An Artificial Neural Network Assisted Dynamic Light Scattering Procedure for Assessing Living Cells Size in Suspension
Abstract
1. Introduction
2. Materials and Methods
2.1. Diluted Yeast Suspension
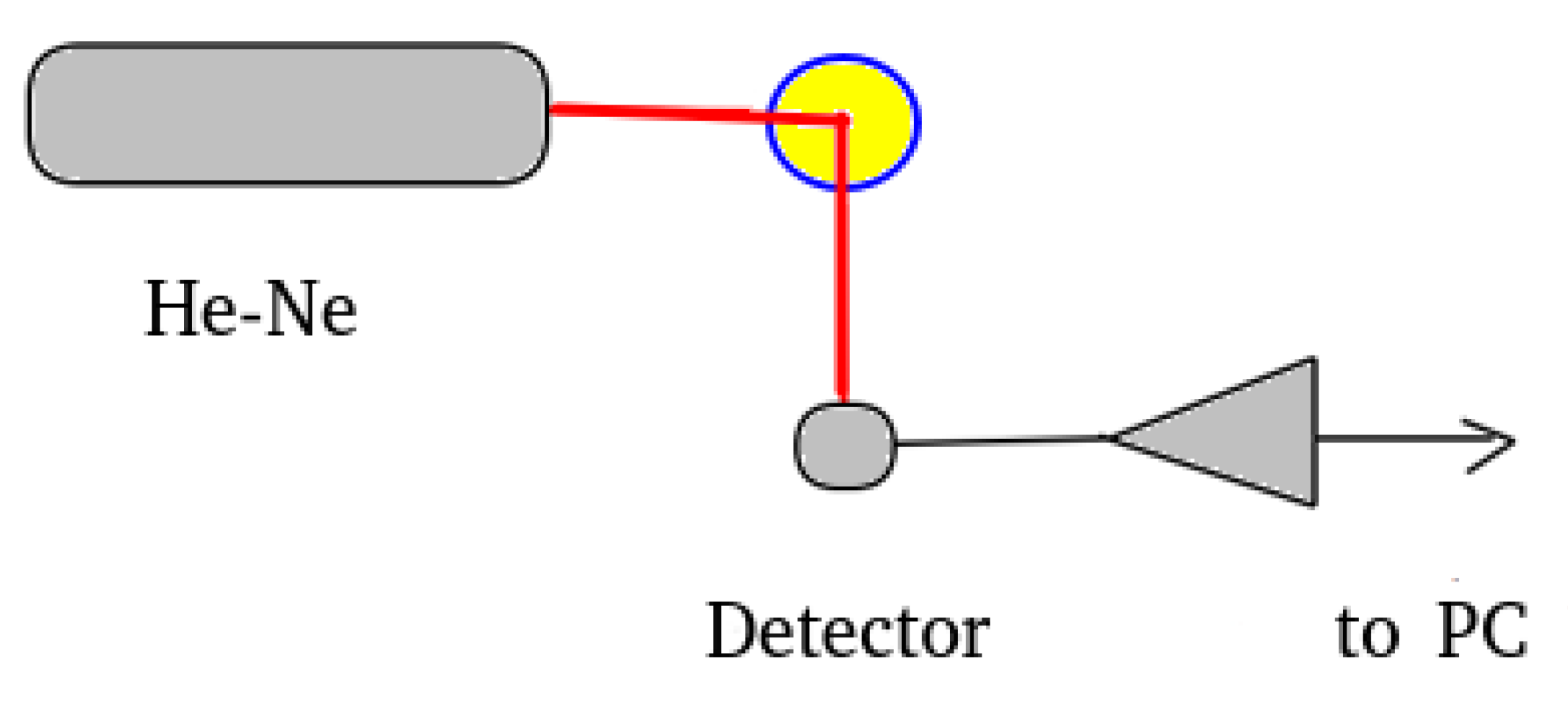
2.2. The Reference DLS Procedure
2.3. The ANN Assisted DLS Time-Series Processing Procedure
2.4. Experimental Procedure and Time-Series Processing
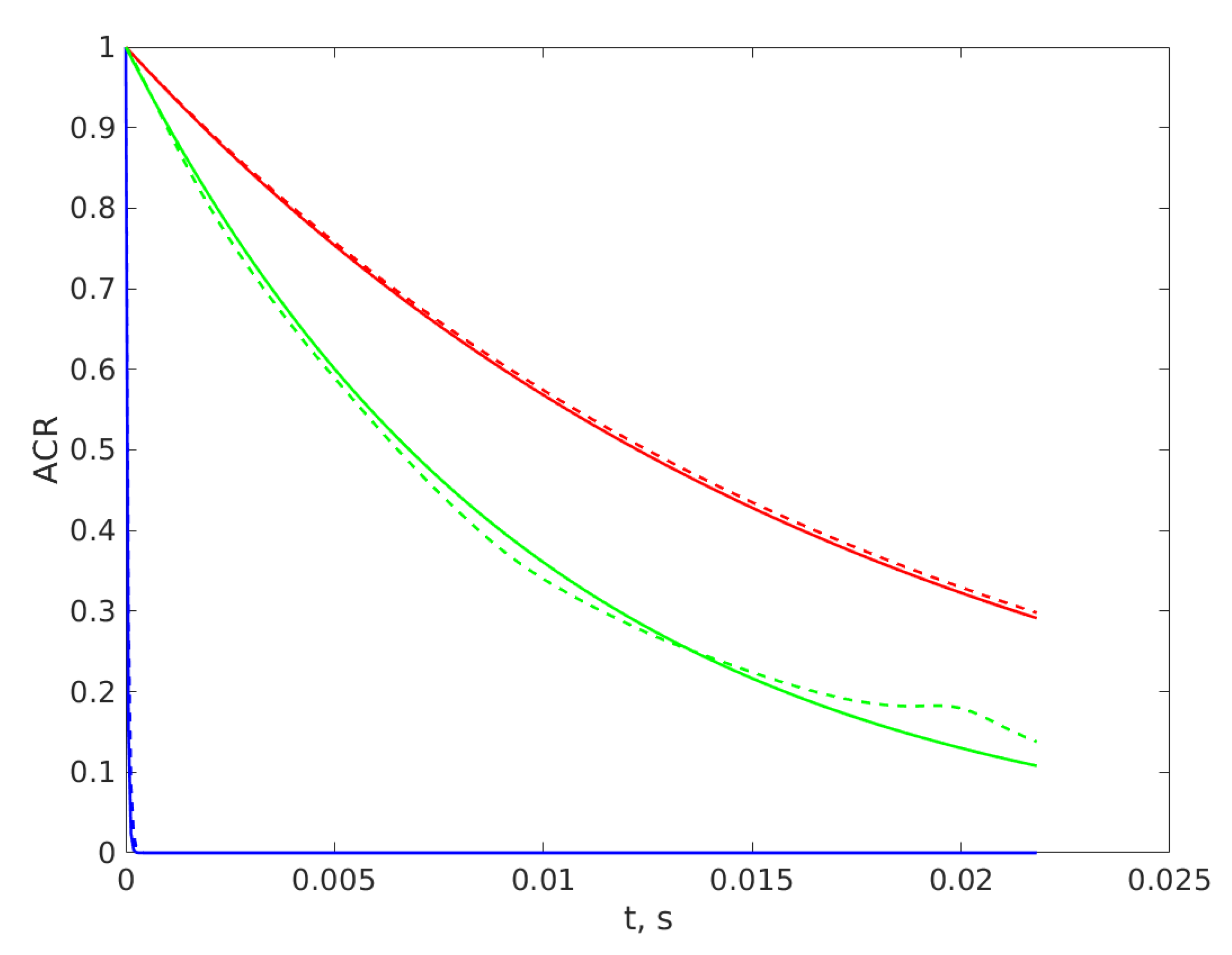
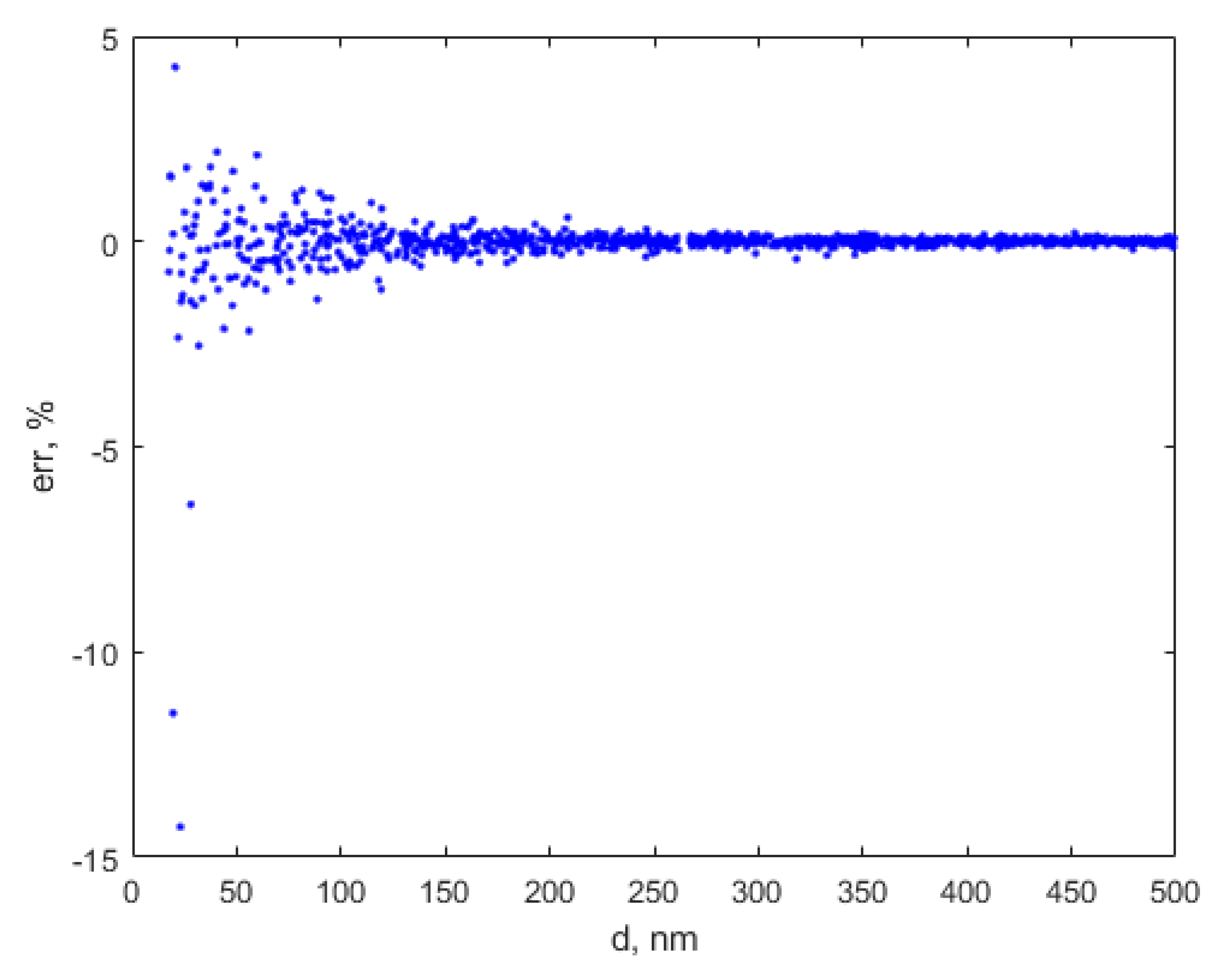
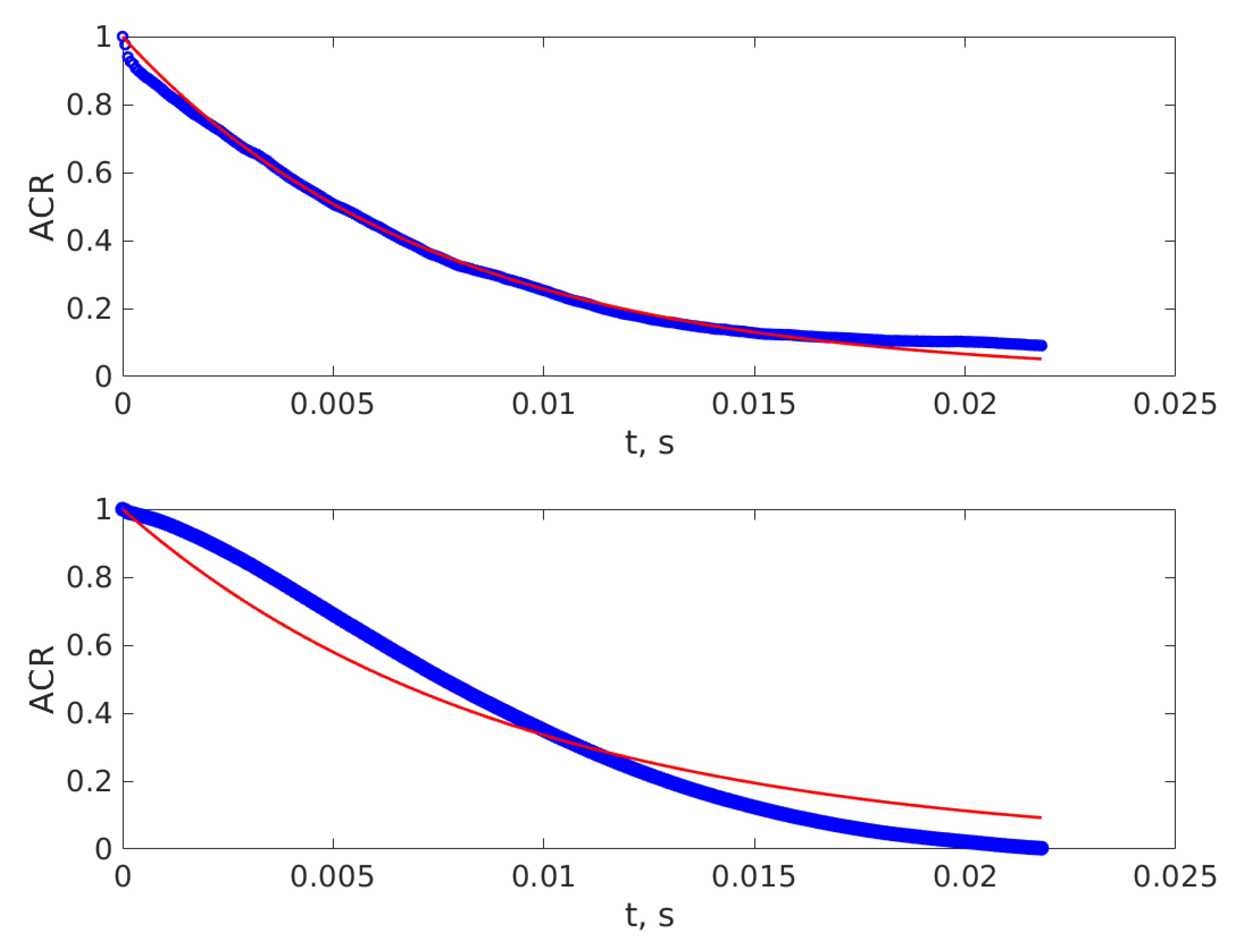
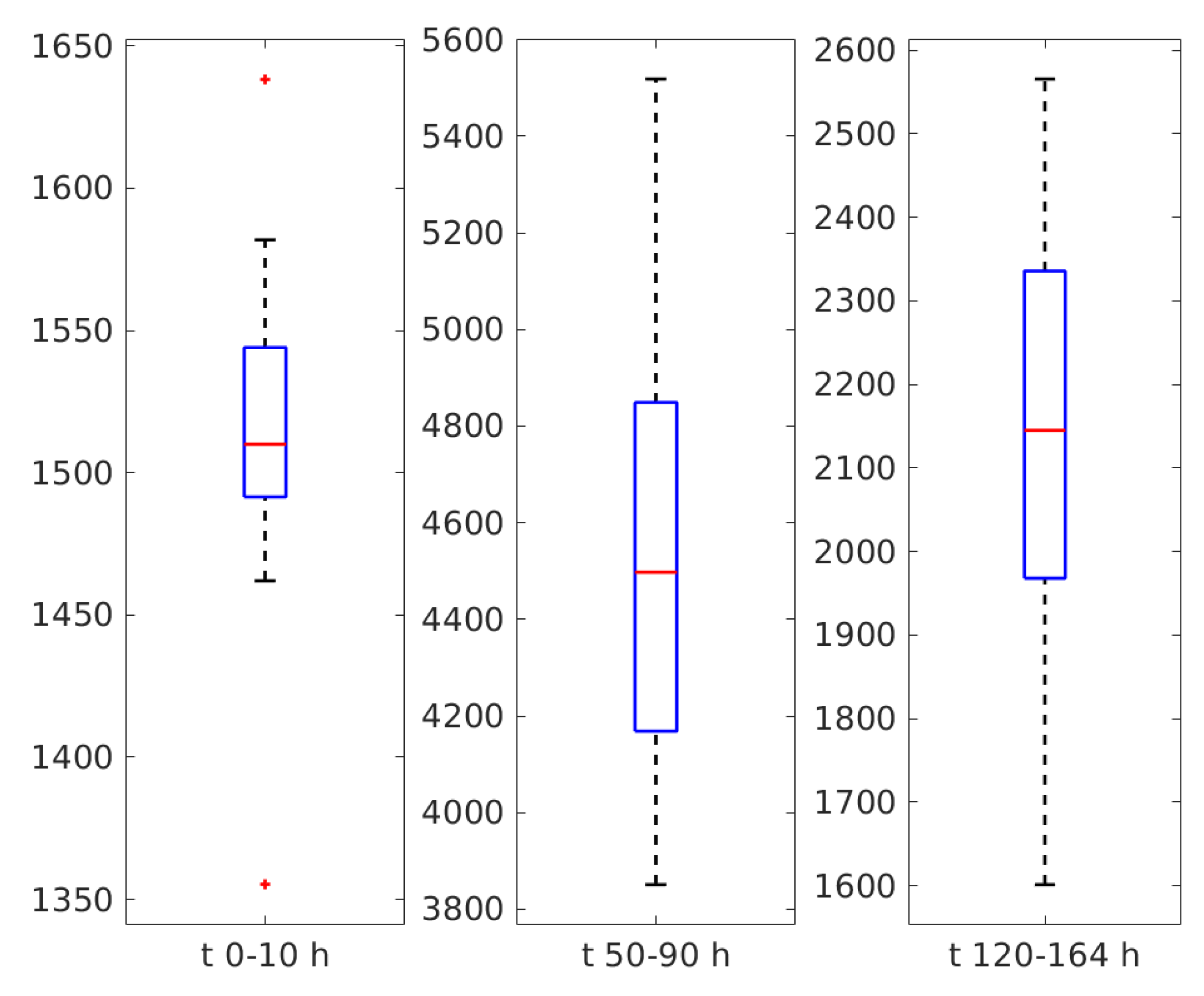
3. Results
4. Discussion and Conclusions
Funding
Conflicts of Interest
References
- Piederrière, Y.; Le Meur, J.; Cariou, J.; Abgrall, J.F.; Blouch, M.T. Particle aggregation monitoring by speckle size measurement; application to blood platelets aggregation. Opt. Express 2004, 12, 4596–4601. [Google Scholar] [CrossRef] [PubMed]
- Piederrière, Y.; Cariou, J.; Guern, Y.; Le Jeune, B.; Le Brun, G.; Lotrian, J. Scattering through fluids: Speckle size measurement and Monte Carlo simulations close to and into the multiple scattering. Opt. Express 2004, 12, 176–188. [Google Scholar] [CrossRef] [PubMed]
- Chicea, D. Speckle size, intensity and contrast measurement application in micron-size particle concentration assessment. Eur. Phys. J. Appl. Phys. 2007, 40, 305–310. [Google Scholar] [CrossRef]
- Clark, N.A. A study of brownian motion using light scattering. Am. J. Phys. 1970, 38, 575. [Google Scholar] [CrossRef]
- Goodman, J.W. Statistical properties of laser speckle patterns. In Laser Speckle and Related Phenomena; Springer: Berlin/Heidelberg, Germany, 1984; pp. 9–75. [Google Scholar]
- Kataoka, T.; Inoue, H.; Endo, K.; Oshikane, Y.; Mori, Y.; Nakano, M.; Wada, K.; An, H. Light scattering by small particles and small defects on the silicon wafer surface. Calculations of scattering light intensity and optical image through a lens. J. Jpn. Soc. Precis. Eng. 2000, 66, 1716–1722. [Google Scholar] [CrossRef]
- Berne, B.J.; Pecora, R. Dynamic Light Scattering: With Applications to Chemistry, Biology, and Physics; Dover Publications: Mineola, NY, USA, 2000; pp. 164–206. [Google Scholar]
- Xu, R. Particle characterization: Light scattering methods. China Particuol. 2003, 1, 271. [Google Scholar] [CrossRef]
- Bhattacharjee, S. DLS and zeta potential—What they are and what they are not? J. Control. Release 2016, 235, 337–351. [Google Scholar] [CrossRef]
- Stetefeld, J.; McKenna, S.A.; Patel, T.R. Dynamic light scattering: A practical guide and applications in biomedical sciences. Biophys. Rev. 2016, 8, 409–427. [Google Scholar] [CrossRef]
- Chicea, D.; Indrea, E.; Cretu, C.M. Assesing Fe3O4 nanoparticle size by DLS, XRD and AFM. J. Optoelectron. Adv. Mater. 2012, 14, 460–466. [Google Scholar]
- Chicea, D. A study of nanoparticle aggregation by coherent light scattering. Curr. Nanosci. 2012, 8, 259–265. [Google Scholar] [CrossRef]
- Gurney, K. An Introduction to Neural Networks; Taylor & Francis e-Library: London, UK, 2004. [Google Scholar]
- Haykin, S. Neural Networks and Learning Machines Third Edition—University Hamilton; Prentice Hall: Ontario, ON, Canada, 2008. [Google Scholar]
- Carrieri, A.H. Neural network pattern recognition by means of differential absorption Mueller matrix spectroscopy. Appl. Opt. 1999, 38, 3759–3766. [Google Scholar] [CrossRef] [PubMed]
- Berdnik, V.V.; Loiko, V.A. Retrieval of size and refractive index of spherical particles by multiangle light scattering: Neural network method application. Appl. Opt. 2009, 48, 6178–6187. [Google Scholar] [CrossRef] [PubMed]
- Berdnik, V.V.; Mukhamedjarov, R.D.; Loiko, V.A. Characterization of optically soft spheroidal particles by multiangle light-scattering data by use of the neural-networks method. Opt. Lett. 2004, 29, 1019–1021. [Google Scholar] [CrossRef] [PubMed]
- Kaye, P.; Hirst, E.; Wang-Thomas, Z. Neural-network-based spatial light-scattering instrument for hazardous airborne fiber detection. Appl. Opt. 1997, 36, 6149–6156. [Google Scholar] [CrossRef]
- Ulanowski, Z.; Wang, Z.; Kaye, P.H.; Ludlow, I.K. Application of neural networks to the inverse light scattering problem for spheres. Appl. Opt. 1998, 37, 4027–4033. [Google Scholar] [CrossRef]
- Chicea, D. Using neural networks for dynamic light scattering time series processing. Meas. Sci. Technol. 2017, 28, 055206. [Google Scholar] [CrossRef]
- Chicea, D.; Rei, S. A fast artificial neural network approach for dynamic light scattering time series processing. Meas. Sci. Technol. 2018, 29, 105201. [Google Scholar] [CrossRef]
- Dong, L.; Chen, X.; Hu, Z. Application of artificial neural networks for the determination of proteins with CPA-pI by rayleigh light scattering technique. J. Lumin. 2007, 124, 85–92. [Google Scholar] [CrossRef]
- He, L.; Kear-Padilla, L.; Lieberman, S.; Andrews, J. Rapid in situ determination of total oil concentration in water using ultraviolet fluorescence and light scattering coupled with artificial neural networks. Anal. Chim. Acta 2003, 478, 245–258. [Google Scholar] [CrossRef]
- Shabanov, A.E.; Petrov, M.N.; Chikitkin, A.V. A multilayer neural network for determination of particle size distribution in dynamic light scattering problem. Comput. Res. Model. 2019, 11, 265–273. [Google Scholar] [CrossRef]
- Zhao, H.; Dreses-Werringloer, U.; Davies, P.; Marambaud, P. Amyloid-beta peptide degradation in cell cultures by mycoplasma contaminants. BMC Res. Notes 2008, 1, 38. [Google Scholar] [CrossRef] [PubMed]
- Schulz-Vogt, H.N.; Jørgensen, B.B. Big bacteria. Annu. Rev. Microbiol. 2001, 55, 105–137. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Fell, J.W. Yeast Systematics and Phylogeny—Implications of Molecular Identification Methods for Studies in Ecology; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- McGovern, P.E.; Hartung, U.; Badler, V.R.; Glusker, D.L.; Exner, L.J. The beginnings of winemaking and viniculture in the ancient Near East and Egypt. Expedition 1997, 39, 3–21. [Google Scholar]
- Cavalieri, D.; McGovern, P.E.; Hartl, D.L.; Mortimer, R.; Polsinelli, M. Evidence for S. cerevisiae fermentation in ancient wine. J. Mol. Evol. 2003, 57, S226–S232. [Google Scholar] [CrossRef]
- McGovern, P.E. Ancient Wine: The Scientific Search for the Origins of Viniculture; Princeton University Press: Princeton, NJ, USA, 2003. [Google Scholar]
- Vaughan-Martini, A.; Martini, A. Saccharomyces meyen ex reess. In The Yeasts; Elsevier B.V.: Amsterdam, The Netherlands, 1998; pp. 358–371. [Google Scholar]
- Botstein, D.; Fink, G. Yeast: An experimental organism for modern biology. Science 1988, 240, 1439–1443. [Google Scholar] [CrossRef]
- Otero, J.M.; Cimini, D.; Patil, K.R.; Poulsen, S.G.; Olsson, L.; Nielsen, J. Industrial systems biology of saccharomyces cerevisiae enables novel succinic acid cell factory. PLoS ONE 2013, 8, e54144. [Google Scholar] [CrossRef]
- Chumnanpuen, P.; Brackmann, C.; Nandy, S.K.; Chatzipapadopoulos, S.; Nielsen, J.; Enejder, A. Lipid biosynthesis monitored at the single-cell level in Saccharomyces cerevisiae. Biotechnol. J. 2011, 7, 594–601. [Google Scholar] [CrossRef]
- Runguphan, W.; Keasling, J.D. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab. Eng. 2014, 21, 103–113. [Google Scholar] [CrossRef]
- Rumble, J.R.; Lide, D.R.; Bruno, T.J. CRC Handbook of Chemistry and Physics, 100th ed.; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Einstein, A. Über die von der molekularkinetischen theorie der wärme geforderte bewegung von in ruhenden flüssigkeiten suspendierten teilchen. Ann. Phys. 1905, 322, 549–560. [Google Scholar] [CrossRef]
- Beale, H.D.; Demuth, H.B.; Hagan, M.T.; DeJesus, O. Neural Network Design. Available online: https://hagan.okstate.edu/NNDesign.pdf (accessed on 5 August 2019).
- Levenberg, K. A method for the solution of certain non-linear problems in least squares. Q. Appl. Math. 1944, 2, 164–168. [Google Scholar] [CrossRef]
- Balasubramanian, M.K.; Bi, E.; Glotzer, M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr. Biol. 2004, 14, R806–R818. [Google Scholar] [CrossRef] [PubMed]
- Yeong, F.M. Severing all ties between mother and daughter: Cell separation in budding yeast. Mol. Microbiol. 2005, 55, 1325–1331. [Google Scholar] [CrossRef] [PubMed]
- Neiman, A.M. Ascospore formation in the yeast saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2005, 69, 565–584. [Google Scholar] [CrossRef] [PubMed]
- Chicea, D. Nanoparticles and nanoparticle aggregates sizing by DLS and AFM. J. Optoelectron. Adv. Mater. 2010, 4, 1310–1315. [Google Scholar]
- Chicea, D. Revealing Fe3O4 nanoparticles aggregation dynamics using dynamic light scattering . Optoelectron. Adv. Mater. Rapid Commun. 2009, 3, 1299–1305. [Google Scholar]
- Provencher, S.W. CONTIN: A General Purpose Constrained Regularization Program for Inverting Noisy Linear Algebraic Integral Equations. Comput. Phys. Commun. 1982, 27, 229–242. [Google Scholar] [CrossRef]

© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chicea, D. An Artificial Neural Network Assisted Dynamic Light Scattering Procedure for Assessing Living Cells Size in Suspension. Sensors 2020, 20, 3425. https://doi.org/10.3390/s20123425
Chicea D. An Artificial Neural Network Assisted Dynamic Light Scattering Procedure for Assessing Living Cells Size in Suspension. Sensors. 2020; 20(12):3425. https://doi.org/10.3390/s20123425
Chicago/Turabian StyleChicea, Dan. 2020. "An Artificial Neural Network Assisted Dynamic Light Scattering Procedure for Assessing Living Cells Size in Suspension" Sensors 20, no. 12: 3425. https://doi.org/10.3390/s20123425
APA StyleChicea, D. (2020). An Artificial Neural Network Assisted Dynamic Light Scattering Procedure for Assessing Living Cells Size in Suspension. Sensors, 20(12), 3425. https://doi.org/10.3390/s20123425




