Point-of-Care Diagnostics of COVID-19: From Current Work to Future Perspectives
Abstract
1. Introduction
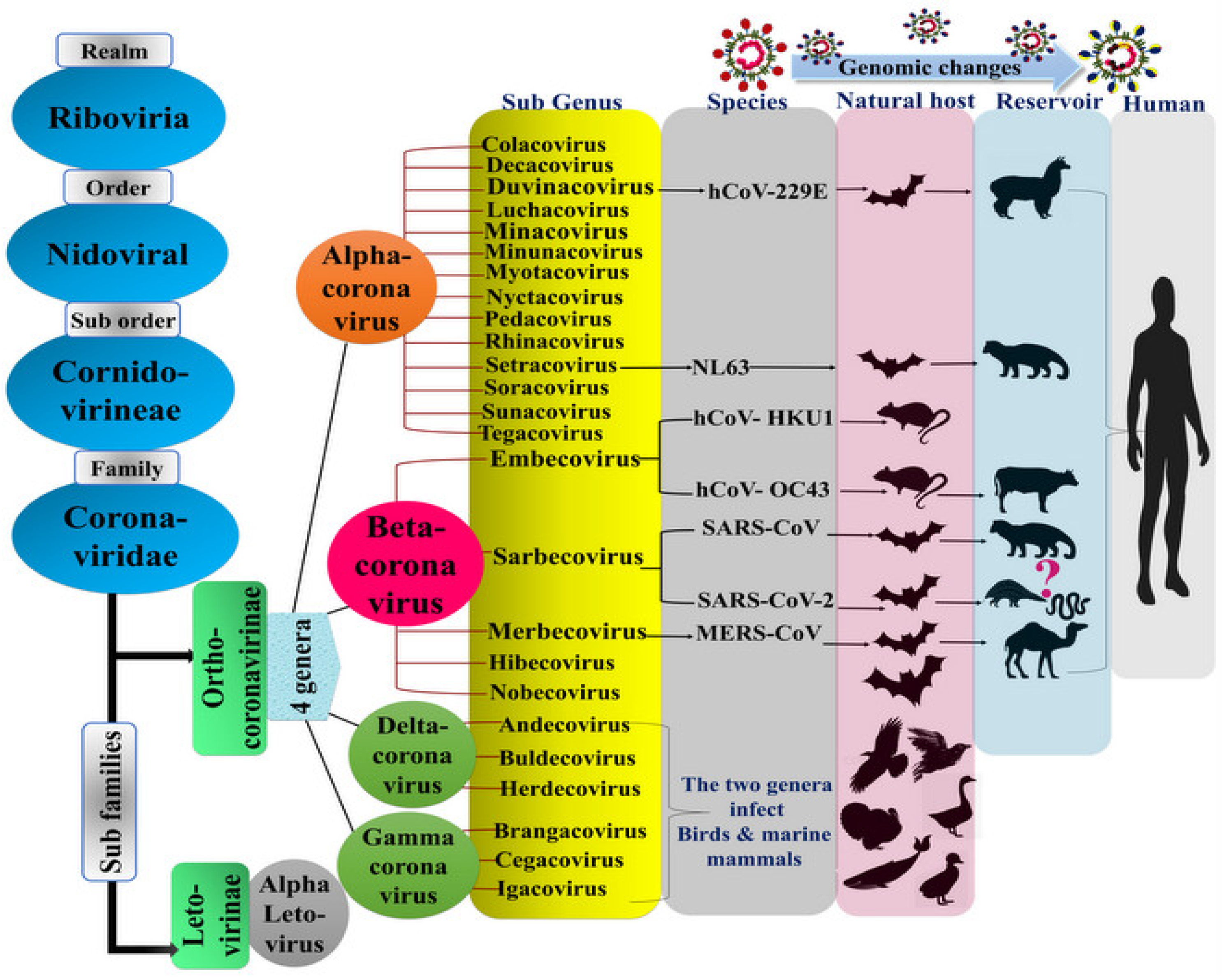
1.1. The Virus Origin and History
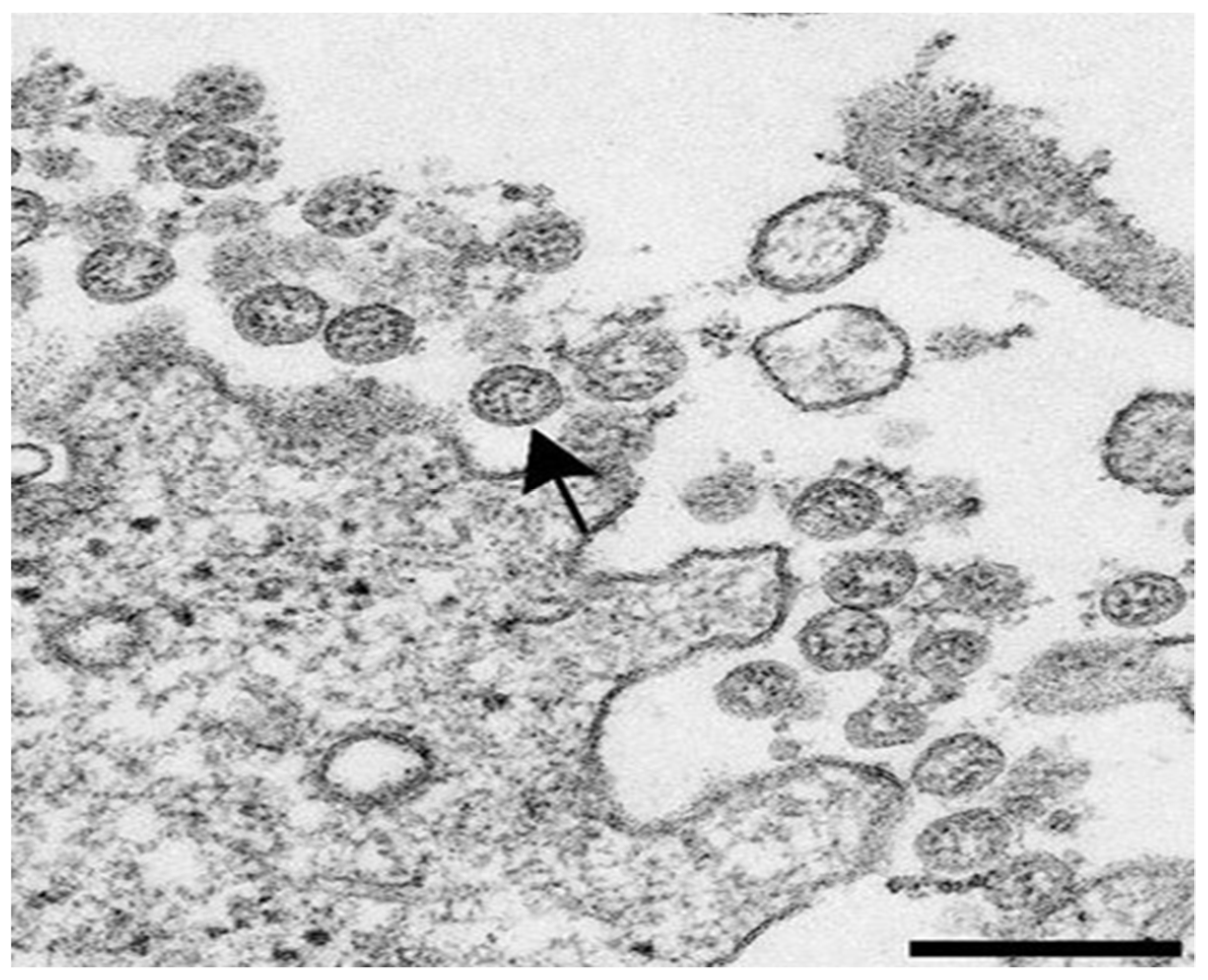
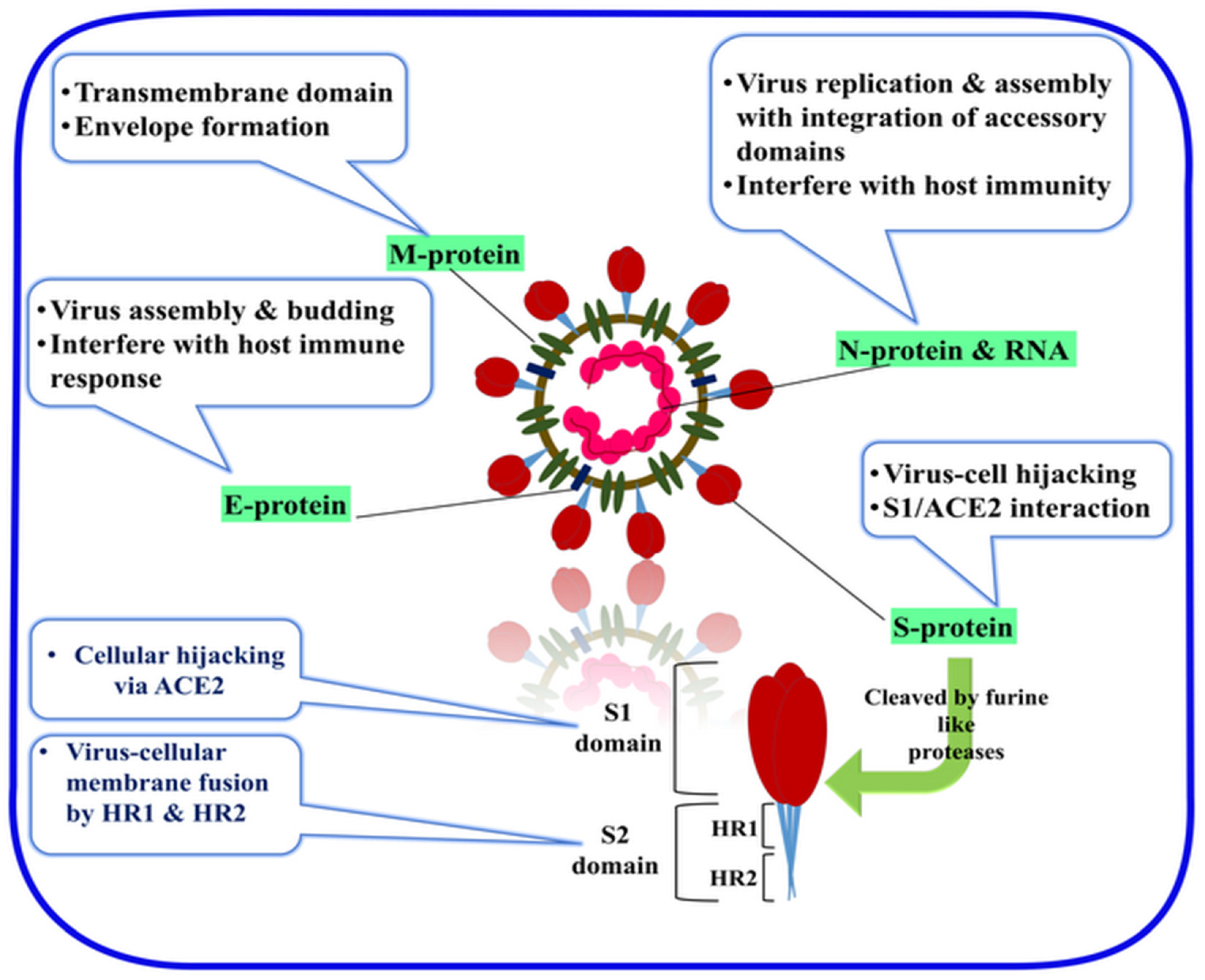
1.2. General Characteristics of the SARS-CoV-2
1.3. Cell Entry and the Role of the S Protein in the Virus Infectivity and Replication
2. Diagnosis of COVID-19
2.1. Laboratory-Based Cell Culture Detection
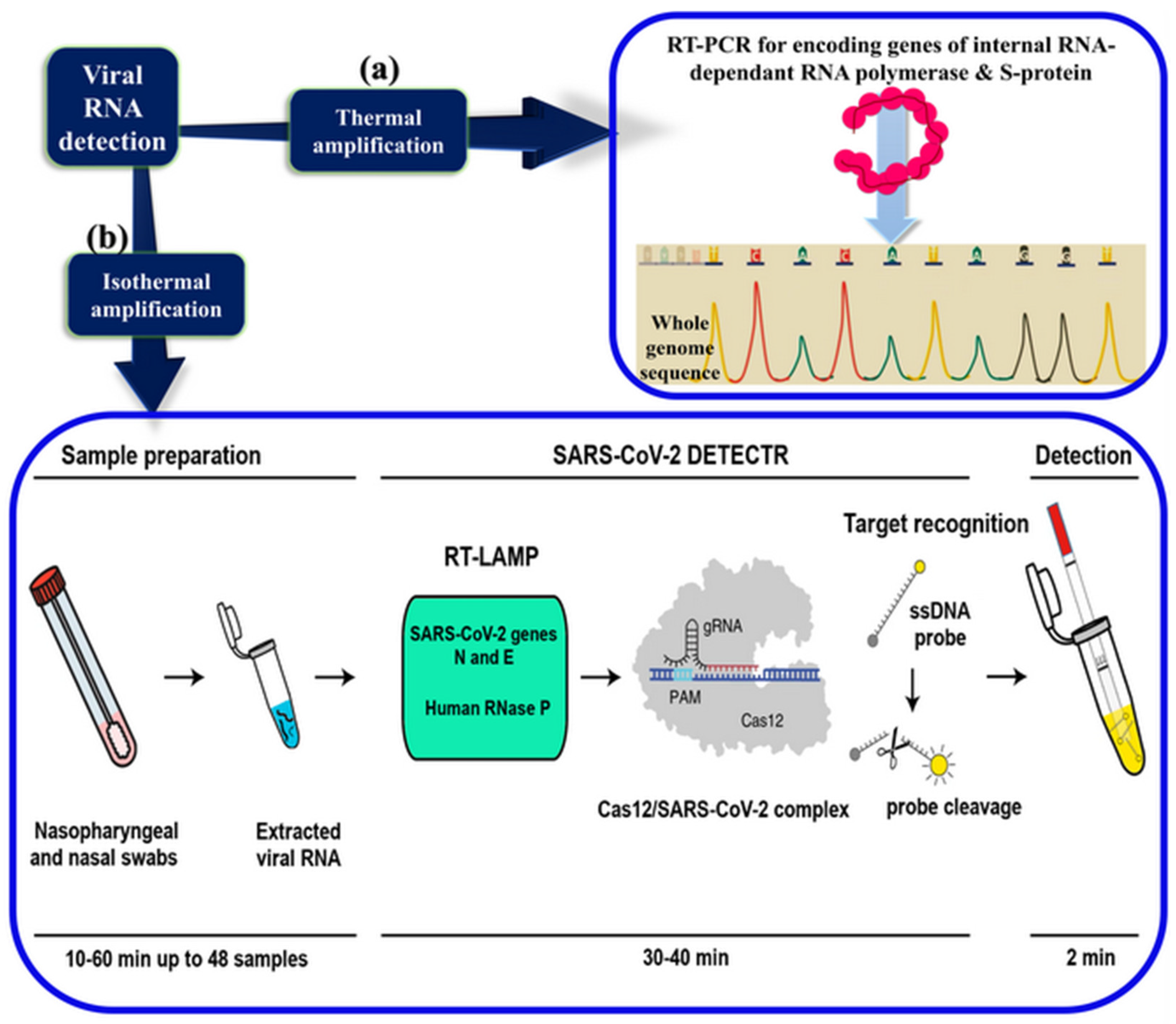
2.2. Laboratory-Based Gene Detection
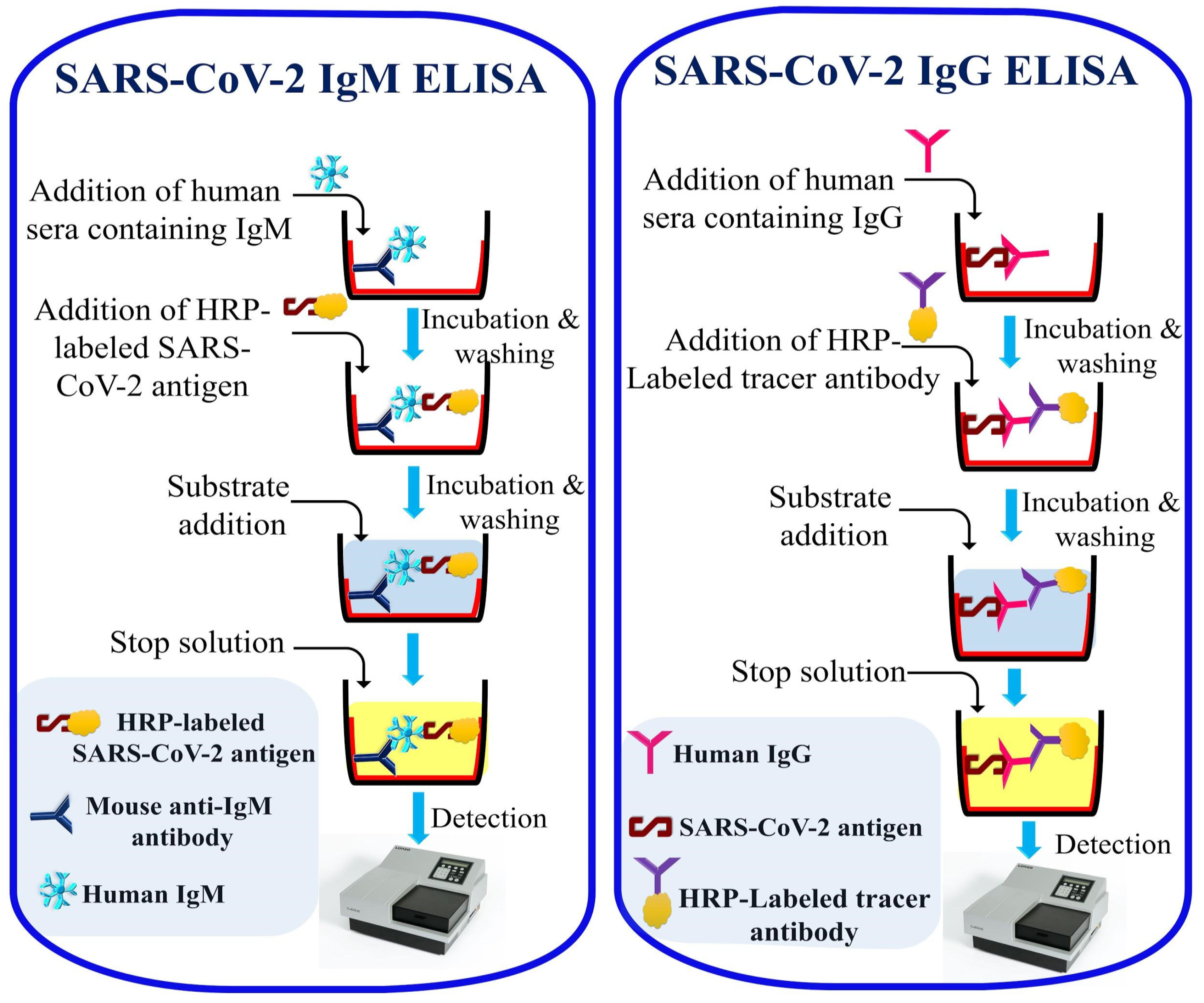
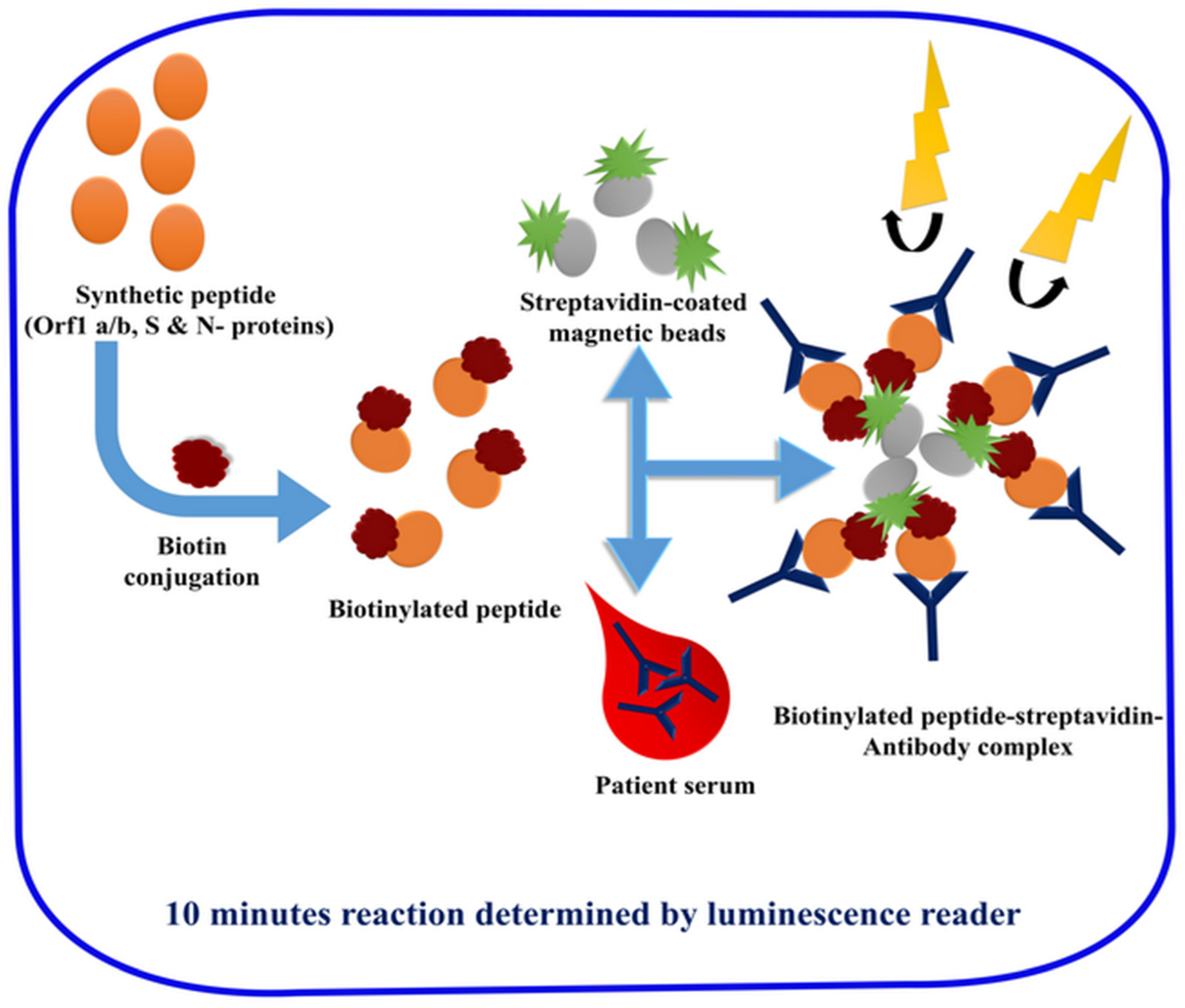
2.3. Laboratory-Based Immunological Detection
2.4. Laboratory-Based Omic Detection
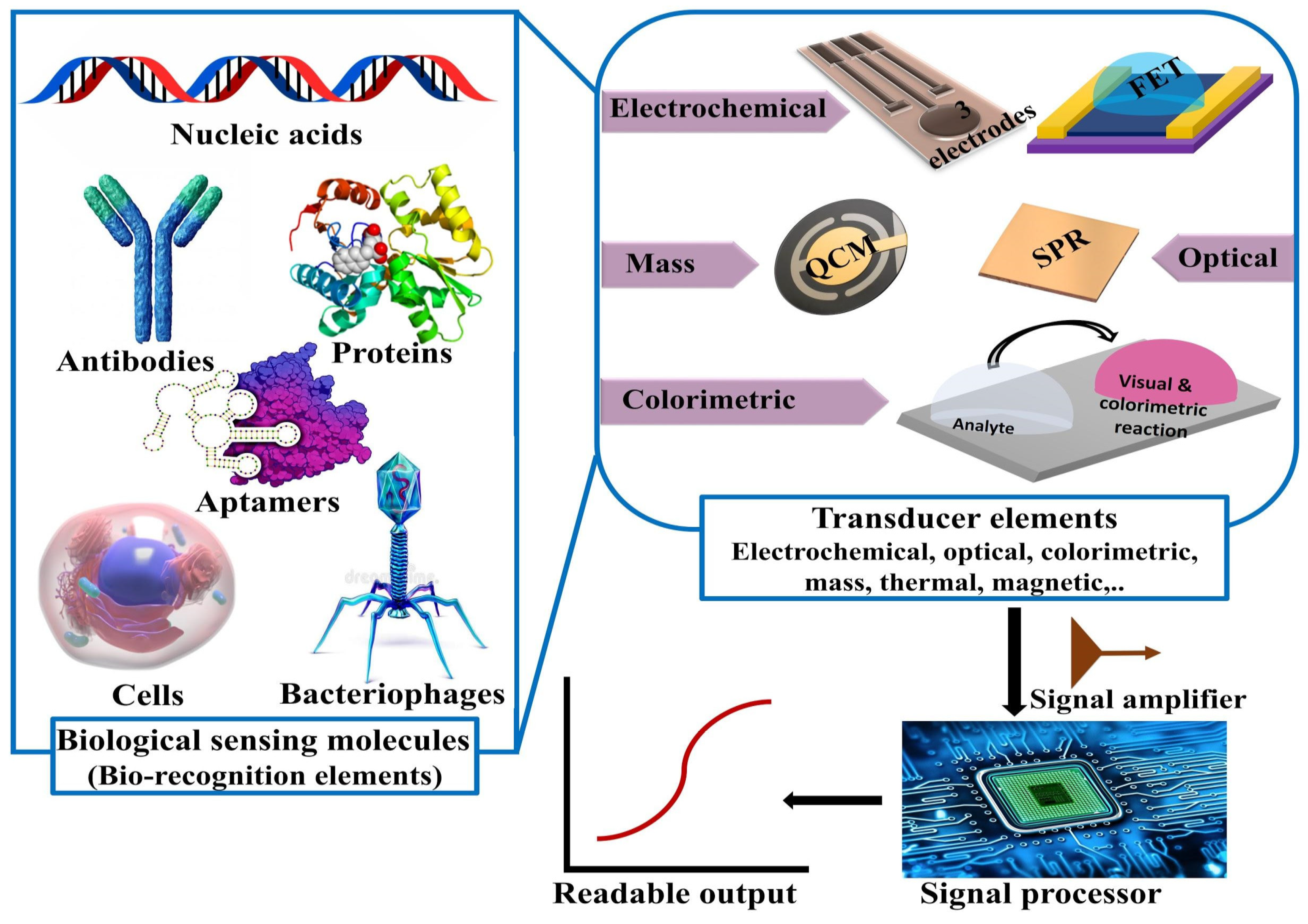
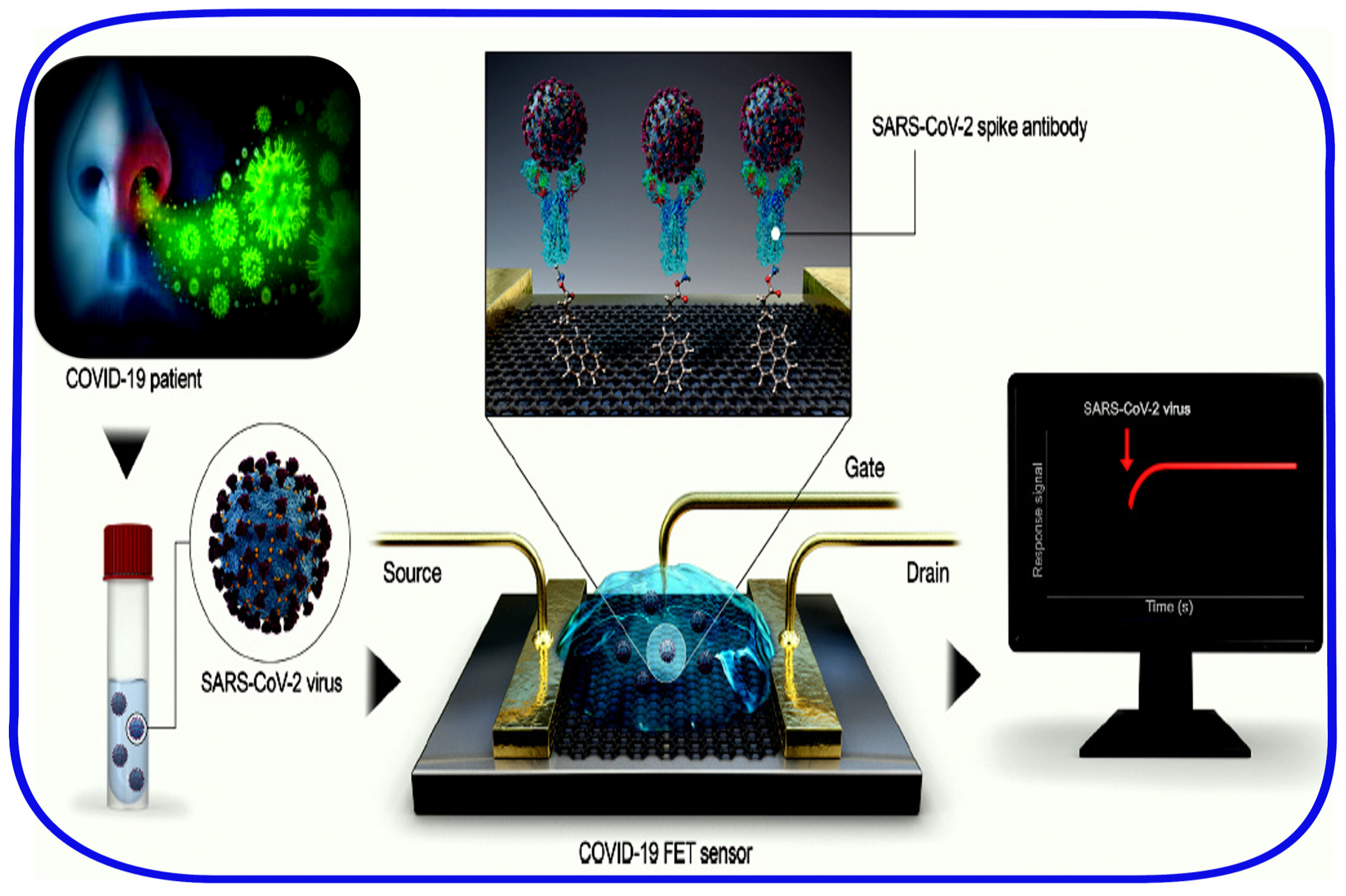
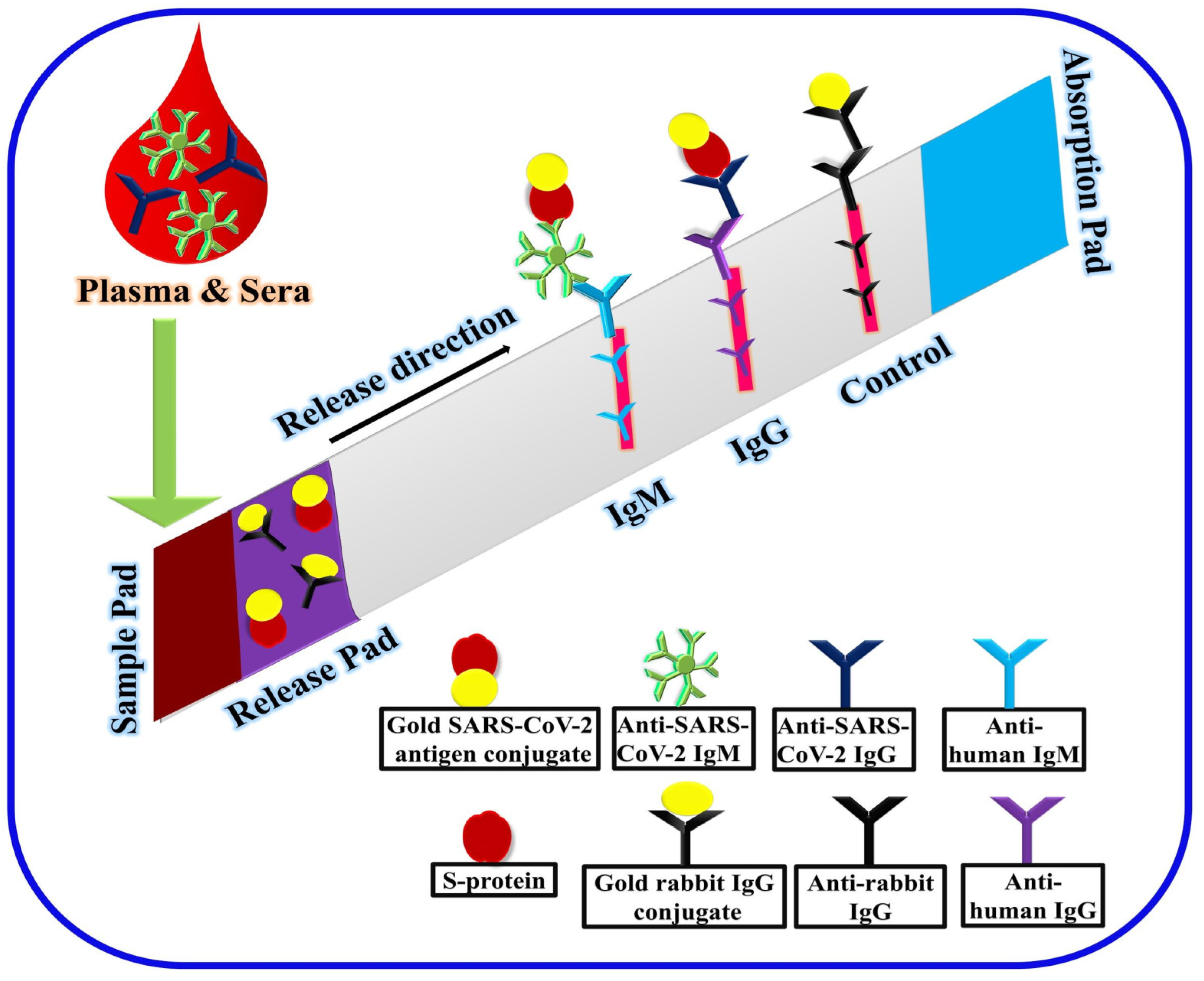
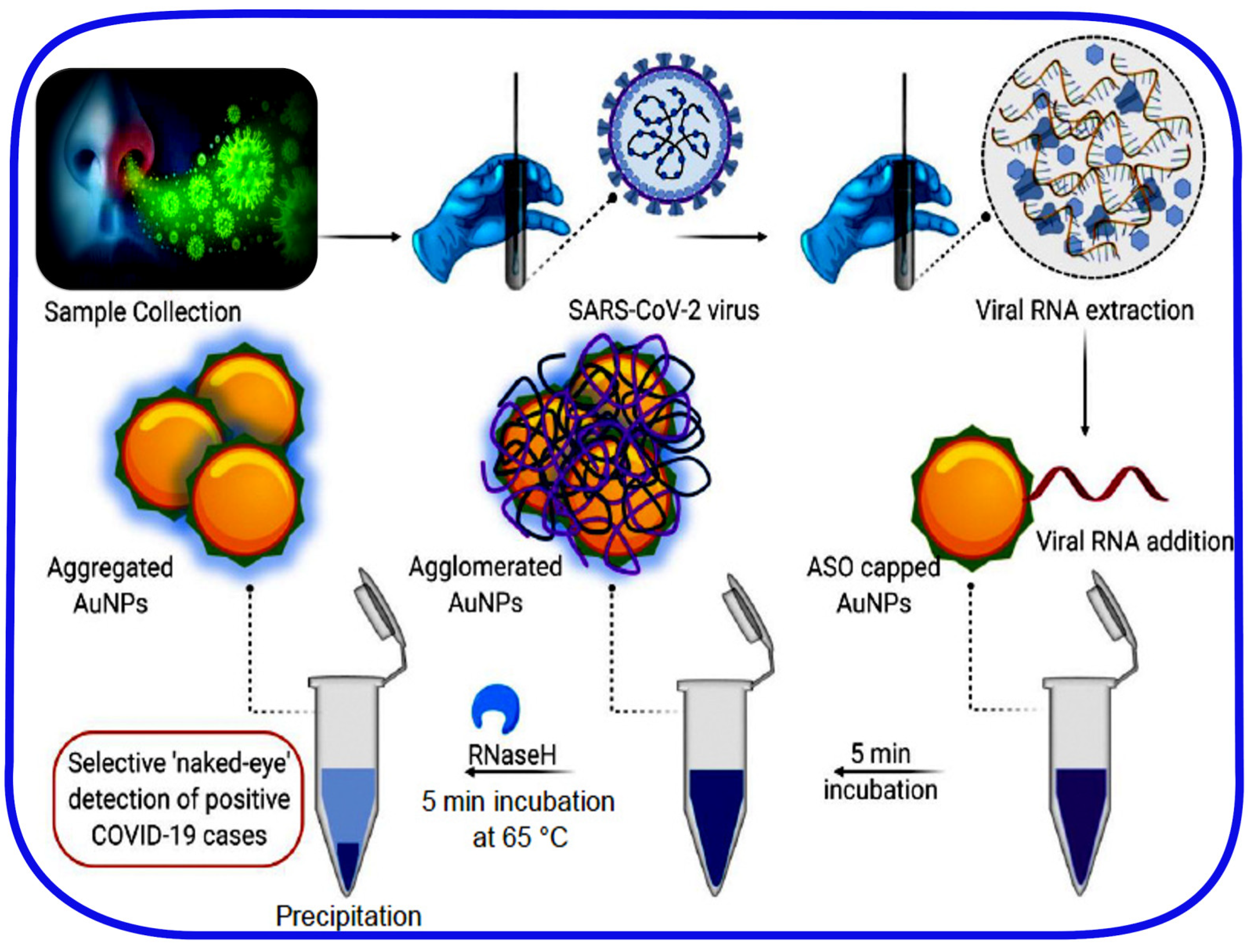
2.5. Point-of-Care and Standalone Biosensing Devices
3. Final Remarks and Future Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Fan, Y.; Zhao, K.; Shi, Z.-L.; Zhou, P. Bat Coronaviruses in China. Viruses 2019, 11, 210. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.-R.; Cao, Q.-D.; Hong, Z.-S.; Tan, Y.-Y.; Chen, S.-D.; Jin, H.-J.; Tan, K.-S.; Wang, D.-Y.; Yan, Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—An update on the status. Mil. Med. Res. 2020, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Forni, D.; Cagliani, R.; Clerici, M.; Sironi, M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017, 25, 35–48. [Google Scholar] [CrossRef] [PubMed]
- WHO. Update 31—Coronavirus Never Before Seen in Humans Is the Cause of SARS. Available online: https://www.who.int/csr/sarsarchive/2003_04_16/en/ (accessed on 20 May 2020).
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Buchholz, U.; Müller, M.A.; Nitsche, A.; Sanewski, A.; Wevering, N.; Bauer-Balci, T.; Bonin, F.; Drosten, C.; Schweiger, B.; Wolff, T.; et al. Contact investigation of a case of human novel coronavirus infection treated in a German hospital, October-November 2012. Eurosurveillance 2013, 18, 20406. [Google Scholar] [PubMed]
- De Groot, R.J.; Baker, S.C.; Baric, R.S.; Brown, C.S.; Drosten, C.; Enjuanes, L.; Fouchier, R.A.M.; Galiano, M.; Gorbalenya, A.E.; Memish, Z.A.; et al. Middle East respiratory syndrome coronavirus (MERS-CoV): Announcement of the Coronavirus Study Group. J. Virol. 2013, 87, 7790–7792. [Google Scholar] [CrossRef] [PubMed]
- Cotten, M.; Watson, S.J.; Zumla, A.I.; Makhdoom, H.Q.; Palser, A.L.; Ong, S.H.; Al Rabeeah, A.A.; Alhakeem, R.F.; Assiri, A.; Al-Tawfiq, J.A.; et al. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. mBio 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Su, S.; Bi, Y.; Wong, G.; Gao, G.F. Bat-Origin Coronaviruses Expand Their Host Range to Pigs. Trends Microbiol. 2018, 26, 466–470. [Google Scholar] [CrossRef] [PubMed]
- De Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef]
- Azhar, E.I.; El-Kafrawy, S.A.; Farraj, S.A.; Hassan, A.M.; Al-Saeed, M.S.; Hashem, A.M.; Madani, T.A. Evidence for Camel-to-Human Transmission of MERS Coronavirus. N. Engl. J. Med. 2014, 370, 2499–2505. [Google Scholar] [CrossRef]
- De Groot, R.J.; Baker, S.C.; Baric, R.S.; Enjuanes, L.; Gorbalenya, A.E.; Holmes, K.V.; Perlman, S.; Poon, L.; Rottier, P.J.M.; Talbot, P.J.; et al. Coronaviridae. In Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier Academic Press: San Diego, CA, USA, 2012; pp. 774–796. [Google Scholar]
- WHO. Novel Coronavirus—China. Available online: http://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/ (accessed on 20 May 2020).
- Chan, J.F.-W.; Kok, K.-H.; Zhu, Z.; Chu, H.; To, K.K.-W.; Yuan, S.; Yuen, K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.K.W.; Pan, Y.; Cheng, S.M.S.; Hui, K.P.Y.; Krishnan, P.; Liu, Y.; Ng, D.Y.M.; Wan, C.K.C.; Yang, P.; Wang, Q.; et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 2020, 66, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Harcourt, J.; Tamin, A.; Lu, X.; Kamili, S.; Sakthivel, S.K.; Murray, J.; Queen, K.; Tao, Y.; Paden, C.R.; Zhang, J.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 from Patient with Coronavirus Disease, United States. Emerg. Infect. Dis. 2020, 26, 1266–1273. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; Zhai, J.; Feng, Y.; Zhou, N.; Zhang, X.; Zou, J.-J.; Li, N.; Guo, Y.; Li, X.; Shen, X.; et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 2020, 583, 286–289. [Google Scholar] [CrossRef]
- Lillie, P.J.; Samson, A.; Li, A.; Adams, K.; Capstick, R.; Barlow, G.D.; Easom, N.; Hamilton, E.; Moss, P.J.; Evans, A.; et al. Novel coronavirus disease (Covid-19): The first two patients in the UK with person to person transmission. J. Infect. 2020, 80, 578–606. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Al-Ahmed, S.H.; Haque, S.; Sah, R.; Tiwari, R.; Malik, Y.S.; Dhama, K.; Yatoo, M.I.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. SARS-CoV-2, SARS-CoV, and MERS-COV: A comparative overview. Infez. Med. 2020, 28, 174–184. [Google Scholar]
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Chowell, G.; Abdirizak, F.; Lee, S.; Lee, J.; Jung, E.; Nishiura, H.; Viboud, C. Transmission characteristics of MERS and SARS in the healthcare setting: A comparative study. BMC Med. 2015, 13, 210. [Google Scholar] [CrossRef]
- Kang, C.K.; Song, K.H.; Choe, P.G.; Park, W.B.; Bang, J.H.; Kim, E.S.; Park, S.W.; Kim, H.B.; Kim, N.J.; Cho, S.I.; et al. Clinical and Epidemiologic Characteristics of Spreaders of Middle East Respiratory Syndrome Coronavirus during the 2015 Outbreak in Korea. J. Korean Med. Sci. 2017, 32, 744–749. [Google Scholar] [CrossRef]
- Pallesen, J.; Wang, N.; Corbett, K.S.; Wrapp, D.; Kirchdoerfer, R.N.; Turner, H.L.; Cottrell, C.A.; Becker, M.M.; Wang, L.; Shi, W.; et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA 2017, 114, E7348–E7357. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Snijder, E.J.; van der Meer, Y.; Zevenhoven-Dobbe, J.; Onderwater, J.J.M.; van der Meulen, J.; Koerten, H.K.; Mommaas, A.M. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006, 80, 5927–5940. [Google Scholar] [CrossRef] [PubMed]
- Perlman, S.; Netland, J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat. Rev. Microbiol. 2009, 7, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Masters, P.S. The Molecular Biology of Coronaviruses. In Advances in Virus Research; Academic Press: Cambridge, MA, USA, 2006; Volume 66, pp. 193–292. [Google Scholar]
- Thiel, V.; Ivanov, K.A.; Putics, Á.; Hertzig, T.; Schelle, B.; Bayer, S.; Weißbrich, B.; Snijder, E.J.; Rabenau, H.; Doerr, H.W.; et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003, 84, 2305–2315. [Google Scholar] [CrossRef]
- Hussain, S.; Pan, J.; Chen, Y.; Yang, Y.; Xu, J.; Peng, Y.; Wu, Y.; Li, Z.; Zhu, Y.; Tien, P.; et al. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J. Virol. 2005, 79, 5288–5295. [Google Scholar] [CrossRef]
- Beniac, D.R.; Andonov, A.; Grudeski, E.; Booth, T.F. Architecture of the SARS coronavirus prefusion spike. Nat. Struct. Mol. Biol. 2006, 13, 751–752. [Google Scholar] [CrossRef]
- Delmas, B.; Laude, H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J. Virol. 1990, 64, 5367–5375. [Google Scholar] [CrossRef]
- Kirchdoerfer, R.N.; Cottrell, C.A.; Wang, N.; Pallesen, J.; Yassine, H.M.; Turner, H.L.; Corbett, K.S.; Graham, B.S.; McLellan, J.S.; Ward, A.B. Pre-fusion structure of a human coronavirus spike protein. Nature 2016, 531, 118–121. [Google Scholar] [CrossRef]
- Walls, A.C.; Tortorici, M.A.; Frenz, B.; Snijder, J.; Li, W.; Rey, F.A.; DiMaio, F.; Bosch, B.-J.; Veesler, D. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016, 23, 899–905. [Google Scholar] [CrossRef]
- Millet, J.K.; Whittaker, G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. USA 2014, 111, 15214–15219. [Google Scholar] [CrossRef]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.W.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.A.; Zaki, A.; Fouchier, R.A.M.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Shi, X.; Jiang, L.; Zhang, S.; Wang, D.; Tong, P.; Guo, D.; Fu, L.; Cui, Y.; Liu, X.; et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013, 23, 986–993. [Google Scholar] [CrossRef]
- Donoghue, M.; Hsieh, F.; Baronas, E.; Godbout, K.; Gosselin, M.; Stagliano, N.; Donovan, M.; Woolf, B.; Robison, K.; Jeyaseelan, R.; et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ. Res. 2000, 87, E1–E9. [Google Scholar] [CrossRef] [PubMed]
- Hamming, I.; Timens, W.; Bulthuis, M.L.C.; Lely, A.T.; Navis, G.J.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kang, Z.; Gong, H.; Xu, D.; Wang, J.; Li, Z.; Li, Z.; Cui, X.; Xiao, J.; Zhan, J.; et al. Digestive system is a potential route of COVID-19: An analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut 2020, 69, 1010–1018. [Google Scholar] [CrossRef]
- Zhang, H.; Kang, Z.; Gong, H.; Xu, D.; Wang, J.; Li, Z.; Cui, X.; Xiao, J.; Meng, T.; Zhou, W.; et al. The digestive system is a potential route of 2019-nCov infection: A bioinformatics analysis based on single-cell transcriptomes. BioRxiv 2020. [Google Scholar] [CrossRef]
- Gui, M.; Song, W.; Zhou, H.; Xu, J.; Chen, S.; Xiang, Y.; Wang, X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017, 27, 119–129. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Glowacka, I.; Bertram, S.; Müller, M.A.; Allen, P.; Soilleux, E.; Pfefferle, S.; Steffen, I.; Tsegaye, T.S.; He, Y.; Gnirss, K.; et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011, 85, 4122–4134. [Google Scholar] [CrossRef]
- Shulla, A.; Heald-Sargent, T.; Subramanya, G.; Zhao, J.; Perlman, S.; Gallagher, T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011, 85, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Vedantham, P.; Lu, K.; Agudelo, J.; Carrion, R.; Nunneley, J.W.; Barnard, D.; Pöhlmann, S.; McKerrow, J.H.; Renslo, A.R.; et al. Protease inhibitors targeting coronavirus and filovirus entry. Antivir. Res. 2015, 116, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Iwata-Yoshikawa, N.; Okamura, T.; Shimizu, Y.; Hasegawa, H.; Takeda, M.; Nagata, N. TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Nal, B.; Chan, C.; Kien, F.; Siu, L.; Tse, J.; Chu, K.; Kam, J.; Staropoli, I.; Crescenzo-Chaigne, B.; Escriou, N.; et al. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005, 86, 1423–1434. [Google Scholar] [CrossRef]
- Neuman, B.W.; Kiss, G.; Kunding, A.H.; Bhella, D.; Baksh, M.F.; Connelly, S.; Droese, B.; Klaus, J.P.; Makino, S.; Sawicki, S.G.; et al. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011, 174, 11–22. [Google Scholar] [CrossRef]
- DeDiego, M.L.; Alvarez, E.; Almazán, F.; Rejas, M.T.; Lamirande, E.; Roberts, A.; Shieh, W.-J.; Zaki, S.R.; Subbarao, K.; Enjuanes, L. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 2007, 81, 1701–1713. [Google Scholar] [CrossRef]
- Nieto-Torres, J.L.; DeDiego, M.L.; Verdiá-Báguena, C.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A.; Fernandez-Delgado, R.; Castaño-Rodriguez, C.; Alcaraz, A.; Torres, J.; Aguilella, V.M.; et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014, 10, e1004077. [Google Scholar] [CrossRef]
- Chang, C.; Sue, S.-C.; Yu, T.; Hsieh, C.-M.; Tsai, C.-K.; Chiang, Y.-C.; Lee, S.; Hsiao, H.; Wu, W.-J.; Chang, W.-L.; et al. Modular organization of SARS coronavirus nucleocapsid protein. J. Biomed. Sci. 2006, 13, 59–72. [Google Scholar] [CrossRef]
- Hurst, K.R.; Koetzner, C.A.; Masters, P.S. Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J. Virol. 2009, 83, 7221–7234. [Google Scholar] [CrossRef]
- Hurst, K.R.; Koetzner, C.A.; Masters, P.S. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J. Virol. 2013, 87, 9159–9172. [Google Scholar] [CrossRef] [PubMed]
- Klausegger, A.; Strobl, B.; Regl, G.; Kaser, A.; Luytjes, W.; Vlasak, R. Identification of a coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza C virus and bovine coronavirus. J. Virol. 1999, 73, 3737–3743. [Google Scholar] [CrossRef] [PubMed]
- Lissenberg, A.; Vrolijk, M.M.; van Vliet, A.L.W.; Langereis, M.A.; de Groot-Mijnes, J.D.F.; Rottier, P.J.M.; de Groot, R.J. Luxury at a cost? Recombinant mouse hepatitis viruses expressing the accessory hemagglutinin esterase protein display reduced fitness in vitro. J. Virol. 2005, 79, 15054–15063. [Google Scholar] [CrossRef] [PubMed]
- Kazi, L.; Lissenberg, A.; Watson, R.; de Groot, R.J.; Weiss, S.R. Expression of hemagglutinin esterase protein from recombinant mouse hepatitis virus enhances neurovirulence. J. Virol. 2005, 79, 15064–15073. [Google Scholar] [CrossRef] [PubMed]
- Algaissi, A.; Hashem, A.M. Evaluation of MERS-CoV Neutralizing Antibodies in Sera Using Live Virus Microneutralization Assay. In MERS Coronavirus: Methods and Protocols; Vijay, R., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; pp. 107–116. ISBN 978-1-07-160211-9. [Google Scholar]
- Lester, S.; Harcourt, J.; Whitt, M.; Al-Abdely, H.M.; Midgley, C.M.; Alkhamis, A.M.; Aziz Jokhdar, H.A.; Assiri, A.M.; Tamin, A.; Thornburg, N. Middle East respiratory coronavirus (MERS-CoV) spike (S) protein vesicular stomatitis virus pseudoparticle neutralization assays offer a reliable alternative to the conventional neutralization assay in human seroepidemiological studies. Access Microbiol. 2019, 1, e000057. [Google Scholar] [CrossRef]
- Matsuyama, S.; Nao, N.; Shirato, K.; Kawase, M.; Saito, S.; Takayama, I.; Nagata, N.; Sekizuka, T.; Katoh, H.; Kato, F.; et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA 2020, 117, 7001–7003. [Google Scholar] [CrossRef] [PubMed]
- Amanat, F.; White, K.M.; Miorin, L.; Strohmeier, S.; McMahon, M.; Meade, P.; Liu, W.-C.; Albrecht, R.A.; Simon, V.; Martinez-Sobrido, L.; et al. An In Vitro Microneutralization Assay for SARS-CoV-2 Serology and Drug Screening. Curr. Protoc. Microbiol. 2020, 58, e108. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Yuan, S.; Kok, K.-H.; To, K.K.-W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.-Y.; Poon, R.W.-S.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef]
- Lewandowski, K.; Xu, Y.; Pullan, S.T.; Lumley, S.F.; Foster, D.; Sanderson, N.; Vaughan, A.; Morgan, M.; Bright, N.; Kavanagh, J.; et al. Metagenomic Nanopore Sequencing of Influenza Virus Direct from Clinical Respiratory Samples. J. Clin. Microbiol. 2019, 58. [Google Scholar] [CrossRef]
- Corman, V.M.; Müller, M.A.; Costabel, U.; Timm, J.; Binger, T.; Meyer, B.; Kreher, P.; Lattwein, E.; Eschbach-Bludau, M.; Nitsche, A.; et al. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Eurosurveillance 2012, 17, 20334. [Google Scholar] [CrossRef]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, P.-R.; Huang, L.-M.; Chen, P.-J.; Kao, C.-L.; Yang, P.-C. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS-associated coronavirus. Clin. Microbiol. Infect. 2004, 10, 1062–1066. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lu, H.; Melino, G.; Boccia, S.; Piacentini, M.; Ricciardi, W.; Wang, Y.; Shi, Y.; Zhu, T. COVID-19 infection: The China and Italy perspectives. Cell Death Dis. 2020, 11, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Wang, T.; Zhang, B.; Luo, Y.; Mao, L.; Wang, F.; Wu, S.; Sun, Z. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin. Transl. Immunol. 2020, 9, e1136. [Google Scholar] [CrossRef]
- Severance, E.G.; Bossis, I.; Dickerson, F.B.; Stallings, C.R.; Origoni, A.E.; Sullens, A.; Yolken, R.H.; Viscidi, R.P. Development of a nucleocapsid-based human coronavirus immunoassay and estimates of individuals exposed to coronavirus in a U.S. metropolitan population. Clin. Vaccine Immunol. CVI 2008, 15, 1805–1810. [Google Scholar] [CrossRef]
- Trivedi, S.U.; Miao, C.; Sanchez, J.E.; Caidi, H.; Tamin, A.; Haynes, L.; Thornburg, N.J. Development and Evaluation of a Multiplexed Immunoassay for Simultaneous Detection of Serum IgG Antibodies to Six Human Coronaviruses. Sci. Rep. 2019, 9, 1390. [Google Scholar] [CrossRef]
- Ju, B.; Zhang, Q.; Ge, X.; Wang, R.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; Tang, X.; Yu, J.; et al. Potent human neutralizing antibodies elicited by SARS-CoV-2 infection. BioRxiv 2020. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Chen, Y.; Wong, E.Y.M.; Chan, K.-H.; Chen, H.; Zhang, L.; Xia, N.; Yuen, K.-Y. Rapid detection of MERS coronavirus-like viruses in bats: Pote1ntial for tracking MERS coronavirus transmission and animal origin. Emerg. Microbes Infect. 2018, 7, 1–7. [Google Scholar] [CrossRef]
- Kasem, S.; Qasim, I.; Al-Hufofi, A.; Hashim, O.; Alkarar, A.; Abu-Obeida, A.; Gaafer, A.; Elfadil, A.; Zaki, A.; Al-Romaihi, A.; et al. Cross-sectional study of MERS-CoV-specific RNA and antibodies in animals that have had contact with MERS patients in Saudi Arabia. J. Infect. Public Health 2018, 11, 331–338. [Google Scholar] [CrossRef]
- Xiang, J.; Yan, M.; Li, H.; Liu, T.; Lin, C.; Huang, S.; Shen, C. Evaluation of Enzyme-Linked Immunoassay and Colloidal Gold- Immunochromatographic Assay Kit for Detection of Novel Coronavirus (SARS-Cov-2) Causing an Outbreak of Pneumonia (COVID-19). MedRxiv 2020. [Google Scholar] [CrossRef]
- Amanat, F.; Stadlbauer, D.; Strohmeier, S.; Nguyen, T.H.O.; Chromikova, V.; McMahon, M.; Jiang, K.; Arunkumar, G.A.; Jurczyszak, D.; Polanco, J.; et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020, 26, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.-F.; Chen, J.; Hu, J.-L.; Long, Q.-X.; Deng, H.-J.; Fan, K.; Liao, P.; Liu, B.-Z.; Wu, G.-C.; Chen, Y.-K.; et al. A Peptide-based Magnetic Chemiluminescence Enzyme Immunoassay for Serological Diagnosis of Coronavirus Disease 2019 (COVID-19). J. Infect. Dis. 2020, 222, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Zambrano, G.; Nastri, F.; Pavone, V.; Lombardi, A.; Chino, M. Use of an Artificial Miniaturized Enzyme in Hydrogen Peroxide Detection by Chemiluminescence. Sensors 2020, 20, 3793. [Google Scholar] [CrossRef]
- Struwe, W.; Emmott, E.; Bailey, M.; Sharon, M.; Sinz, A.; Corrales, F.J.; Thalassinos, K.; Braybrook, J.; Mills, C.; Barran, P. The COVID-19 MS Coalition—Accelerating diagnostics, prognostics, and treatment. Lancet 2020, 395, 1761–1762. [Google Scholar] [CrossRef]
- Orsburn, B.C.; Jenkins, C.; Miller, S.M.; Neely, B.A.; Bumpus, N.N. In silico approach toward the identification of unique peptides from viral protein infection: Application to COVID-19. BioRxiv 2020. [Google Scholar] [CrossRef]
- Bezstarosti, K.; Lamers, M.M.; Haagmans, B.L.; Demmers, J.A.A. Targeted Proteomics for the Detection of SARS-CoV-2 Proteins. BioRxiv 2020. [Google Scholar] [CrossRef]
- Ihling, C.; Tänzler, D.; Hagemann, S.; Kehlen, A.; Hüttelmaier, S.; Sinz, A. Mass Spectrometric Identification of SARS-CoV-2 Proteins from Gargle Solution Samples of COVID-19 Patients. BioRxiv 2020. [Google Scholar] [CrossRef]
- Nikolaev, E.N.; Indeykina, M.I.; Brzhozovskiy, A.G.; Bugrova, A.E.; Kononikhin, A.S.; Starodubtseva, N.L.; Petrotchenko, E.V.; Kovalev, G.; Borchers, C.H.; Sukhikh, G.T. Mass Spectrometric detection of SARS-CoV-2 virus in scrapings of the epithelium of the nasopharynx of infected patients via Nucleocapsid N protein. BioRxiv 2020. [Google Scholar] [CrossRef]
- Messner, C.B.; Demichev, V.; Wendisch, D.; Michalick, L.; White, M.; Freiwald, A.; Textoris-Taube, K.; Vernardis, S.I.; Egger, A.-S.; Kreidl, M.; et al. Clinical classifiers of COVID-19 infection from novel ultra-high-throughput proteomics. MedRxiv 2020. [Google Scholar] [CrossRef]
- Mustafa, F.; Hassan, R.Y.A.; Andreescu, S. Multifunctional Nanotechnology-Enabled Sensors for Rapid Capture and Detection of Pathogens. Sensors 2017, 17, 2121. [Google Scholar] [CrossRef]
- Hassan, R.Y.A.; Wollenberger, U. Direct Determination of Bacterial Cell Viability Using Carbon Nanotubes Modified Screen-printed Electrodes. Electroanalysis 2019, 31, 1112–1117. [Google Scholar] [CrossRef]
- Kaushik, A.; Yndart, A.; Kumar, S.; Jayant, R.D.; Vashist, A.; Brown, A.N.; Li, C.-Z.; Nair, M. A sensitive electrochemical immunosensor for label-free detection of Zika-virus protein. Sci. Rep. 2018, 8, 9700. [Google Scholar] [CrossRef] [PubMed]
- Layqah, L.A.; Eissa, S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Mikrochim. Acta 2019, 186, 224. [Google Scholar] [CrossRef] [PubMed]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2020. [Google Scholar] [CrossRef]
- Moitra, P.; Alafeef, M.; Dighe, K.; Frieman, M.B.; Pan, D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14, 7617–7627. [Google Scholar] [CrossRef]
- Tai, D.-F.; Lin, C.-Y.; Wu, T.-Z.; Chen, L.-K. Recognition of dengue virus protein using epitope-mediated molecularly imprinted film. Anal. Chem. 2005, 77, 5140–5143. [Google Scholar] [CrossRef]
- Lu, C.-H.; Zhang, Y.; Tang, S.-F.; Fang, Z.-B.; Yang, H.-H.; Chen, X.; Chen, G.-N. Sensing HIV related protein using epitope imprinted hydrophilic polymer coated quartz crystal microbalance. Biosens. Bioelectron. 2012, 31, 439–444. [Google Scholar] [CrossRef]
- Hussein, H.A.; Hassan, R.Y.A.; El Nashar, R.M.; Khalil, S.A.; Salem, S.A.; El-Sherbiny, I.M. Designing and fabrication of new VIP biosensor for the rapid and selective detection of foot-and-mouth disease virus (FMDV). Biosens. Bioelectron. 2019, 141, 111467. [Google Scholar] [CrossRef]
- Sangma, C.; Lieberzeit, P.A.; Sukjee, W. H5N1 Virus Plastic Antibody Based on Molecularly Imprinted Polymers. In Synthetic Antibodies: Methods and Protocols; Tiller, T., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; pp. 381–388. ISBN 978-1-4939-6857-2. [Google Scholar]
- Yang, B.; Gong, H.; Chen, C.; Chen, X.; Cai, C. A virus resonance light scattering sensor based on mussel-inspired molecularly imprinted polymers for high sensitive and high selective detection of Hepatitis A Virus. Biosens. Bioelectron. 2017, 87, 679–685. [Google Scholar] [CrossRef]
- Yang, Y.-J.; Li, J.-Y.; Liu, X.-W.; Zhang, J.-Y.; Liu, Y.-R.; Li, B. A non-biological method for screening active components against influenza virus from traditional Chinese medicine by coupling a LC column with oseltamivir molecularly imprinted polymers. PLoS ONE 2013, 8, e84458. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, A.; Piovesana, S.; Zenezini Chiozzi, R.; Montone, C.M.; Bossi, A.M.; Laganà, A. Does the protein corona take over the selectivity of molecularly imprinted nanoparticles? The biological challenges to recognition. J. Proteom. 2020, 219, 103736. [Google Scholar] [CrossRef] [PubMed]
- Schirhagl, R.; Ren, K.; Zare, R.N. Surface-imprinted polymers in microfluidic devices. Sci. China Chem. 2012, 55, 469–483. [Google Scholar] [CrossRef]
- Yang, J.C.; Shin, H.-K.; Hong, S.W.; Park, J.Y. Lithographically patterned molecularly imprinted polymer for gravimetric detection of trace atrazine. Sens. Actuators B Chem. 2015, 216, 476–481. [Google Scholar] [CrossRef]
- Li, F.; Chen, W.; Zhang, S. Development of DNA electrochemical biosensor based on covalent immobilization of probe DNA by direct coupling of sol–gel and self-assembly technologies. Biosens. Bioelectron. 2008, 24, 781–786. [Google Scholar] [CrossRef]
- Tancharoen, C.; Sukjee, W.; Thepparit, C.; Jaimipuk, T.; Auewarakul, P.; Thitithanyanont, A.; Sangma, C. Electrochemical Biosensor Based on Surface Imprinting for Zika Virus Detection in Serum. ACS Sens. 2019, 4, 69–75. [Google Scholar] [CrossRef]
- Gast, M.; Kühner, S.; Sobek, H.; Mizaikoff, B. Understanding the viral load during the synthesis and after rebinding of virus imprinted particles via real-time quantitative PCR. Analyst 2018, 143, 2616–2622. [Google Scholar] [CrossRef]
- Liang, C.; Wang, H.; He, K.; Chen, C.; Chen, X.; Gong, H.; Cai, C. A virus-MIPs fluorescent sensor based on FRET for highly sensitive detection of JEV. Talanta 2016, 160, 360–366. [Google Scholar] [CrossRef]
- Nandy Chatterjee, T.; Bandyopadhyay, R. A Molecularly Imprinted Polymer-Based Technology for Rapid Testing of COVID-19. Trans. Indian Natl. Acad. Eng. 2020, 1–4. [Google Scholar] [CrossRef]
- Parisi, O.I.; Dattilo, M.; Patitucci, F.; Malivindi, R.; Pezzi, V.; Perrotta, I.; Ruffo, M.; Amone, F.; Puoci, F. “Monoclonal-type” plastic antibodies for SARS-CoV-2 based on Molecularly Imprinted Polymers. BioRxiv 2020. [Google Scholar] [CrossRef]
- D’Alonzo, D.; De Fenza, M.; Pavone, V. COVID-19 and pneumonia: A role for the uPA/uPAR system. Drug Discov. Today 2020. [Google Scholar] [CrossRef]
- Eskandarifar, A.; Rasouli, M.A.; Mansouri, M.; Moosavi, S.; Fotoohi, A. Validity of glucose measurements in the blood by a glucometer reagent strip in critically ill infants. Diabetes Metab. Syndr. 2019, 13, 464–466. [Google Scholar] [CrossRef]
- Kim, H.N.; Moon, K.C.; Yoon, S.-Y. Performance evaluation of three i-SENS glucometers using arterial blood samples compared with the YSI 2300 Glucose Analyzer. J. Clin. Lab. Anal. 2020, e23356. [Google Scholar] [CrossRef]
- Bahadır, E.B.; Sezgintürk, M.K. Applications of commercial biosensors in clinical, food, environmental, and biothreat/biowarfare analyses. Anal. Biochem. 2015, 478, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.M.-H. Over-the-Counter Biosensors: Past, Present, and Future. Sensors 2008, 8, 5535–5559. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Campbell, A.S.; de Ávila, B.E.-F.; Wang, J. Wearable biosensors for healthcare monitoring. Nat. Biotechnol. 2019, 37, 389–406. [Google Scholar] [CrossRef] [PubMed]
- Cetrangolo, G.P.; Gori, C.; Rusko, J.; Terreri, S.; Manco, G.; Cimmino, A.; Febbraio, F. Determination of Picomolar Concentrations of Paraoxon in Human Urine by Fluorescence-Based Enzymatic Assay. Sensors 2019, 19, 4852. [Google Scholar] [CrossRef]
- Cetrangolo, G.P.; Rusko, J.; Gori, C.; Carullo, P.; Manco, G.; Chino, M.; Febbraio, F. Highly Sensitive Detection of Chemically Modified Thio-Organophosphates by an Enzymatic Biosensing Device: An Automated Robotic Approach. Sensors 2020, 20, 1365. [Google Scholar] [CrossRef]
- Febbraio, F. Biochemical strategies for the detection and detoxification of toxic chemicals in the environment. World J. Biol. Chem. 2017, 8, 13–20. [Google Scholar] [CrossRef]
- Carullo, P.; Cetrangolo, G.P.; Mandrich, L.; Manco, G.; Febbraio, F. Fluorescence Spectroscopy Approaches for the Development of a Real-Time Organophosphate Detection System Using an Enzymatic Sensor. Sensors 2015, 15, 3932–3951. [Google Scholar] [CrossRef]
- Carullo, P.; Chino, M.; Cetrangolo, G.P.; Terreri, S.; Lombardi, A.; Manco, G.; Cimmino, A.; Febbraio, F. Direct detection of organophosphate compounds in water by a fluorescence-based biosensing device. Sens. Actuators B Chem. 2018, 255, 3257–3266. [Google Scholar] [CrossRef]
- Porzio, E.; Bettazzi, F.; Mandrich, L.; Del Giudice, I.; Restaino, O.F.; Laschi, S.; Febbraio, F.; De Luca, V.; Borzacchiello, M.G.; Carusone, T.M.; et al. Innovative Biocatalysts as Tools to Detect and Inactivate Nerve Agents. Sci. Rep. 2018, 8, 13773. [Google Scholar] [CrossRef]
- Mavrikou, S.; Moschopoulou, G.; Tsekouras, V.; Kintzios, S. Development of a Portable, Ultra-Rapid and Ultra-Sensitive Cell-Based Biosensor for the Direct Detection of the SARS-CoV-2 S1 Spike Protein Antigen. Sensors 2020, 20, 3121. [Google Scholar] [CrossRef] [PubMed]
- Korendovych, I.V.; DeGrado, W.F. De novo protein design, a retrospective. Q. Rev. Biophys. 2020, 53. [Google Scholar] [CrossRef] [PubMed]
- Baker, E.G.; Bartlett, G.J.; Porter Goff, K.L.; Woolfson, D.N. Miniprotein Design: Past, Present, and Prospects. Acc. Chem. Res. 2017, 50, 2085–2092. [Google Scholar] [CrossRef]
- Huang, P.-S.; Boyken, S.E.; Baker, D. The coming of age of de novo protein design. Nature 2016, 537, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, A.; Pirro, F.; Maglio, O.; Chino, M.; DeGrado, W.F. De Novo Design of Four-Helix Bundle Metalloproteins: One Scaffold, Diverse Reactivities. Acc. Chem. Res. 2019, 52, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Chino, M.; Leone, L.; Zambrano, G.; Pirro, F.; D’Alonzo, D.; Firpo, V.; Aref, D.; Lista, L.; Maglio, O.; Nastri, F.; et al. Oxidation catalysis by iron and manganese porphyrins within enzyme-like cages. Biopolymers 2018, 109, e23107. [Google Scholar] [CrossRef]
- Sesterhenn, F.; Yang, C.; Bonet, J.; Cramer, J.T.; Wen, X.; Wang, Y.; Chiang, C.-I.; Abriata, L.A.; Kucharska, I.; Castoro, G.; et al. De novo protein design enables the precise induction of RSV-neutralizing antibodies. Science 2020, 368. [Google Scholar] [CrossRef]
- Caserta, G.; Chino, M.; Firpo, V.; Zambrano, G.; Leone, L.; D’Alonzo, D.; Nastri, F.; Maglio, O.; Pavone, V.; Lombardi, A. Enhancement of Peroxidase Activity in Artificial Mimochrome VI Catalysts through Rational Design. ChemBioChem 2018, 19, 1823–1826. [Google Scholar] [CrossRef]
- Chino, M.; Leone, L.; Maglio, O.; D’Alonzo, D.; Pirro, F.; Pavone, V.; Nastri, F.; Lombardi, A. A De Novo Heterodimeric Due Ferri Protein Minimizes the Release of Reactive Intermediates in Dioxygen-Dependent Oxidation. Angew. Chem. Int. Ed. 2017, 56, 15580–15583. [Google Scholar] [CrossRef] [PubMed]
- Chino, M.; Leone, L.; Maglio, O.; Lombardi, A. Designing Covalently Linked Heterodimeric Four-Helix Bundles. Methods Enzymol. 2016, 580, 471–499. [Google Scholar] [CrossRef] [PubMed]

| Detection | Target Analyte | Source | Time | LOD | Sensitivity | Selectivity | On Market | Ref. |
|---|---|---|---|---|---|---|---|---|
| Cell culture | ||||||||
| CPE 1 | Virus | Swabs 4 | 2–3 days | - | - | - | No | 63 |
| MNA 2 | Human IgM and IgG | Plasma/Serum | 2–4 days | 10,000 TCID50/mL | - | 100% | No | 63 |
| Genetic | ||||||||
| RT-PCR | Viral RNA | Swabs 4 | 3–4 h | 1 copy/μL of input 7 | 95% | 100%- | Yes | 64 |
| CRISPR-Cas12 assay | E gene and N gene | Swabs 4 | 30–45 min | 10 copies/μL of input | 95% | - | No | 67 |
| Immunological | ||||||||
| ELISA | Human IgM and IgG | Plasma/Serum | 3–4 h | - | 44.4% 8 82.5% 9 | 100% | Yes | 76 |
| Chemiluminescent Immunoassay | Human IgM and IgG in a separate assays | Serum | 48 min | - | 100% 9 | 57.2% 8 71.4% 9 | Yes | 78 |
| Proteomic * | ||||||||
| LC-MS | Virus proteins | Swabs 4 | 1–2 h | 10,000 particles | - | - | No | 82 |
| Biosensig device | ||||||||
| FET-based biosensor | SARS-CoV-2 S-protein | Swabs 5 Cultured virus-cell suspension | 3–10 min 6 | 242 copies/mL | - | 100% | No | 90 |
| GICA | Human IgM and IgG | Plasma/Serum | 15 min | - | 88.66% | 90.63% | Yes | 91 |
| ASOs-Capped Plasmonic Nanoparticles 3 | SARS-CoV-2 N-protein | Swabs 4 | ~1 h | 0.18 ng/µL | - | 100% | No | 92 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hussein, H.A.; Hassan, R.Y.A.; Chino, M.; Febbraio, F. Point-of-Care Diagnostics of COVID-19: From Current Work to Future Perspectives. Sensors 2020, 20, 4289. https://doi.org/10.3390/s20154289
Hussein HA, Hassan RYA, Chino M, Febbraio F. Point-of-Care Diagnostics of COVID-19: From Current Work to Future Perspectives. Sensors. 2020; 20(15):4289. https://doi.org/10.3390/s20154289
Chicago/Turabian StyleHussein, Heba A., Rabeay Y. A. Hassan, Marco Chino, and Ferdinando Febbraio. 2020. "Point-of-Care Diagnostics of COVID-19: From Current Work to Future Perspectives" Sensors 20, no. 15: 4289. https://doi.org/10.3390/s20154289
APA StyleHussein, H. A., Hassan, R. Y. A., Chino, M., & Febbraio, F. (2020). Point-of-Care Diagnostics of COVID-19: From Current Work to Future Perspectives. Sensors, 20(15), 4289. https://doi.org/10.3390/s20154289








