion matrices; while other (3D CNN [
11,
49], RNN [
14]) have some share of underperforming models, which have lower accuracy and distinct confusion matrices. The latter effect is responsible for both a lower mean score and the increase in standard deviation. In comparison to a set of scenarios discussed in the previous paragraph, there is a visible change not only in magnitude of errors for individual classes (see
Table 4), but also in their order—some classes which had lower error rate now have a higher one. For example, while the
blood class has a range of errors 9.9–15.6%, the
poster paint has a range of 1.8–2.5% and
tomato concentrate 16.4–26.4%. In those images the scene background is composed of objects with complex spectra, which forms spectral mixtures with classes to be recognized, making class spectra more diverse and challenging to model. This phenomenon is known to be present in many hyperspectral images [
21], and the enhanced scene complexity translates into lower scores.
The inductive classification (HIC experiment) is a significantly more challenging scenario, where different images are used for training and testing. In some cases, e.g.,
F(2k)→E(7), the difference can be in: scene composition (
frame vs
comparison scene), sample characteristics (two day old samples for training, seven day old for testing) and acquisition equipment (SPECIM vs SOC-710 camera). One of the tested inductive cases is
F(2k)↔F(2), where the same frame scene is independently imaged with two different cameras. The mean scores are lower than HTC
F/frame, but higher than HTC
E/comparison scene. Some networks have their score lowered by particular classes (e.g., 3D CNN [
11] with
tomato concentrate,
errors or RNN [
14] with
artificial blood,
, see
Table 5). Some networks (3D CNN [
11] in particular) have higher percentage of less performing models, which are responsible for lower scores. The change in acquisition device produces distinct, in relation to other scenarios, patterns in the results. The training on higher quality device (SPECIM,
F(2k)) produces better scores than the opposite scenario. Both the MLP and the reference SVM classifier achieve high accuracy in this case.
The final case—training on
F/frame and testing on
E/comparison scene—is viewed by the authors as a true application-related test of hyperspectral classification. The
F/frame image simulates laboratory-taken image, which is then used to investigate a sample forensic scene. This adds the variation of classes spectra and lighting on top of complexities already present in the
comparison scene. The first observation is in that both the reference SVM classifier and the simple MLP architecture are outperformed by more complex models such as RNN [
14] or 3D CNN [
49]. In this setting the errors of classes
ketchup and
artificial blood again dominate (42.7–84.6% and 76.7–85.3% respectively). The
blood class, detection of which is interesting from an application perspective, has a range of errors 29.4–44.8%. The confusion matrices show large variations and no noticeable patterns.
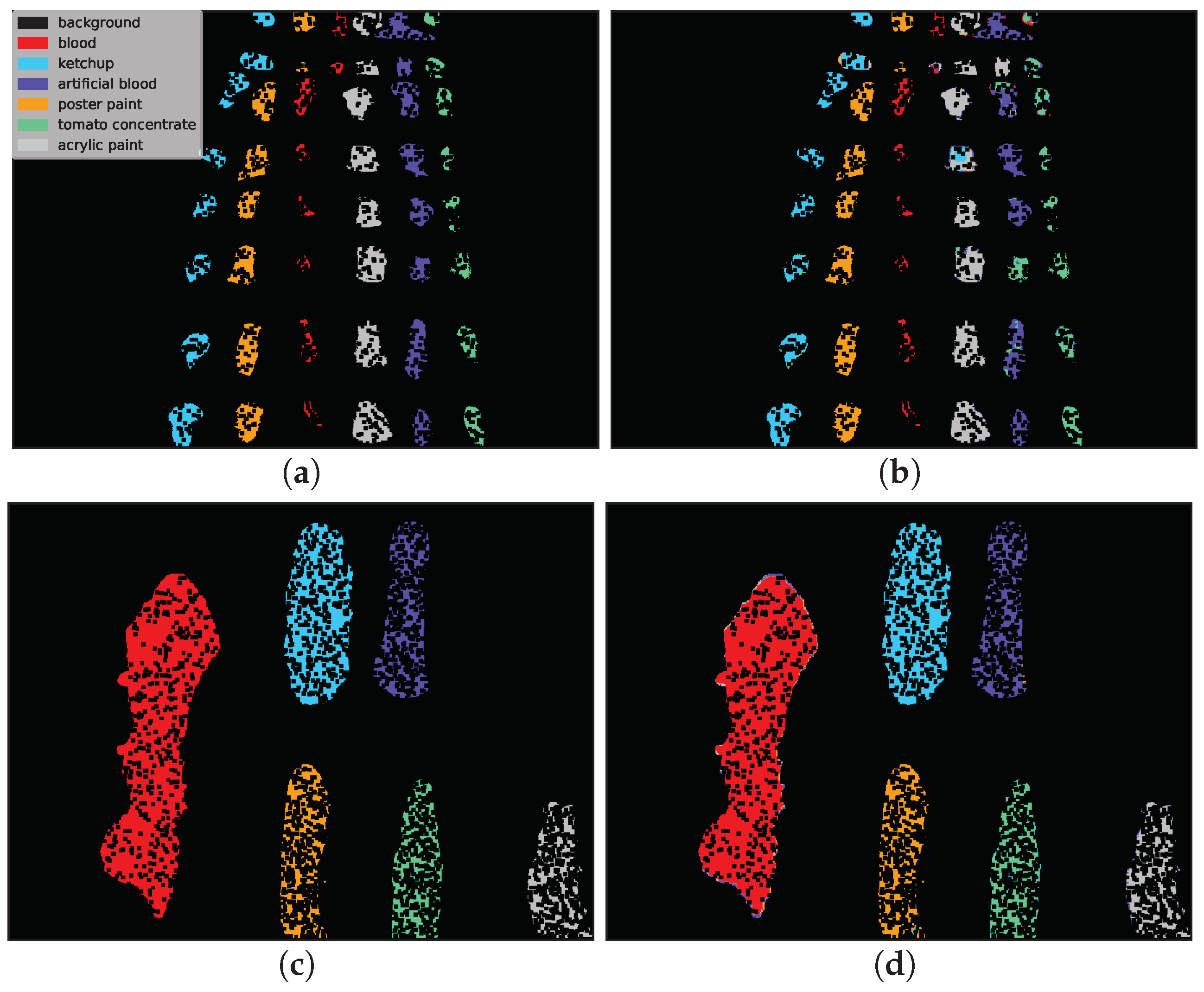
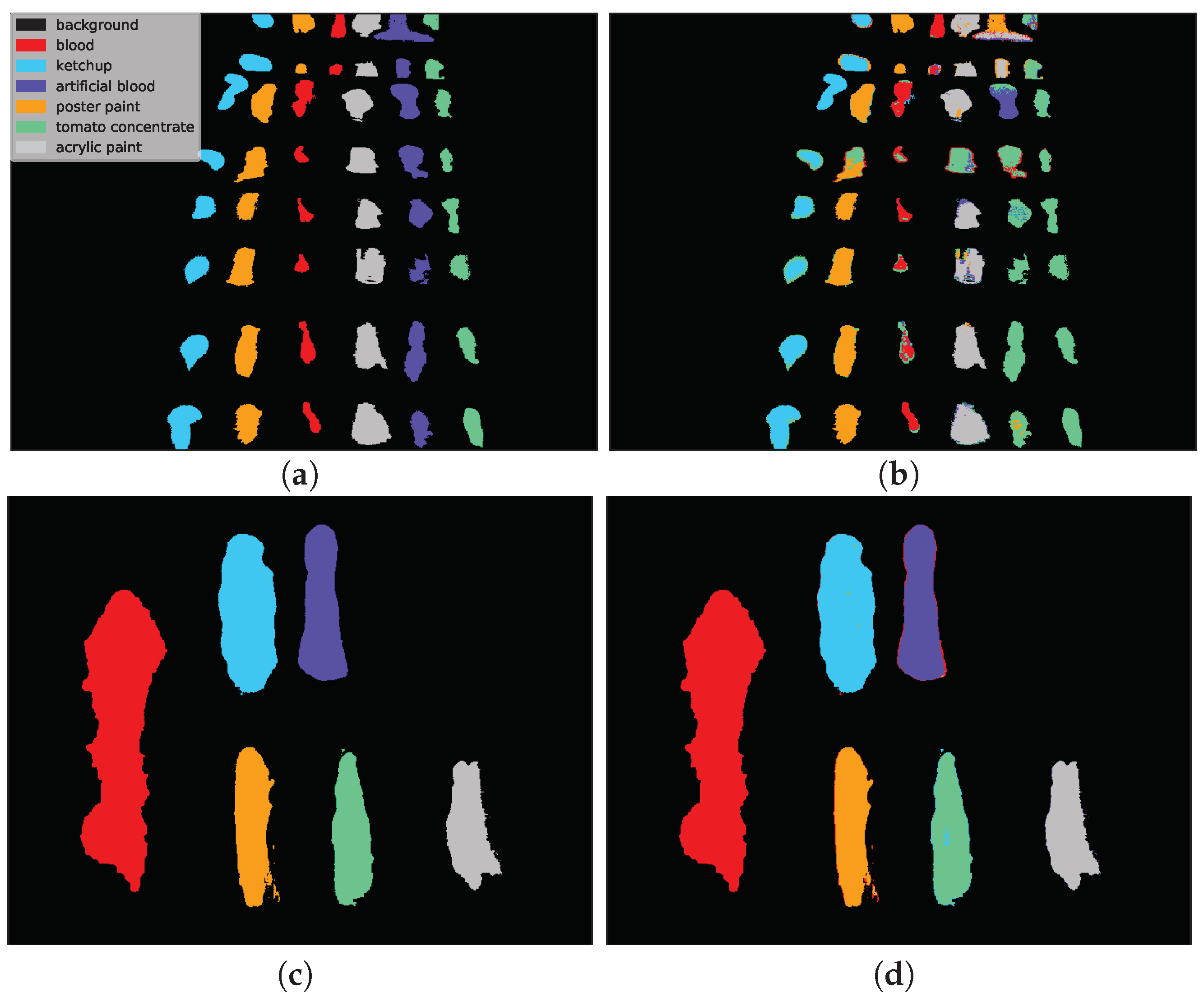
Example classification maps for different architectures in the HTC and HIC scenarios are presented in
Figure 12 and
Figure 13, respectively.
Figure 12 presents example predictions for the HTC scenario: 1D CNN [
10] for
E(1) image and 2D CNN [
24] for
F(1) image. Note that in the HTC scenario only a subset of available pixels could be used as a test set (see
Section 3.2) which results in ‘holes’ in classification maps. In the first row we observe misclassifications of
artificial blood class, where some pixels were incorrectly recognized as
tomato concentrate and
acrylic paint. The example in the second row was easier to classify and errors were mainly concentrated on the borders of substances (uncertain areas where spectra are likely to be mixed).
Figure 13 presents the HIC scenario. The first row is related to
F(1) → E(1) case and presents a sample run for RNN [
14] network. One can observe that almost all pixels of
artificial blood class were incorrectly classified as
tomato concentrate,
acrylic paint or
poster paint. The prediction presented in the second row i.e., 3D CNN [
49] network for
F(2) → F(2k) scenario is highly accurate except for some border pixels.