Breast Cancer Histopathology Image Classification Using an Ensemble of Deep Learning Models
Abstract
1. Introduction
2. Related Work
3. Materials and Methods
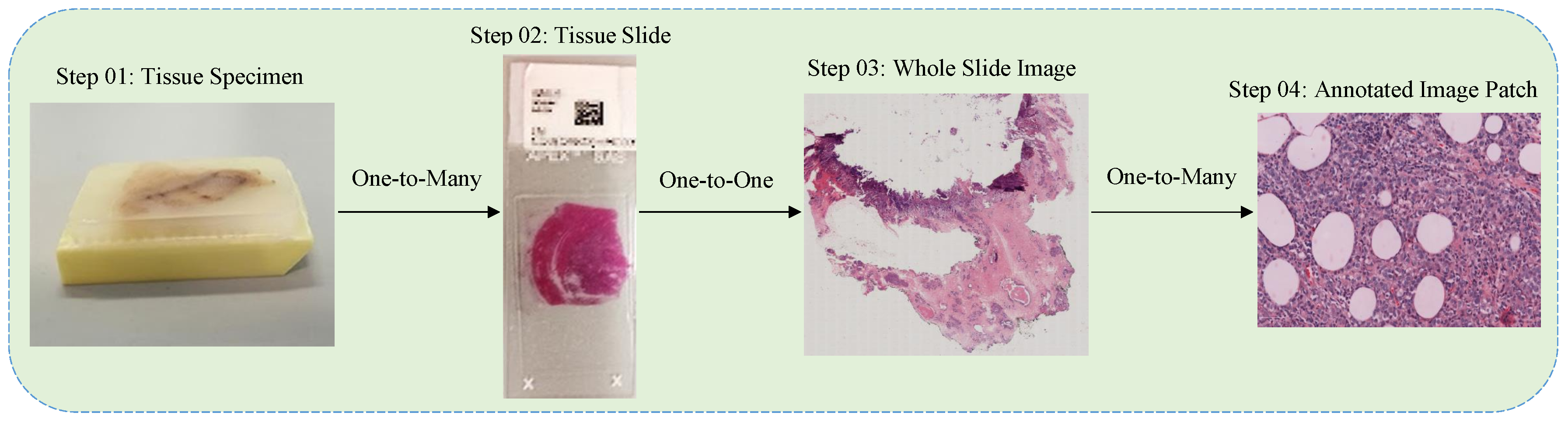
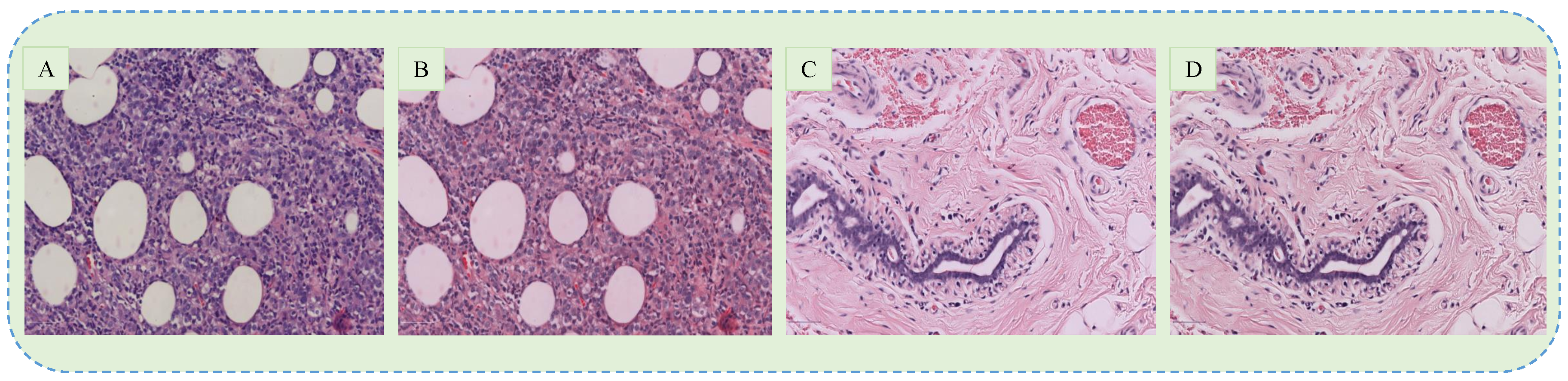
3.1. Data Collection
3.2. Preprocessing
3.3. Training Criteria
3.4. Data Augmentation
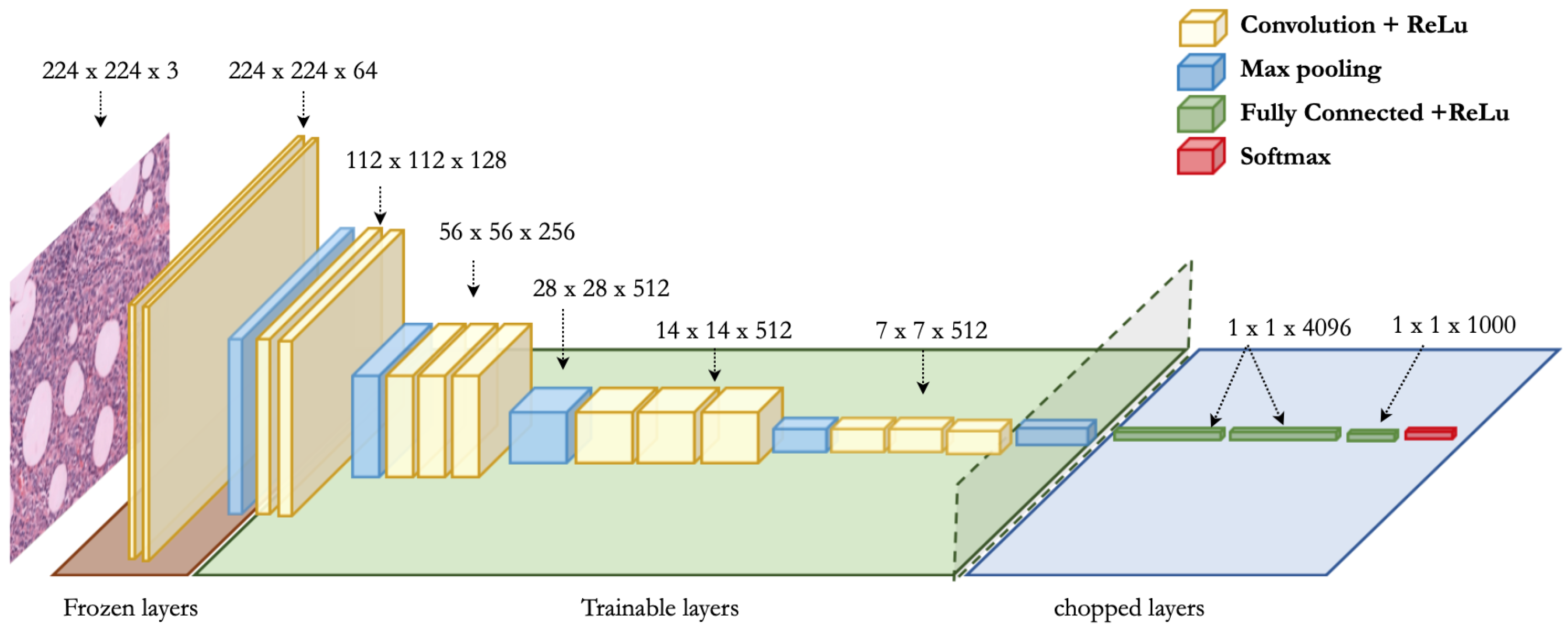
3.5. VGG Architecture
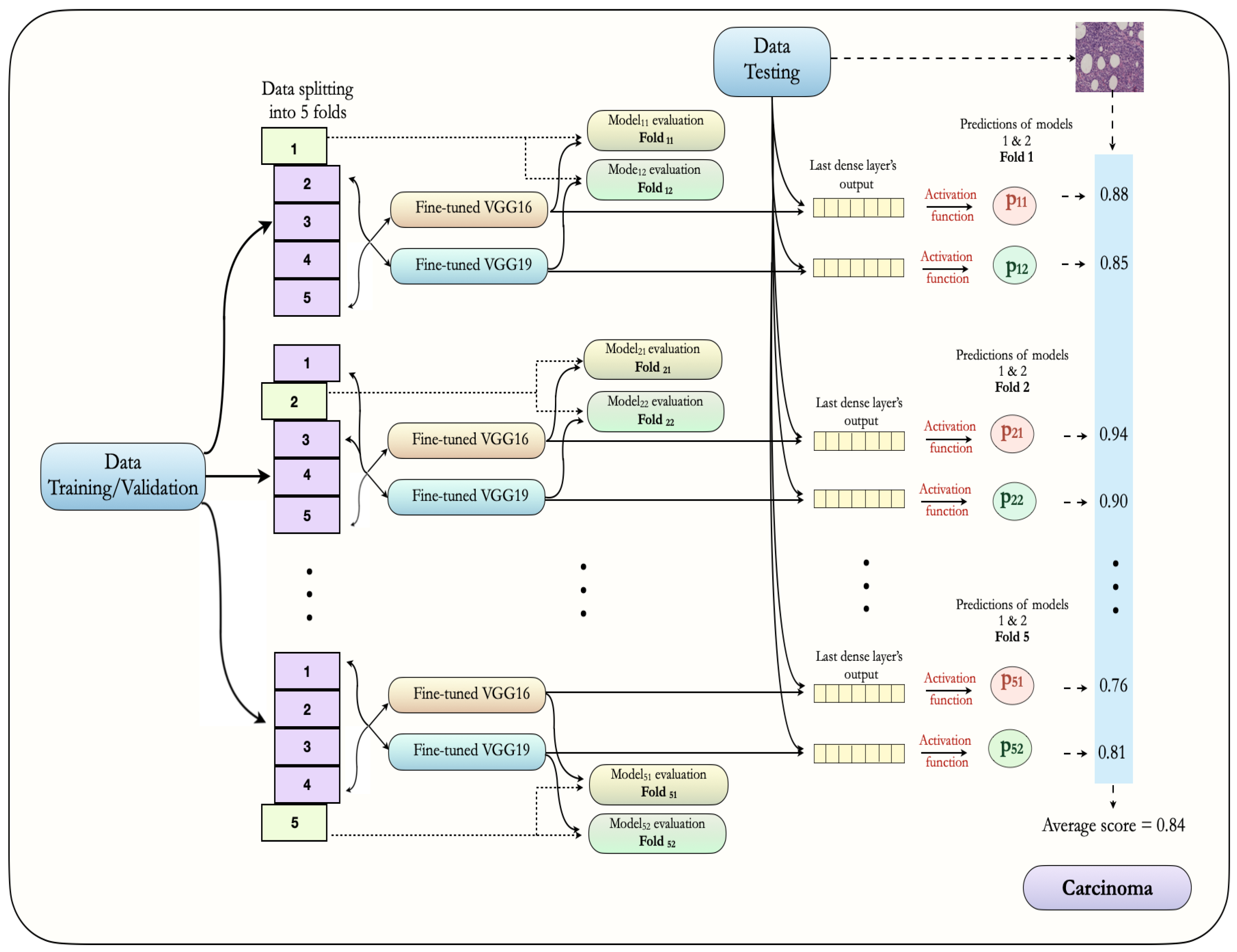
3.6. Proposed Ensemble Approach
4. Experimental Setup
4.1. Implementation
4.2. Evaluation Metrics
- Precision: It quantifies exactness of a model, and represents the ratio of carcinoma images accurately classified out of the union of predicted same-class images.
- Sensitivity: Sensitivity, also called “recall” computes completeness of a model. It represents the ratio of images accurately classified as carcinoma out of the total number of carcinoma images.
- Accuracy: It evaluates correctness of a model, and is the ratio of the number of images accurately classified out of the total number of testing images.
- F1-score: It represents the harmonic average of precision and recall, and is usually used for the optimization of a model towards either precision or recall.
4.3. Hyperparameter Tuning
5. Results and Discussion
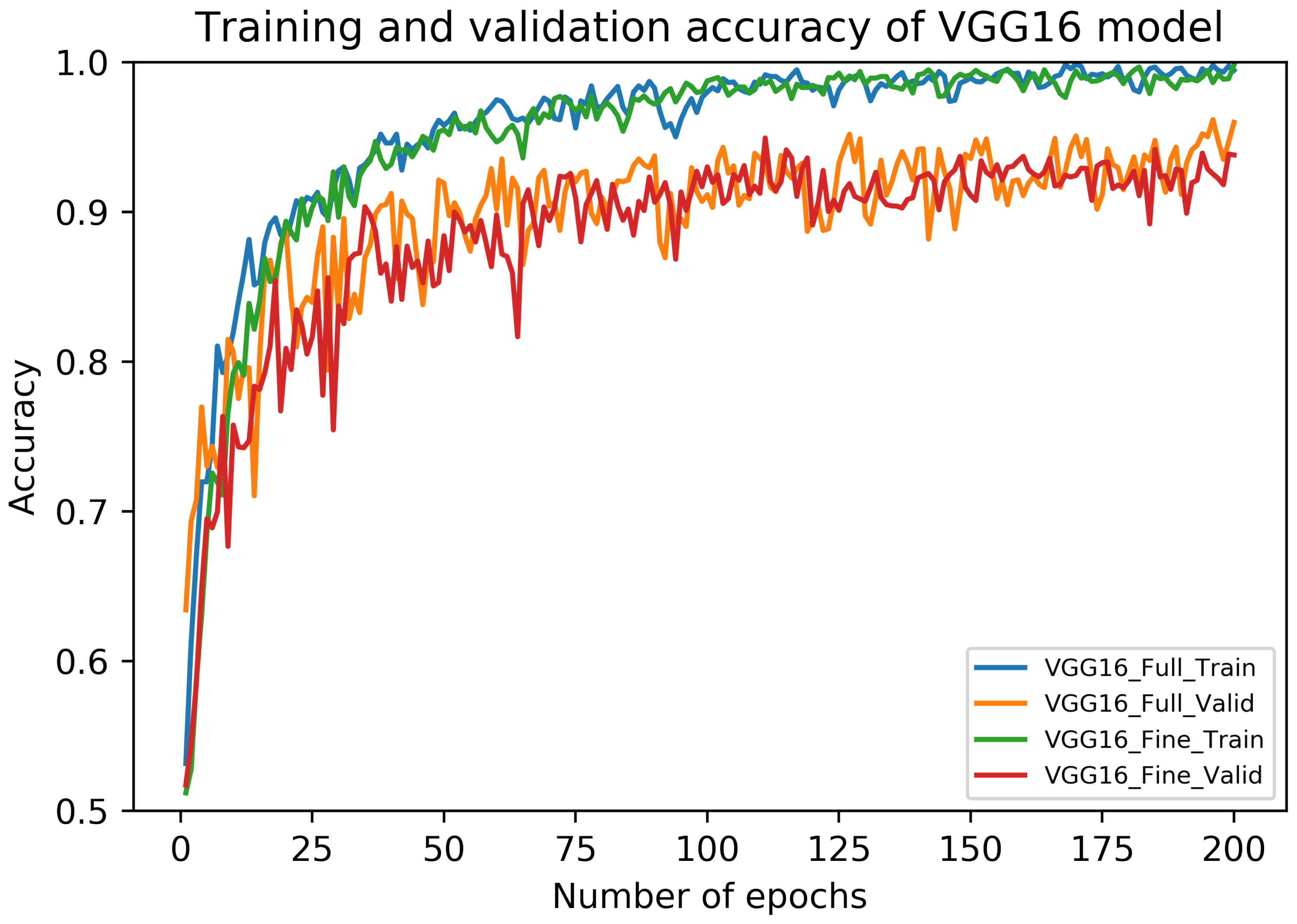
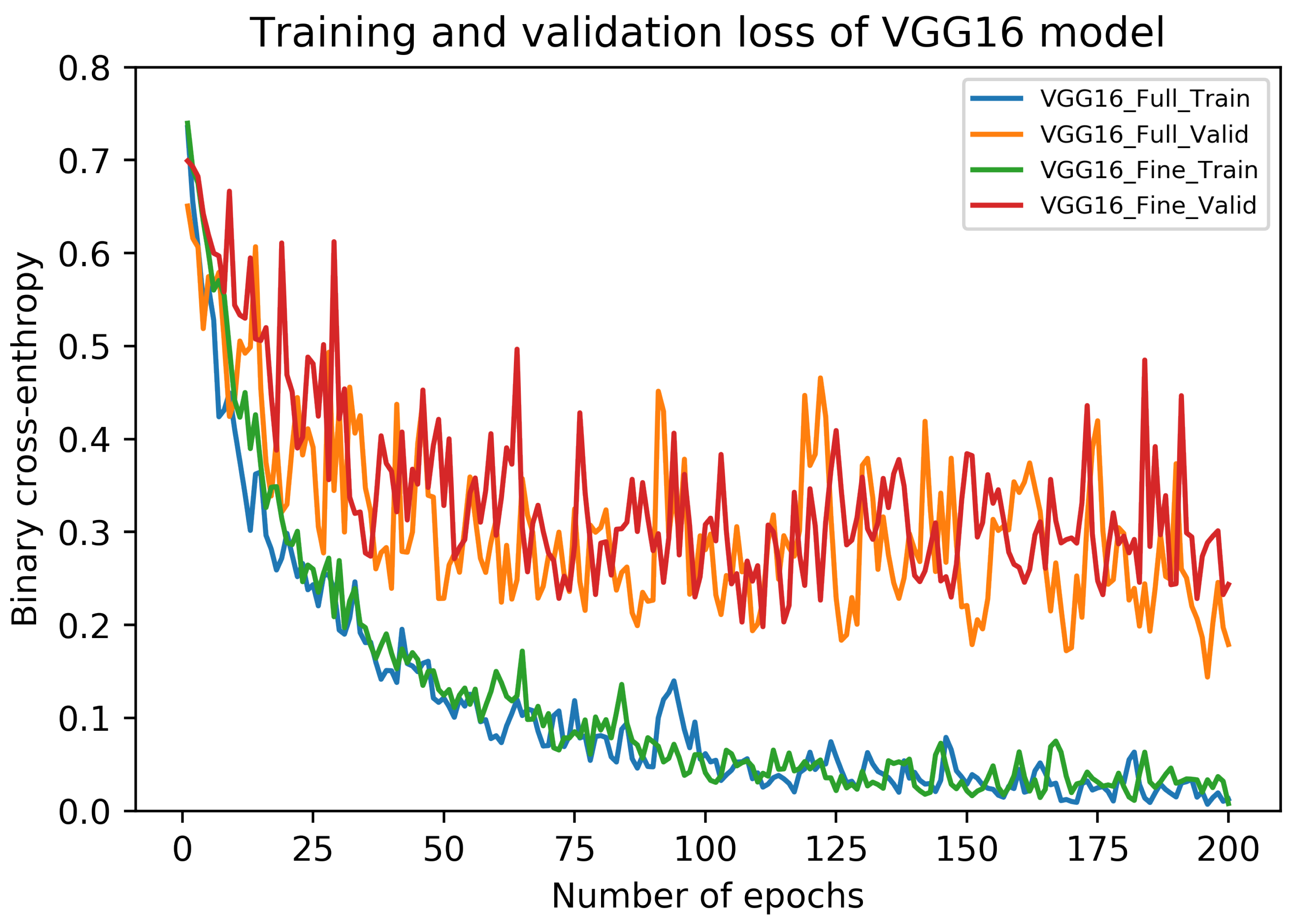
5.1. Results of VGG16 Architecture
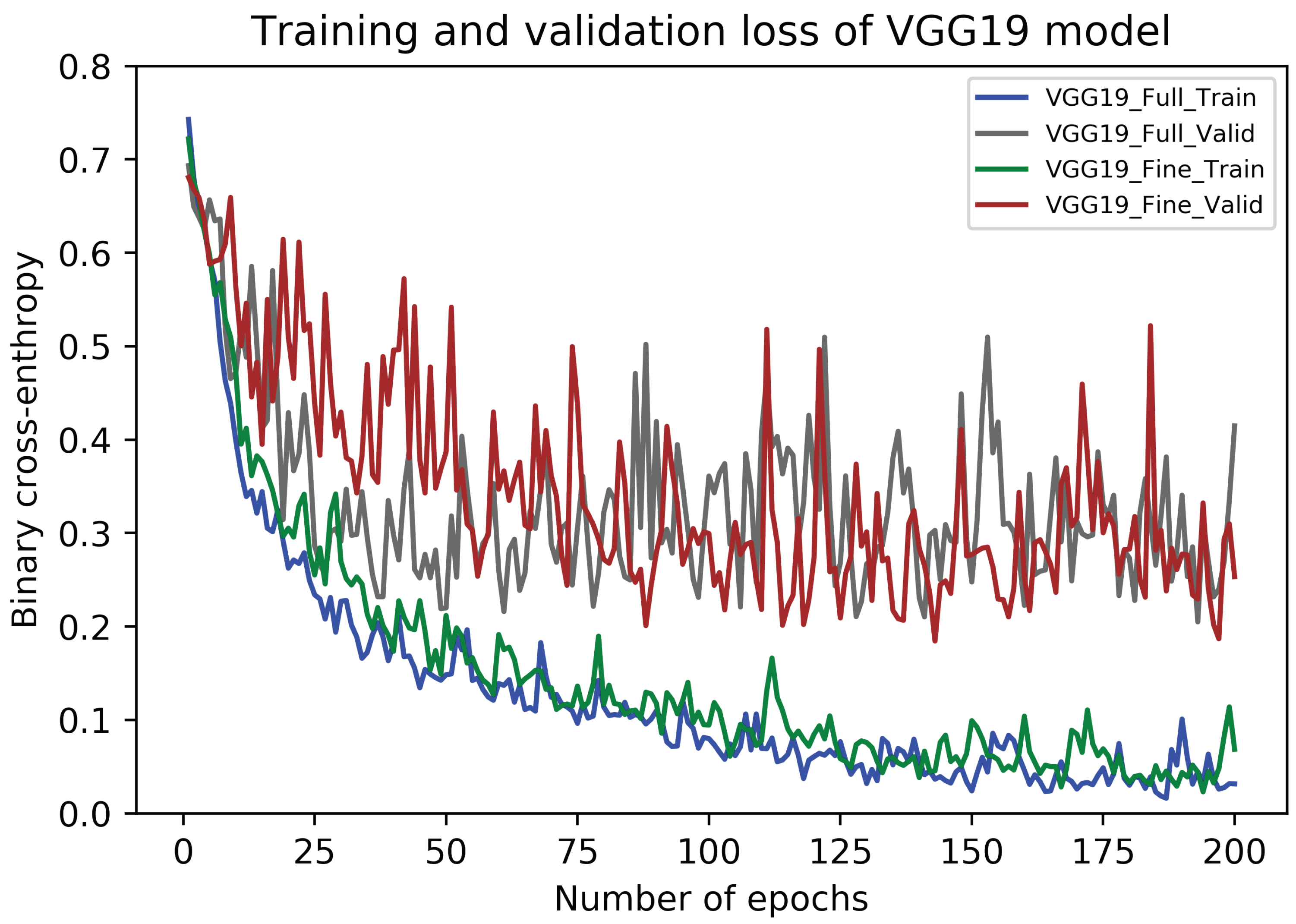
5.2. Results of VGG19 Architecture
5.3. Results of Ensemble VGG16 and VGG19
5.4. Discussion
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| GBD | Global Burden of Disease |
| MRI | Magnetic Resonance Imaging |
| CNN | Convolutional Neural Network |
| LSTM | Long Short Term Memory |
| KNN | K-Nearest Neighbor |
| NB | Naive Bayes |
| DT | Decision Tree |
| SVM | Support Vector Machine |
| H&E | Hematoxylin and Eosin |
| SVD | Singular Value Decomposition |
| OD | Optical Density |
| WSI | Whole Slides Image |
| ER | Estrogen Receptor |
| PR | Progesterone Receptor |
| HER2 | Human Epidermal growth factor Receptor-2 |
References
- Fitzmaurice, C. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017: A systematic analysis for the global burden of disease study. JAMA Oncol. 2019, 5, 1749–1768. [Google Scholar]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Weigelt, B.; Geyer, F.C.; Reis-Filho, J.S. Histological types of breast cancer: How special are they? Mol. Oncol. 2010, 4, 192–208. [Google Scholar] [CrossRef] [PubMed]
- Dromain, C.; Boyer, B.; Ferré, R.; Canale, S.; Delaloge, S.; Balleyguier, C. Computed-aided diagnosis (CAD) in the detection of breast cancer. Eur. J. Radiol. 2013, 82, 417–423. [Google Scholar] [CrossRef]
- Wang, L. Early Diagnosis of Breast Cancer. Sensors 2017, 17, 1572. [Google Scholar] [CrossRef]
- Veta, M.; Pluim, J.P.W.; Diest, P.J.V.; Viergever, M.A. Breast Cancer Histopathology Image Analysis: A Review. IEEE Trans. Biomed. Eng. 2014, 61, 1400–1411. [Google Scholar] [CrossRef]
- Dimitriou, N.; Arandjelović, O.; Caie, P.D. Deep Learning for Whole Slide Image Analysis: An Overview. Front. Med. 2019, 6. [Google Scholar] [CrossRef]
- Elmore, J.G.; Longton, G.M.; Carney, P.A.; Geller, B.M.; Onega, T.; Tosteson, A.N.A.; Nelson, H.D.; Pepe, M.S.; Allison, K.H.; Schnitt, S.J.; et al. Diagnostic Concordance among Pathologists Interpreting Breast Biopsy Specimens. JAMA 2015, 313, 1122. [Google Scholar] [CrossRef]
- Aswathy, M.; Jagannath, M. Detection of breast cancer on digital histopathology images: Present status and future possibilities. Inform. Med. Unlocked 2017, 8, 74–79. [Google Scholar] [CrossRef]
- Robertson, S.; Azizpour, H.; Smith, K.; Hartman, J. Digital image analysis in breast pathology—From image processing techniques to artificial intelligence. Transl. Res. 2018, 194, 19–35. [Google Scholar] [CrossRef]
- Lowe, D. Object recognition from local scale-invariant features. In Proceedings of the Seventh IEEE International Conference on Computer Vision, Corfu, Greece, 20–25 September 1999. [Google Scholar]
- Bay, H.; Tuytelaars, T.; Gool, L.V. SURF: Speeded Up Robust Features. In Proceedings of the Computer Vision—ECCV 2006 Lecture Notes in Computer Science, Graz, Austria, 7–13 May 2006; pp. 404–417. [Google Scholar]
- Ojala, T.; Pietikainen, M.; Maenpaa, T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 971–987. [Google Scholar] [CrossRef]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Laak, J.A.V.D.; Ginneken, B.V.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Zahia, S.; Zapirain, M.B.G.; Sevillano, X.; González, A.; Kim, P.J.; Elmaghraby, A. Pressure injury image analysis with machine learning techniques: A systematic review on previous and possible future methods. Artif. Intell. Med. 2020, 102, 101742. [Google Scholar] [CrossRef]
- Bour, A.; Castillo-Olea, C.; Garcia-Zapirain, B.; Zahia, S. Automatic colon polyp classification using Convolutional Neural Network: A Case Study at Basque Country. In Proceedings of the 2019 IEEE International Symposium on Signal Processing and Information Technology (ISSPIT), Louisville, KY, USA, 10–12 December 2019. [Google Scholar]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Proceedings of the Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the International Conference on Learning Representations (ICLR), San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Kowal, M.; Filipczuk, P.; Obuchowicz, A.; Korbicz, J.; Monczak, R. Computer-aided diagnosis of breast cancer based on fine needle biopsy microscopic images. Comput. Biol. Med. 2013, 43, 1563–1572. [Google Scholar] [CrossRef]
- Filipczuk, P.; Fevens, T.; Krzyzak, A.; Monczak, R. Computer-Aided Breast Cancer Diagnosis Based on the Analysis of Cytological Images of Fine Needle Biopsies. IEEE Trans. Med. Imaging 2013, 32, 2169–2178. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, B.; Coenen, F.; Lu, W. Breast cancer diagnosis from biopsy images with highly reliable random subspace classifier ensembles. Mach. Vis. Appl. 2012, 24, 1405–1420. [Google Scholar] [CrossRef]
- Bengio, Y.; Courville, A.; Vincent, P. Representation Learning: A Review and New Perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1798–1828. [Google Scholar] [CrossRef]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. A Dataset for Breast Cancer Histopathological Image Classification. IEEE Trans. Biomed. Eng. 2016, 63, 1455–1462. [Google Scholar] [CrossRef]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. Breast cancer histopathological image classification using Convolutional Neural Networks. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016. [Google Scholar]
- Bayramoglu, N.; Kannala, J.; Heikkila, J. Deep learning for magnification independent breast cancer histopathology image classification. In Proceedings of the 2016 23rd International Conference on Pattern Recognition (ICPR), Cancún, Mexico, 4–8 December 2016. [Google Scholar]
- Araújo, T.; Aresta, G.; Castro, E.; Rouco, J.; Aguiar, P.; Eloy, C.; Polónia, A.; Campilho, A. Classification of breast cancer histology images using Convolutional Neural Networks. PLoS ONE 2017, 12, e0177544. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Ren, F.; Wang, Z.; Wang, L.; Zhang, T.; Liu, Y.; Rao, X.; Zheng, C.; Zhang, F. Breast cancer histopathological image classification using a hybrid deep neural network. Methods 2020, 173, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; Mcart, D.G.; Dunne, P.D.; Mcquaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Macenko, M.; Niethammer, M.; Marron, J.S.; Borland, D.; Woosley, J.T.; Guan, X.; Schmitt, C.; Thomas, N.E. A method for normalizing histology slides for quantitative analysis. In Proceedings of the 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Boston, MA, USA, 28 June–1 July 2009. [Google Scholar]
- Bianconi, F.; Kather, J.N.; Reyes-Aldasoro, C.C. Evaluation of Colour Pre-processing on Patch-Based Classification of H&E-Stained Images. In European Congress on Digital Pathology; Springer: Cham, Switzerland, 2019; pp. 56–64. [Google Scholar]
- Yao, H.; Zhang, X.; Zhou, X.; Liu, S. Parallel Structure Deep Neural Network Using CNN and RNN with an Attention Mechanism for Breast Cancer Histology Image Classification. Cancers 2019, 11, 1901. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. Keras: Deep Learning Library. 2015. Available online: https://keras.io/api/preprocessing/image/ (accessed on 24 January 2020).
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; The MIT Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Xie, J.; Liu, R.; Luttrell, J.; Zhang, C. Deep Learning Based Analysis of Histopathological Images of Breast Cancer. Front. Genet. 2019, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Bardou, D.; Zhang, K.; Ahmad, S.M. Classification of Breast Cancer Based on Histology Images Using Convolutional Neural Networks. IEEE Access 2018, 6, 24680–24693. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Han, Z.; Wei, B.; Zheng, Y.; Yin, Y.; Li, K.; Li, S. Breast Cancer Multi-classification from Histopathological Images with Structured Deep Learning Model. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nahid, A.-A.; Mehrabi, M.A.; Kong, Y. Histopathological Breast Cancer Image Classification by Deep Neural Network Techniques Guided by Local Clustering. BioMed Res. Int. 2018, 2018, 2362108. [Google Scholar] [CrossRef] [PubMed]
- Deniz, E.; Şengür, A.; Kadiroğlu, Z.; Guo, Y.; Bajaj, V.; Budak, Ü. Transfer learning based histopathologic image classification for breast cancer detection. Health Inf. Sci. Syst. 2018, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Benhammou, Y.; Achchab, B.; Herrera, F.; Tabik, S. BreakHis based breast cancer automatic diagnosis using deep learning: Taxonomy, survey and insights. Neurocomputing 2020, 375, 9–24. [Google Scholar] [CrossRef]

| Images | Quantity | Color Model | Staining |
|---|---|---|---|
| carcinoma | 437 | RGB | H & E |
| non-carcinoma | 408 | RGB | H & E |
| Total | 845 | RGB | H & E |
| No. of Images | Percentage | |
|---|---|---|
| Training | 540 | 64% |
| Validation | 135 | 16% |
| Test | 170 | 20% |
| Total | 845 | 100% |
| Parameters of Image Augmentation | Values |
|---|---|
| Zoom range | 0.2 |
| Rotation range | 40 |
| Width shift range | 0.2 |
| Height shift range | 0.2 |
| Horizontal flip | True |
| Fill mode | Reflect |
| Hyperparameters | VGG16 with Data Augmentation | VGG19 with Data Augmentation |
|---|---|---|
| Train approach | 5-fold cross-validation | 5-fold cross-validation |
| Optimizer | Adam | Adam |
| Loss function | Binary cross-entropy | Binary cross-entropy |
| Learning rate | ||
| Batch size | 32 | 32 |
| Convolution | with stride 1 | with stride 1 |
| Padding | Same | Same |
| Pooling | max-pooling with stride 2 | max-pooling with stride 2 |
| Epochs | 200 | 200 |
| Drop out | 0.3 | 0.3 |
| Regularizer | N/A | N/A |
| Architecture | Fully-trained and Fine-tuned | Fully-trained and Fine-tuned |
| Architecture | Folds | Confusion Matrices | Performance Evaluation (%) | Average (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Predict → Actual ↓ | NC | C | Precision | Recall | F1 | Test | Acc. | F1 | ||
| Fully-Trained VGG16 | Fold 1 | non-carcinoma | 75 | 7 | 97.40 | 91.46 | 94.34 | 82 | 94.71 | 94.70 |
| carcinoma | 2 | 86 | 92.47 | 97.73 | 95.03 | 88 | ||||
| Fold 2 | non-carcinoma | 65 | 17 | 90.28 | 79.27 | 84.42 | 82 | 85.88 | 85.80 | |
| carcinoma | 7 | 81 | 82.65 | 92.05 | 87.10 | 88 | ||||
| Fold 3 | non-carcinoma | 73 | 9 | 93.59 | 89.02 | 91.25 | 82 | 91.76 | 91.75 | |
| carcinoma | 5 | 83 | 90.22 | 94.32 | 92.22 | 88 | ||||
| Fold 4 | non-carcinoma | 70 | 12 | 95.89 | 85.37 | 90.32 | 82 | 91.18 | 91.13 | |
| carcinoma | 3 | 85 | 87.63 | 96.59 | 91.89 | 88 | ||||
| Fold 5 | non-carcinoma | 78 | 4 | 91.76 | 95.12 | 93.41 | 82 | 93.53 | 93.53 | |
| carcinoma | 7 | 81 | 95.29 | 92.05 | 93.64 | 88 | ||||
| Avg. | non-carcinoma | – | – | 93.78 | 88.05 | 90.75 | 82 | 91.41 | 91.38 | |
| carcinoma | – | – | 89.65 | 94.55 | 91.98 | 88 | ||||
| Fine-Tuned VGG16 | Fold 1 | non-carcinoma | 67 | 15 | 87.01 | 81.71 | 84.28 | 82 | 85.29 | 85.27 |
| carcinoma | 10 | 78 | 83.87 | 88.64 | 86.19 | 88 | ||||
| Fold 2 | non-carcinoma | 74 | 8 | 92.50 | 90.24 | 91.36 | 82 | 91.76 | 91.76 | |
| carcinoma | 6 | 82 | 91.11 | 93.18 | 92.13 | 88 | ||||
| Fold 3 | non-carcinoma | 76 | 6 | 95.00 | 92.68 | 93.83 | 82 | 94.12 | 94.11 | |
| carcinoma | 4 | 84 | 93.33 | 95.45 | 94.38 | 88 | ||||
| Fold 4 | non-carcinoma | 73 | 9 | 96.05 | 89.02 | 92.41 | 82 | 92.94 | 92.92 | |
| carcinoma | 3 | 85 | 90.43 | 96.59 | 93.41 | 88 | ||||
| Fold 5 | non-carcinoma | 75 | 7 | 96.15 | 91.46 | 93.75 | 82 | 94.12 | 94.11 | |
| carcinoma | 3 | 85 | 92.39 | 96.59 | 94.44 | 88 | ||||
| Avg. | non-carcinoma | – | – | 93.34 | 89.02 | 91.13 | 82 | 91.67 | 91.63 | |
| carcinoma | – | – | 90.23 | 94.09 | 92.11 | 88 | ||||
| Model | Single Training Time | 5-Fold Training Time | Prediction Time |
|---|---|---|---|
| Fully-trained VGG16 | 17 min 50 s | 89 min | 30 s |
| Fine-tuned VGG16 | 17 min 25 s | 87 min | 31 s |
| Fully-trained VGG19 | 20 min 40 s | 103 min | 35 s |
| Fine-tuned VGG19 | 19 min 55 s | 99 min | 36 s |
| Architecture | Folds | Confusion Matrices | Performance Evaluation (%) | Average (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Predict → Actual ↓ | NC | C | Precision | Recall | F1 | Test | Acc. | F1 | ||
| Fully-Trained VGG19 | Fold 1 | non-carcinoma | 66 | 16 | 98.51 | 80.49 | 88.59 | 82 | 90.00 | 89.89 |
| carcinoma | 1 | 87 | 84.47 | 98.86 | 91.10 | 88 | ||||
| Fold 2 | non-carcinoma | 71 | 11 | 94.67 | 86.59 | 90.45 | 82 | 91.18 | 91.15 | |
| carcinoma | 4 | 84 | 88.42 | 95.45 | 91.80 | 88 | ||||
| Fold 3 | non-carcinoma | 69 | 13 | 98.57 | 84.15 | 90.79 | 82 | 91.76 | 91.70 | |
| carcinoma | 1 | 87 | 87.00 | 98.86 | 92.55 | 88 | ||||
| Fold 4 | non-carcinoma | 69 | 13 | 90.79 | 84.15 | 87.34 | 82 | 88.24 | 88.21 | |
| carcinoma | 7 | 81 | 86.17 | 92.05 | 89.01 | 88 | ||||
| Fold 5 | non-carcinoma | 73 | 9 | 91.25 | 89.02 | 90.12 | 82 | 90.59 | 90.58 | |
| carcinoma | 7 | 81 | 90.00 | 92.05 | 91.01 | 88 | ||||
| Avg. | non-carcinoma | – | – | 94.76 | 84.88 | 89.46 | 82 | 90.35 | 90.31 | |
| carcinoma | – | – | 87.21 | 95.45 | 91.09 | 88 | ||||
| Fine-Tuned VGG19 | Fold 1 | non-carcinoma | 64 | 18 | 98.46 | 78.05 | 87.07 | 82 | 88.82 | 88.67 |
| carcinoma | 1 | 87 | 82.86 | 98.86 | 90.16 | 88 | ||||
| Fold 2 | non-carcinoma | 75 | 7 | 93.75 | 91.46 | 92.59 | 82 | 92.94 | 92.94 | |
| carcinoma | 5 | 83 | 92.22 | 94.32 | 93.26 | 88 | ||||
| Fold 3 | non-carcinoma | 75 | 7 | 97.40 | 91.46 | 94.34 | 82 | 94.71 | 94.70 | |
| carcinoma | 2 | 86 | 92.47 | 97.73 | 95.03 | 88 | ||||
| Fold 4 | non-carcinoma | 76 | 6 | 96.20 | 92.68 | 94.41 | 82 | 94.71 | 94.70 | |
| carcinoma | 3 | 85 | 93.41 | 96.59 | 94.97 | 88 | ||||
| Fold 5 | non-carcinoma | 71 | 11 | 89.87 | 86.59 | 88.20 | 82 | 88.82 | 88.81 | |
| carcinoma | 8 | 80 | 87.91 | 90.91 | 89.39 | 88 | ||||
| Avg. | non-carcinoma | – | – | 95.14 | 88.05 | 91.32 | 82 | 92.00 | 91.96 | |
| carcinoma | – | – | 89.77 | 95.68 | 92.56 | 88 | ||||
| Ensemble Method | Confusion Matrices | Performance Evaluation (%) | Average (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Predict → Actual ↓ | NC | C | Precision | Recall | F1 | Test | Accuracy | F1 | |
| Full-Trained | non-carcinoma | 73 | 9 | 97.33 | 89.02 | 92.99 | 82 | 93.53 | 93.51 |
| VGG16+VGG19 | carcinoma | 2 | 86 | 90.53 | 97.73 | 93.99 | 88 | ||
| Fine-Tuned | non-carcinoma | 76 | 6 | 97.44 | 92.68 | 95.00 | 82 | 95.29 | 95.29 |
| VGG16+VGG19 | carcinoma | 2 | 86 | 93.48 | 97.73 | 95.56 | 88 | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hameed, Z.; Zahia, S.; Garcia-Zapirain, B.; Javier Aguirre, J.; María Vanegas, A. Breast Cancer Histopathology Image Classification Using an Ensemble of Deep Learning Models. Sensors 2020, 20, 4373. https://doi.org/10.3390/s20164373
Hameed Z, Zahia S, Garcia-Zapirain B, Javier Aguirre J, María Vanegas A. Breast Cancer Histopathology Image Classification Using an Ensemble of Deep Learning Models. Sensors. 2020; 20(16):4373. https://doi.org/10.3390/s20164373
Chicago/Turabian StyleHameed, Zabit, Sofia Zahia, Begonya Garcia-Zapirain, José Javier Aguirre, and Ana María Vanegas. 2020. "Breast Cancer Histopathology Image Classification Using an Ensemble of Deep Learning Models" Sensors 20, no. 16: 4373. https://doi.org/10.3390/s20164373
APA StyleHameed, Z., Zahia, S., Garcia-Zapirain, B., Javier Aguirre, J., & María Vanegas, A. (2020). Breast Cancer Histopathology Image Classification Using an Ensemble of Deep Learning Models. Sensors, 20(16), 4373. https://doi.org/10.3390/s20164373






